Abstract
The blaCMY -2/4-carrying IncB/O/K-like plasmids of seven Escherichia coli strains from poultry, poultry meat and human urine samples were examined using comparative analysis of whole plasmid sequences. The incompatibility group was determined by analysis of the incRNAI region and conjugation assays with strains containing the IncK and IncB/O reference plasmids. Strains were additionally characterized using MLST and MIC determination. The complete DNA sequences of all plasmids showed an average nucleotide identity of 91.3%. Plasmids were detected in E. coli sequence type (ST) 131, ST38, ST420, ST1431, ST1564 and belonged to a new plasmid variant (IncK2) within the IncK and IncB/O groups. Notably, one E. coli from poultry meat and one from human contained the same plasmid. The presence of a common recently recognized IncK2 plasmid in diverse E. coli from human urine isolates and poultry meat production suggests that the IncK2 plasmids originated from a common progenitor and have the capability to spread to genetically diverse E. coli in different reservoirs. This discovery is alarming and stresses the need of rapidly introducing strict hygiene measures throughout the food chain, limiting the spread of such plasmids in the human settings.
Keywords: ESBL, pAmpC, food, antibiotic resistance, animals
Introduction
The worldwide dissemination of Enterobacteriaceae along with resistance plasmids carrying β-lactamase-encoding genes (bla) is posing an increasing threat to the public health system (Mathers et al., 2015). Poultry and poultry meat have been revealed as a large reservoir of extended-spectrum β-lactamases (ESBL) and plasmid-mediated AmpC (pAmpC) enzymes in several countries (Endimiani et al., 2012; Dierikx et al., 2013; Nilsson et al., 2014; Hansen et al., 2016; Mo et al., 2016). These resistance genes are carried by various mobile genetic elements; primarily located on plasmids which are self-replicating extrachromosomal elements (Couturier et al., 1988; Carattoli, 2009). Such plasmids not only vary in their size and compatibility to other plasmid groups, but also in their capability of transferring to various bacterial species (Carattoli, 2009, 2013).
The involvement of plasmids in the zoonotic spread of β-lactamase- and carbapenemase-encoding genes between human and animal reservoirs has already been emphasized based on the presence of similar plasmids in Escherichia coli from humans, poultry, and poultry meat (Leverstein-van Hall et al., 2011; Börjesson et al., 2013; Seiffert et al., 2013b; Voets et al., 2013; Guerra et al., 2014; Wang et al., 2014). However, there is a lack of firm evidence since there are only few completely sequenced and assembled plasmids to demonstrate the spread and exchange of the same plasmids in E. coli of human and poultry origin.
By comparing datasets from poultry, poultry meat, and clinical human isolates from Switzerland, we noticed that the pAmpC blaCMY -2 and blaCMY -4 genes were associated with plasmids belonging to the IncB/O/K groups (Endimiani et al., 2012; Seiffert et al., 2013a; Vogt et al., 2014). Other studies investigating pAmpC-producing E. coli have detected the nearly identical kind of plasmid in healthy humans (Baudry et al., 2009; Porres-Osante et al., 2015; Hansen et al., 2016), poultry (Dierikx et al., 2010; Hansen et al., 2016; Mo et al., 2016), poultry meat (Egervärn et al., 2014), and dogs (Hansen et al., 2016). This prompted us to perform an in-depth analysis of blaCMY -2/4-carrying IncB/O/K plasmids from E. coli isolated in three settings in Switzerland to determine their genetic relatedness, revealing a novel IncK-group variant, the IncK2.
Materials and Methods
Bacterial Strains
All strains were obtained from previous studies characterizing 3rd-generation cephalosporin-resistant E. coli (3GC-R-Ec) from poultry, poultry meat, and humans isolated at the Institute of Veterinary Bacteriology and at the Laboratory of Microbiology of the Institute of Infectious Diseases, University of Bern, Switzerland (Endimiani et al., 2012; Seiffert et al., 2013a; Vogt et al., 2014). Strains were collected between 2011 and 2014 (Table 1) and were selected based on the presence of blaCMY -2/4 by PCR/DNA sequencing (Seiffert et al., 2013a; Vogt et al., 2014) and on ambiguous results with the PCR-based replicon typing (PBRT) of plasmids, giving cross-reaction with either IncK and/or IncB/O PCRs (Carattoli et al., 2005).
Table 1.
Origin, characteristics and resistance profile of the original E. coli strains and of the E. coli strains transformed with the blaCMY -2/4 containing IncK2 plasmids.
| Strain | Origin (date of isolation) | Sequence type | Antibiotic resistance profile | Reference |
|---|---|---|---|---|
| 4809.66 | Human UTI, hospital (2011) | ST1431 | FOX CPD CRO CTX CAZ CFZ CEF CIP NAL TMP SMZ | Seiffert et al., 2013a |
| 5312.29 | Human UTI, hospital (2014) | ST131 | FOX CPD CRO CTX CAZ CFZ CEF PIP/TAZ NAL | This study |
| DV10 | Poultry retail meat, MPP from slaughterhouse A (Apr 2013) | ST38 | FOX CPD CRO CTX CAZ CFZ CEF NAL | Vogt et al., 2014 |
| DV45 | Poultry retail meat, MPP from slaughterhouse A (Apr 2013) | ST1564 | FOX CPD CRO CTX CAZ CFZ CEF TMP SMZ NAL TET KAN | Vogt et al., 2014 |
| MSA1088 | Broiler cloaca, slaughterhouse A (Dec 2012) | ST38 | FOX CPD CRO CTX CAZ CFZ CEF TMP TET | Endimiani et al., 2012; this study |
| MSA992 | Broiler cloaca, slaughterhouse A (Nov 2012) | ST420 | FOX CPD CRO CTX CAZ CFZ CEF PIP/TAZ NAL | Endimiani et al., 2012; this study |
| MSA970 | Broiler cloaca, slaughterhouse B (Nov 2012) | ST420 | FOX CPD CRO CTX CAZ CFZ CEF PIP/TAZ NAL | Endimiani et al., 2012; this study |
| SNS4809.66 | DH10B transformed with blaCMY -2 plasmid p4809.66 | – | FOX CPD CRO CTX CAZ CFZ CEF | This study |
| SNS5312.29 | DH10B transformed with blaCMY -2 plasmid p5312.29 | – | FOX CPD CRO CTX CAZ CFZ CEF | This study |
| ALC74 | TOP10 transformed with blaCMY -4 plasmid pDV10 | – | FOX CPD CRO CTX CAZ CFZ CEF | This study |
| ALC76 | TOP10 transformed with blaCMY -2 plasmid pDV45 | – | FOX CPD CRO CTX CAZ CFZ CEF | This study |
| TMSA1088 | TOP10 transformed with blaCMY -2 plasmid pTMSA1088 | – | FOX CPD CRO CTX CAZ CFZ CEF | This study |
| TMSA992 | TOP10 transformed with blaCMY -2 plasmid pTMSA992 | – | FOX CPD CRO CTX CAZ CFZ CEF PIP/TAZ NAL | This study |
| TMSA970 | TOP10 transformed with blaCMY -2 plasmid pTMSA970 | – | FOX CPD CRO CTX CAZ CFZ CEF PIP/TAZ | This study |
| TOP10 | One Shot® TOP10 ElectrocompTM cells | – | Invitrogen, Life Technologies | |
| DH10B | ElectroMax DH10B competent cells | – | Invitrogen, Life Technologies |
The three isolates from broiler cloacae were from three different herds. The three isolates from poultry meat were from three different lots originating from the meat packing plant (MPP) of slaughterhouse A; The three isolates from human infections were from patients hospitalized at the same hospital. CEF, cephalotin; CFZ, cefazolin; FEP, cefepime; CTX, cefotaxime; CTX-CLA, cefotaxime/clavulanic acid; FOX, cefoxitin; CPD, cefpodoxime; CAZ, ceftazidime; CRO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; CST, colistin; GEN, gentamicin; IPM, imipenem; KAN, kanamycin; MEM, meropenem; NAL, nalidixic acid; TZP, piperacillin/tazobactam; STR, streptomycin; SMZ, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim.
Plasmids were transformed by electroporation into either E. coli ElectroMax DH10B competent cells or into E. coli One Shot® TOP10 Electrocomp cells (Invitrogen, Life Technologies) as previously described (Seiffert et al., 2013a; Vogt et al., 2014). The MIC of antibiotics were obtained from donors and transformants using the microdilution Sensititre panels ESB1F and EUMVS2 (Trek Diagnostics) and interpreted following the CLSI guidelines (CLSI, 2016). Sequence types (ST) were assigned following the multi-locus sequence typing (MLST) Achtman scheme1.
Whole Plasmid Sequencing (WPS)
Plasmids were purified using the Genopure Plasmid Midi Kit (Roche Diagnostic) according to the manufacturer’s procedures and used for Whole-Plasmid Sequencing (WPS). WPS was performed using the 454-Junior Genome Sequencer procedure (Roche Diagnostic). The libraries of plasmid DNA were constructed using the GS FLX Titanium Rapid Library Preparation Kit.
De novo Assembly of DNA Reads, Gap-Closure and Annotation
Contigs with at least a 70-fold coverage were obtained using the GS de novo Assembler software (Roche Diagnostics). Scaffolding was first done in silico by using the 454 ReadStatus output file to identify reads with overlapping adjacent contigs. The assembly and gap-closure, as well as insertion/deletion events, single nucleotide polymorphisms and shufflons were confirmed and verified by PCR followed by Sanger DNA sequencing. The annotation was done using the Artemis Version 8 (Sanger Institute) in combination with a pairwise alignment using a BLASTN and BLASTP homology search2.
Phylogenetic Analysis of IncB/O/K Plasmids
For the phylogenetic analysis of the IncB/O/K-like plasmids, two fully sequenced plasmids, namely pCT belonging to IncK (GenBank acc. No. FN868832.1; Cottell et al., 2011) and p3521 belonging to IncB (GU256641; Papagiannitsis et al., 2011) were included as reference. These plasmids were chosen as they represent IncK and IncB plasmids whose complete nucleotide sequence is available in the GenBank. The phylogenetic trees and the homology matrices were constructed by UPGMA method and Jukes-Cantor correction [Multiple alignment (open gap penalty (OG): 100%, unit gap penalty (UG): 0%, gap penalty: 100%), 2000 Bootstrap trials] using Bionumerics 7.6 (Applied Maths, Kortrijk, Belgium).
The analyses were performed using the full length sequences of the IncK2 plasmids with and without shufflon region. The deleted shufflon region started at the shufflon-specific DNA recombinase (rci) and ended with the last shufflon gene which varies from plasmid to plasmid (position 62686–65450 in pTMSA970). In the case of the reference IncK and IncB plasmids, in addition to the shufflon region, the 3,144-bp blaCTX-M-14-carrying element was removed from the IncK reference plasmid (position 68837–71981 in FN868832) and the 26,548-bp large blaACC-4 containing element was removed from the IncB reference plasmid (position 59569–86117 in GU256641).
Conjugation for Incompatibility Testing
Mating experiments were performed crossing the transformant strain containing plasmid p5312.29 (strain SNS5312.29) or pTMSA970 (strain TMSA970) with reference plasmids Rhh72 (IncB/O), and R387 (IncK; provided by the Istituto Superiore di Sanità, Rome, Italy) which are used for incompatibility testing (Couturier et al., 1988). Conjugations were performed on Luria-Bertani (LB) agar solid media streaking two crossing lines, one for each mating strain, and incubating the mating plates overnight at 37°C.
Donor and transconjugant colonies were counted after plating 10-fold serial dilutions on LB plates containing 50 μg/ml of ampicillin (as resistance marker for p5312.29 and pTMSA970), 50 μg/ml kanamycin (as resistance marker for Rhh72), 25 μg/ml chloramphenicol (as resistance marker for R387), 50 μg/ml of ampicillin plus 50 μg/ml kanamycin, and 50 μg/ml of ampicillin plus 25 μg/ml chloramphenicol.
Stability of the plasmids within the conjugant bacterial cells was determined as follows: three independent colonies resulting from each mating and exhibiting the double resistance (ampicillin/kanamycin, or ampicillin/chloramphenicol) were grown 24 h at 37°C in 50 ml of LB without antibiotics. Ten-fold dilutions of the conjugant cultures were plated on LB plates containing either ampicillin, kanamycin or chloramphenicol alone, and on plates containing both ampicillin and kanamycin, or both ampicillin and chloramphenicol. Colonies were counted after an overnight incubation at 37°C. Stability was determined as the ratio between the number of ampicillin-resistant colonies and the number of ampicillin/kanamycin-resistant colonies for the p5312.29 × Rhh72 and pTMSA970 × Rhh72 transconjugants, and between the number of ampicillin-resistant colonies and the number of ampicillin/chloramphenicol-resistant colonies for the p5312.29 × R387 and pTMSA970 × R387 transconjugants, respectively.
Nucleotide Sequence Accession Numbers
The complete nucleotide sequence of the seven IncK2 plasmids have been deposited into the GenBank database under accession numbers KR905385 (p5312.29), KR905389 (p4809.66), KR905387 (pTMSA992), KR905388 (pTMSA970), KR905386 (pTMSA1088), KR905384 (pDV45), and KR905390 (pDV10).
Results and Discussion
Plasmid Sequencing
Whole plasmid sequencing of seven blaCMY -2/4-containing plasmids [poultry cloacae: pTMSA970, pTMSA992, pTMSA1088; poultry meat: pDV10, pDV45; human urine isolates: p4809.66, p5312.29] revealed the presence of highly related plasmids in both genetically related and diverse E. coli from poultry (ST38, ST420), poultry meat (ST38, ST1564) and human clinical samples (ST131, ST1431).
Plasmids mainly differed from each other within the highly variable shufflon region, and a region situated between impC and nikA (Figure 1). A major deletion of approximately 6 kb, that starts after yfbA and ends before psiB, distinguished plasmids pDV10, pTMSA1088, pTMSA970, pTMSA992 from the larger plasmids pDV45, p4809.66, and p5312.29 (Figure 1).
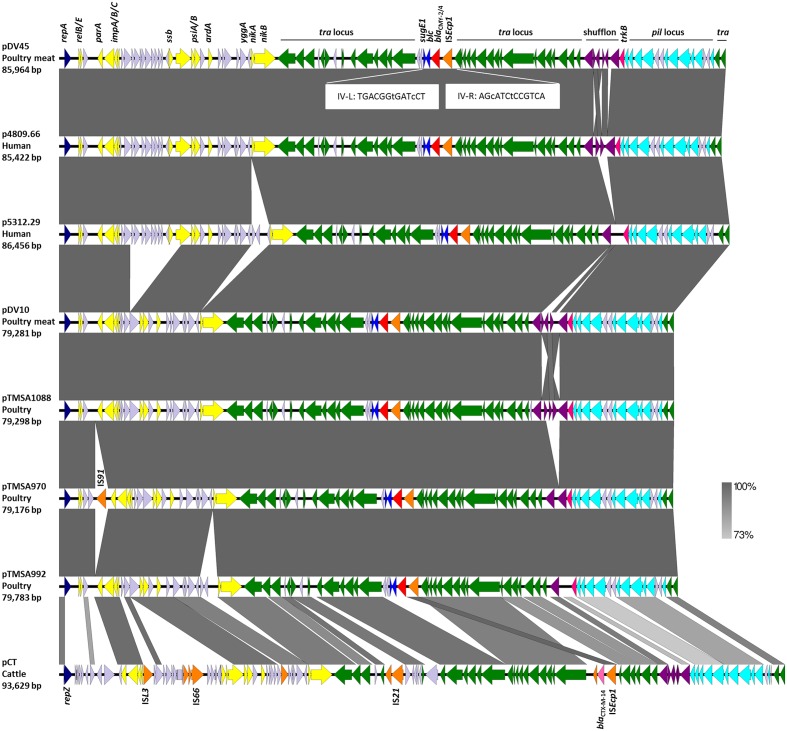
FIGURE 1.
Structural features of seven fully sequenced IncK2 plasmids encoding CMY-2/4 β-lactamases and of the IncK plasmid pCT. The open reading frames are illustrated by arrows pointing in direction of their respective orientation. The figure was generated with EasyFig 2.1 (Sullivan et al., 2011). Imperfect inverted repeats (IV-R and IV-L) are shown in white boxes. The color code is as follows: The tra and trb transfer loci are in green, antibiotic resistance genes in red (blaCMY -2/4) and pink (blaCMY -2/4), insertion sequence element (IS) family in orange, replicase genes in dark blue, partitioning genes and toxin/anti-toxin and other stabilization systems in yellow, shufflon-associated genes in violet, pilus genes in pale blue, sugE1 and blc genes in blue, and genes with unknown function are in lavender. Important genes and loci are indicated above the first plasmid sequence. Plasmid sequences of pTMSA970 and pCT carry additional IS elements which are indicated in the figure. The black lines above the figure indicate the different loci and the shufflon-region.
Another fragment of 2.3 kb situated between yggA and nikA distinguished plasmids p5312.29 and pTMSA992 from the other plasmids (Figure 1). Plasmid pTMSA970 also had an extra insertion of a previously unreported IS-element which is closely related to ISSbo1 (GenBank acc. no. CP001062) and belongs to the IS91 family. Integrations or losses of these regions might have occurred as single events during the transfer of the plasmids from one bacterial host to the other rather than during a longer evolutionary process. Outside of these regions, the plasmid sequences were virtually identical, only differing by a few SNPs, which have all been confirmed by Sanger sequencing.
Of note, except for the variable shufflon regions, plasmid pDV45 from E. coli from poultry meat was identical to plasmid p4809.66 from human E. coli, and plasmid pDV10 E. coli from poultry meat was identical to plasmid pTMSA1088 from poultry (Figure 1), which strongly suggests transfer of plasmids between genetically diverse E. coli along the food chain.
The variable region between impC and nikA also harbors, along with many genes encoding for proteins with unknown function the single-stranded DNA-binding (Ssb) protein. One of the main functions of the Ssb protein is binding of ssDNA and subsequent protection from degradation by nucleases (Meyer and Laine, 1990). The direct consequence of the loss of Ssb for the IncB/O/K-group plasmids is difficult to determine and needs further investigations.
The shufflon is a biological switch that controls the C-terminal end of the PilV proteins via site-specific recombination. This mechanism has been described in R64 as well as in pCT where the shufflon determines the recipient specificity in liquid mating despite being part of a minor component of the thin pilus (Komano, 1999; Cottell et al., 2014). The shufflon of R64 consists of four invertible DNA fragments embedded with seven 19-bp repeat sequences. These 19-bp repeats can be subdivided into a 7-bp core-site region where the DNA crossover occurs and the highly conserved 12-bp right arm region (Komano, 1999; Brouwer et al., 2015).
The shufflon of the seven plasmids is highly diverse and contains between two and eight recombination sites. Four of the 19-bp repeat sequences found in R64 were also identified in the plasmids. Interestingly, the 7-bp core-site region is identical to the ones identified in the IncI1/I2 plasmids. The terminology of 19-bp repeat sequences follows the designation of the repeats in R64 (Komano, 1999) (i.e., repeat-e: GTGCCAATCCGGTtcgTGG; repeat-f: GTGCCAATCCGGTatcTGG; repeat-g: GTGCCAATCCGGTGtgtGG; repeat-h: GTGCCAATCCGGTttgTGG; letters in italic represent the core-site, small letters for the variable bases). The three plasmids pTMSA970, pTMSA992, and p5312.29 have only two repeat-e sequences, followed by p4809.66 with four repeat sequences in the following order: e-e-e-g. The most complex shufflons with eight repeat sequences each were detected in pDV10 (g-g-e-e-e-e-h-f), pDV45 (e-e-e-e-g-g-h-f), and pTMSA1088 (e-e-h-f-e-e-g-g). Although, no apparent correlation between the numbers of recombination sites could be detected (neither with the size of the plasmid nor with their origin), the observation of such great diversity may have an impact on the mating preferences of the various plasmids.
Incompatibility Assays
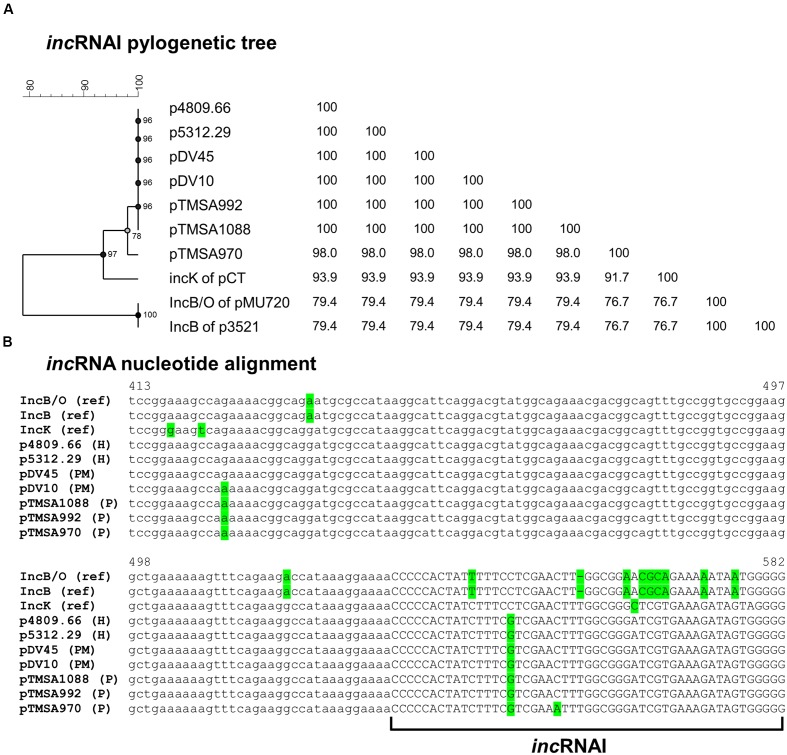
Sequence analysis of the region upstream of the repA including the incRNAI revealed that the DNA sequences of the seven plasmids diverged from the previously reported incRNAI of the IncK, IncB, and IncB/O plasmids (Figure 2).
FIGURE 2.
(A) Phylogenetic tree and (B) sequence alignment of the incRNAI-II of the IncB/O/K-group plasmids including references. The sequences were extracted from the IncK2 plasmids as well as from the reference plasmids of IncB and IncK. The IncB/O incRNAI-II reference sequence was derived from the paper of Couturier et al. (1988). Due to the absence of the complete nucleotide sequence of the IncB/O plasmid, pMU720 was not included into Figure 3. The phylogenetic tree was constructed by UPGMA method and Jukes-Cantor correction [Multiple alignment (open gap penalty (OG): 100%, unit gap penalty (UG): 0%, gap penalty: 100%), 2000 Bootstraps] using Bionumerics 7.6 (Applied Maths, Kortrijk, Belgium). H, human urine sample; P, poultry; PM, poultry meat.
This region before the repA is highly conserved among the specific plasmid Inc groups and is crucial for the incompatibility behavior among plasmids of the same group (Couturier et al., 1988; Praszkier et al., 1991). The key element for this behavior is the incRNAI that forms a stem and loop structure and negatively regulates the transcription of repA. Consequently, if two plasmids of the same or similar Inc group(s) reside in the same bacterial cell the incRNAIs hybridize with each other and prohibit translation (Couturier et al., 1988). Additionally, a phylogenetic tree constructed using only the incRNAI sequences containing the stem-and loop structure strengthened the previous observations that the seven plasmids are highly similar to each other, and are more closely related to the IncK group than the IncB/O groups (Figure 2).
The divergent incRNAI sequences suggested that these plasmids could be compatible with the IncK and IncB/O reference plasmids. Therefore, incompatibility assays were performed crossing SNS5312.29 and TMSA970 strains with both IncB/O Rhh72 and IncK R387 reference strains, respectively. Transconjugants were obtained from all mating experiments, but at different frequency. Both SNS5312.29 and TMSA970 showed higher frequencies of conjugation when crossed with IncB/O (1 × 10-5 transconjugant/donor cell) than when crossed with the IncK (1 × 10-7 transconjugant/donor cell) reference plasmid. This suggests that the plasmids under study are more closely related to IncK than to IncB/O. Furthermore, IncB transconjugants demonstrated higher stability (>90%) when grown in liquid medium without antibiotics then the IncK transconjugants (>50%). Based on both conjugation results and incRNAI sequence analysis, the plasmids were assigned to a novel variant of the IncK group designated as IncK2.
Phylogenetic Analysis of the IncK2 Plasmids
The 4,198-bp long blaCMY -2/4-like-carrying mobile element (ISEcp1-blaCMY -2/4-blc-sugE1) which is flanked by two 13-bp imperfect inverted repeats (IV-R and IV-L) was integrated between the traT and traU in all seven plasmids. Integration of ISEcp1-blaCMY -2/4-blc-sugE1 at the same site inside the tra locus is a solid indication that all the IncK2 plasmids analyzed have a common ancestor (Figure 1).
The same structure (ISEcp1-blaCMY -2/4-blc) has already been reported in IncK plasmid from E. coli of human origin in Spain and from E. coli from chicken meat in Norway (Porres-Osante et al., 2015; Mo et al., 2016), as well as in IncA/C plasmids from Salmonella enterica subsp. enterica serotype Newport in the USA (Cao et al., 2015). Notably, the only blaCMY -4-carrying plasmid pDV10 found has also developed from the same progenitor as the other plasmids and was subjected to subsequent evolution via a Trp221Arg substitution in CMY-2.
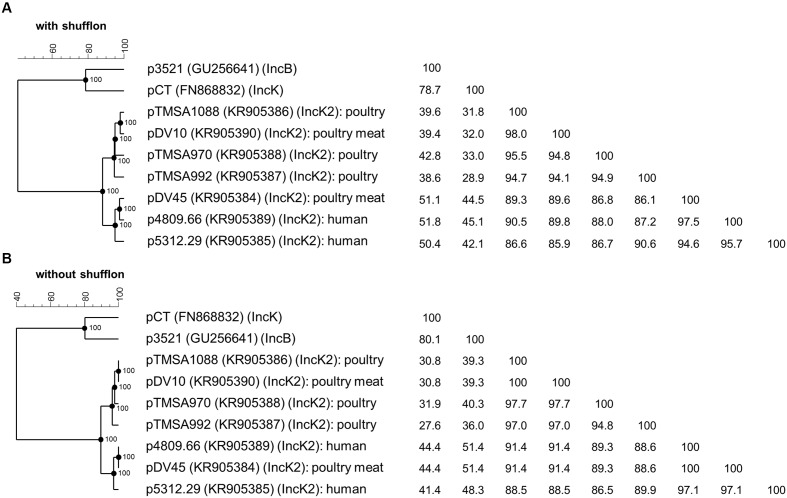
The phylogenetic analysis of the IncK2 plasmids supports the hypothesis of a common ancestor. All the IncK2 plasmids share a nucleotide identity ranging from 86.1 to 98.0% which increases to 86.5 to 100% once the shufflon region has been removed. This similarity is remarkable given that the percentage of identity for the two reference plasmids IncK and IncB is maximally 78.7% and without the shufflon region 80.1% (Figure 3). The IncK2 plasmids differed from the IncK plasmid pCT not only by their incompatibility and replication gene, but also in their structure and base composition (Figure 1).
FIGURE 3.
Phylogenetic analysis and identity matrix of the IncK2 group of plasmids of E. coli from poultry, poultry meat, and human. The analysis includes the plasmids of the novel IncK2 group with (A) and without shufflon (B) as well as core sequences of the reference plasmids pCT of the IncK group and p3521 of the IncB group; the reference plasmid pMU720 of the IncB/O group was not included in the tree due the absence of complete plasmid sequence. GenBank accession numbers are indicated in parentheses. The phylogenetic trees and the homology matrices were obtained by UPGMA method and Jukes-Cantor correction [Multiple alignment (open gap penalty (OG): 100%, unit gap penalty (UG): 0%, gap penalty: 100%), 2000 Bootstraps] using Bionumerics 7.6 (Applied Maths, Kortrijk, Belgium).
Taken together, these results lead to the conclusion that the IncK2 plasmids are an overall highly homogenous cluster although they are distributed in genetically diverse E. coli isolated from different sources and timeframes. The phylogenetic analysis points toward a common ancestor that has subsequently spread between the various reservoirs. Indeed, the most striking characteristic of the IncK2 group is its occurrence in different reservoirs (including poultry, poultry-meat, and humans isolates) from which they were isolated. There is strong evidence that all IncK2 plasmids originated from a common source due to: (i) the identical integration site of the blaCMY-carrying mobile element; (ii) the overall 91.3% identity among all the plasmids.
Publications from Denmark, Japan, the Netherlands, Norway, Spain, and Sweden investigating pAmpC-producing E. coli isolates from healthy and hospitalized humans as well as strains from poultry and poultry-meat have reported a high prevalence of blaCMY -2 in both reservoirs (Dierikx et al., 2010; Börjesson et al., 2013; Voets et al., 2013; de Been et al., 2014; Egervärn et al., 2014; Huijbers et al., 2015; Porres-Osante et al., 2015; Hansen et al., 2016; Mo et al., 2016). These publications were all using the PBRT kit for characterization of their blaCMY -2-carrying plasmids, whereby a substantial part of them was either identified as IncK plasmids or as IncB/O plasmids.
Based on the present study, it has to be noted that the IncK2 plasmid-carrying strains may have been falsely identified as positive for the IncK or IncB/O plasmid groups using the PBRT. Indeed, sequence alignment of our IncK2 plasmids with DNA sequences from the GenBank revealed that such plasmids have already been identified in E. coli from poultry origin in different countries. One plasmid (GenBank acc. no. JXMX01000007.1) isolated from E. coli strain 53C from retail chicken meat in the Netherlands (de Been et al., 2014) was almost identical to pDV45 (also from chicken meat) with 98.2% nucleotide identity and 100% without considering the shufflon. Another plasmid, pNVI1292, from E. coli strain 2012-01-1292 (GenBank acc. no. KU312044) from retail chicken meat in Norway (Mo et al., 2016) was nearly identical to pDV10 (also from poultry meat) and to pTMSA1088 (poultry) with 98.7 and 97.8% nucleotide identity, respectively, and 100% without considering the shufflon. Of note, strain 2012-01-1292 also belonged to ST38 like both strains harboring pDV10 and to pTMSA1088 indicating that specific clones may also contribute to the spread of the IncK2 plasmids.
The novel IncK2 group seems to be an emerging epidemic plasmid with a potential for dissemination in E. coli from different reservoirs including human and poultry. To our knowledge, this is the first time that a highly conserved plasmid family is described in isolates from human clinical samples, poultry, and poultry meat all coming from the same geographic region. However, further epidemiological studies are necessary to assess the scope of the spread of this plasmid in both human and animals. Nevertheless, the presence of a common plasmid in three different settings is alarming and should stimulate rapid introduction of adequate measures in the meat production chain to limit the spread of drug-resistant plasmids.
Author Contributions
Conception and design (VP, AE, SNS, AC); acquisition of data (VP, AE, SNS, AC, ACo, SS); analysis and interpretation of data (VP, AE, SNS, AC); drafting the work (VP, AE, SNS, AC); critical revison for important intellectual content (all authors); final approval of the version to be published (all authors); agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved (all authors).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Matt Riley and Stephen Kania for advice and support allocated to VP during his sabbatical leave from March to July 2015 at the Department of Biomedical and Diagnostic Sciences, College of Veterinary Medicine, University of Tennessee, Knoxville.
Funding. This work was supported by Grant 1.12.06 from the Federal Food Safety and Veterinary Office (FSVO) and internal funds of Institute of Infectious Diseases (IFIK) and Institute of Veterinary Bacteriology (IVB), University of Bern.
References
- Baudry P. J., Mataseje L., Zhanel G. G., Hoban D. J., Mulvey M. R. (2009). Characterization of plasmids encoding CMY-2 AmpC β-lactamases from Escherichia coli in Canadian intensive care units. Diagn. Microbiol. Infect. Dis. 65 379–383. 10.1016/j.diagmicrobio.2009.08.011 [DOI] [PubMed] [Google Scholar]
- Börjesson S., Jernberg C., Brolund A., Edquist P., Finn M., Landén A., et al. (2013). Characterization of plasmid-mediated AmpC-producing E. coli from Swedish broilers and association with human clinical isolates. Clin. Microbiol. Infect. 19 E309–E311. 10.1111/1469-0691.12192 [DOI] [PubMed] [Google Scholar]
- Brouwer M. S., Tagg K. A., Mevius D. J., Iredell J. R., Bossers A., Smith H. E., et al. (2015). IncI shufflons: assembly issues in the next-generation sequencing era. Plasmid 80 111–117. 10.1016/j.plasmid.2015.04.009 [DOI] [PubMed] [Google Scholar]
- Cao G., Allard M. W., Hoffmann M., Monday S. R., Muruvanda T., Luo Y., et al. (2015). Complete sequences of six IncA/C plasmids of multidrug-resistant Salmonella enterica subsp. enterica Serotype Newport. Genome Announc. 3 e00027–15. 10.1128/genomeA.00027-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents. Chemother. 53 2227–2238. 10.1128/AAC.01707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303 298–304. 10.1016/j.ijmm.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing; twenty-sixth informational supplement M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI). [Google Scholar]
- Cottell J. L., Saw H. T., Webber M. A., Piddock L. J. (2014). Functional genomics to identify the factors contributing to successful persistence and global spread of an antibiotic resistance plasmid. BMC Microbiol. 14:168 10.1186/1471-2180-14-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottell J. L., Webber M. A., Coldham N. G., Taylor D. L., Cerdeño-Tárraga A. M., Hauser H., et al. (2011). Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerg. Infect. Dis. 17 645–652. 10.3201/eid1704.101009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M., Bex F., Bergquist P. L., Maas W. K. (1988). Identification and classification of bacterial plasmids. Microbiol. Rev. 52 375–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Been M., Lanza V. F., de Toro M., Scharringa J., Dohmen W., Du Y., et al. (2014). Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 10:e1004776 10.1371/journal.pgen.1004776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierikx C., Essen-Zandbergen A., Veldman K., Smith H., Mevius D. (2010). Increased detection of extended spectrum β-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 145 273–278. 10.1016/j.vetmic.2010.03.019 [DOI] [PubMed] [Google Scholar]
- Dierikx C. M., van der Goot J. A., Smith H. E., Kant A., Mevius D. J. (2013). Presence of ESBL/AmpC -producing Escherichia coli in the broiler production pyramid: a descriptive study. PLoS ONE 8:e79005 10.1371/journal.pone.0079005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egervärn M., Börjesson S., Byfors S., Finn M., Kaipe C., Englund S., et al. (2014).Escherichia coli with extended-spectrum β-lactamases or transferable AmpC β-lactamases and Salmonella on meat imported into Sweden. Int. J. Food Microbiol. 171 8–14. 10.1016/j.ijfoodmicro.2013.11.005 [DOI] [PubMed] [Google Scholar]
- Endimiani A., Rossano A., Kunz D., Overesch G., Perreten V. (2012). First countrywide survey of third-generation cephalosporin-resistant Escherichia coli from broilers, swine, and cattle in Switzerland. Diagn. Microbiol. Infect. Dis. 73 31–38. 10.1016/j.diagmicrobio.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Guerra B., Fischer J., Helmuth R. (2014). An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet. Microbiol. 171 290–297. 10.1016/j.vetmic.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Hansen K. H., Bortolaia V., Nielsen C. A., Nielsen J. B., Schønning K., Agersø Y., et al. (2016). Host-specific patterns of genetic diversity among IncI1-Iγ and IncK plasmids encoding CMY-2 β-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl. Environ. Microbiol. 82 4705–4714. 10.1128/AEM.00495-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers P. M., van Hoek A. H., Graat E. A., Haenen A. P., Florijn A., Hengeveld P. D., et al. (2015). Methicillin-resistant Staphylococcus aureus and extended-spectrum and AmpC β-lactamase-producing Escherichia coli in broilers and in people living and/or working on organic broiler farms. Vet. Microbiol. 176 120–125. 10.1016/j.vetmic.2014.12.010 [DOI] [PubMed] [Google Scholar]
- Komano T. (1999). Shufflons: multiple inversion systems and integrons. Annu. Rev. Genet. 33 171–191. 10.1146/annurev.genet.33.1.171 [DOI] [PubMed] [Google Scholar]
- Leverstein-van Hall M. A., Dierikx C. M., Cohen S. J., Voets G. M., van den Munckhof M. P., van Essen-Zandbergen A., et al. (2011). Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17 873–880. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- Mathers A. J., Peirano G., Pitout J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28 565–591. 10.1128/CMR.00116-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. R., Laine P. S. (1990). The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 54 342–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo S. S., Slettemeås J. S., Berg E. S., Norström M., Sunde M. (2016). Plasmid and host strain characteristics of Escherichia coli resistant to extended-spectrum cephalosporins in the Norwegian broiler production. PLoS ONE 11:e0154019 10.1371/journal.pone.0154019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O., Börjesson S., Landén A., Bengtsson B. (2014). Vertical transmission of Escherichia coli carrying plasmid-mediated AmpC (pAmpC) through the broiler production pyramid. J. Antimicrob. Chemother. 69 1497–1500. 10.1093/jac/dku030 [DOI] [PubMed] [Google Scholar]
- Papagiannitsis C. C., Tzouvelekis L. S., Kotsakis S. D., Tzelepi E., Miriagou V. (2011). Sequence of pR3521, an IncB plasmid from Escherichia coli encoding ACC-4, SCO-1, and TEM-1 β-lactamases. Antimicrob. Agents Chemother. 55 376–381. 10.1128/AAC.00875-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porres-Osante N., Saenz Y., Somalo S., Torres C. (2015). Characterization of β-lactamases in faecal Enterobacteriaceae recovered from healthy humans in Spain: focusing on AmpC polymorphisms. Microb. Ecol. 70 132–140. 10.1007/s00248-014-0544-9 [DOI] [PubMed] [Google Scholar]
- Praszkier J., Wei T., Siemering K., Pittard J. (1991). Comparative analysis of the replication regions of IncB, IncK, and IncZ plasmids. J. Bacteriol. 173 2393–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiffert S. N., Hilty M., Kronenberg A., Droz S., Perreten V., Endimiani A. (2013a). Extended-spectrum cephalosporin-resistant Escherichia coli in community, specialized outpatient clinic and hospital settings in Switzerland. J. Antimicrob. Chemother. 68 2249–2254. 10.1093/jac/dkt208 [DOI] [PubMed] [Google Scholar]
- Seiffert S. N., Hilty M., Perreten V., Endimiani A. (2013b). Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug. Resist. Updat. 16 22–45. 10.1016/j.drup.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voets G. M., Fluit A. C., Scharringa J., Schapendonk C., van den M. T., Leverstein-van Hall M. A., et al. (2013). Identical plasmid AmpC β-lactamase genes and plasmid types in E. coli isolates from patients and poultry meat in the Netherlands. Int. J. Food. Microbiol. 167 359–362. 10.1016/j.ijfoodmicro.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Vogt D., Overesch G., Endimiani A., Collaud A., Thomann A., Perreten V. (2014). Occurence and genetic characteristics of third-generation cephalosporin-restitant Escherichia coli in Swiss retail meat. Microb. Drug. Resist. 20 485–494. 10.1089/mdr.2013.0210 [DOI] [PubMed] [Google Scholar]
- Wang J., Stephan R., Power K., Yan Q., Hächler H., Fanning S. (2014). Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J. Antimicrob. Chemother. 69 2658–2668. 10.1093/jac/dku206 [DOI] [PubMed] [Google Scholar]