ABSTRACT
Many autotrophic microorganisms are likely to adapt to scarcity in dissolved inorganic carbon (DIC; CO2 + HCO3− + CO32−) with CO2 concentrating mechanisms (CCM) that actively transport DIC across the cell membrane to facilitate carbon fixation. Surprisingly, DIC transport has been well studied among cyanobacteria and microalgae only. The deep-sea vent gammaproteobacterial chemolithoautotroph Thiomicrospira crunogena has a low-DIC inducible CCM, though the mechanism for uptake is unclear, as homologs to cyanobacterial transporters are absent. To identify the components of this CCM, proteomes of T. crunogena cultivated under low- and high-DIC conditions were compared. Fourteen proteins, including those comprising carboxysomes, were at least 4-fold more abundant under low-DIC conditions. One of these proteins was encoded by Tcr_0854; strains carrying mutated copies of this gene, as well as the adjacent Tcr_0853, required elevated DIC for growth. Strains carrying mutated copies of Tcr_0853 and Tcr_0854 overexpressed carboxysomes and had diminished ability to accumulate intracellular DIC. Based on reverse transcription (RT)-PCR, Tcr_0853 and Tcr_0854 were cotranscribed and upregulated under low-DIC conditions. The Tcr_0853-encoded protein was predicted to have 13 transmembrane helices. Given the mutant phenotypes described above, Tcr_0853 and Tcr_0854 may encode a two-subunit DIC transporter that belongs to a previously undescribed transporter family, though it is widespread among autotrophs from multiple phyla.
IMPORTANCE DIC uptake and fixation by autotrophs are the primary input of inorganic carbon into the biosphere. The mechanism for dissolved inorganic carbon uptake has been characterized only for cyanobacteria despite the importance of DIC uptake by autotrophic microorganisms from many phyla among the Bacteria and Archaea. In this work, proteins necessary for dissolved inorganic carbon utilization in the deep-sea vent chemolithoautotroph T. crunogena were identified, and two of these may be able to form a novel transporter. Homologs of these proteins are present in 14 phyla in Bacteria and also in one phylum of Archaea, the Euryarchaeota. Many organisms carrying these homologs are autotrophs, suggesting a role in facilitating dissolved inorganic carbon uptake and fixation well beyond the genus Thiomicrospira.
KEYWORDS: autotroph, bicarbonate transporter, carbon concentrating mechanism, carbon fixation, chemolithoautotroph, hydrothermal vent
INTRODUCTION
Autotrophic organisms of extraordinary physiological and phylogenetic diversity are responsible for the synthesis of the organic carbon necessary to sustain food webs. Befitting the broad phylogenetic distribution of autotrophic organisms within all three domains of life, multiple pathways are responsible for fixing and assimilating cytoplasmic dissolved inorganic carbon (DIC; consisting of CO2 + HCO3− + CO32−) (1). The mechanisms for the delivery of DIC from an aqueous environment, across the cell envelope, and to the cytoplasm are also likely to be extremely diverse but to date have been well characterized only among the cyanobacteria (2), some microalgae, and aquatic plants (3). Given that active DIC uptake facilitates rapid growth by autotrophs when DIC concentrations are low (4), uptake rates could have an impact on food web productivity.
Members of the genus Thiomicrospira contribute to food webs at hydrothermal vents, mud flats, and other sulfide-rich habitats worldwide (5, 6). These chemolithoautotrophic gammaproteobacteria are primarily mesophilic and can use reduced sulfur compounds (e.g., hydrogen sulfide and thiosulfate) and sometimes hydrogen gas as electron donors (7); oxygen is their terminal electron acceptor (8, 9). They fix CO2 via the Calvin-Benson-Bassham cycle (10).
Habitat heterogeneity at deep-sea hydrothermal vents complicates rapid chemolithoautotrophic growth. Warm (up to 50°C) hydrothermal fluid emitted from fissures in the basalt provides sulfide and other electron donors, while cold bottom water provides oxygen (11, 12). Dilute hydrothermal fluid also carries high concentrations of DIC (up to 6 mM), with an acidic pH as low as 5 that elevates CO2 to greater than 1 mM (12). Due to the differences in temperature, the two sources of water mix turbulently as they meet, and microorganisms attached to surfaces at the vents alternate between two conditions, warm (abundant reductant and CO2) and cold (abundant oxidant and low CO2) (13).
Despite the temporally disjointed supply of nutrients in its hydrothermal vent habitat, Thiomicrospira crunogena, a common isolate, is one of the fastest-growing autotrophic microorganisms (14). It is capable of sustaining this rapid growth in the presence of very low concentrations of DIC by inducing higher whole-cell affinity for DIC and raising its intracellular DIC pool up to 100-fold higher than the extracellular pool (15). These responses to low-DIC conditions indicate that T. crunogena has a CO2 concentrating mechanism (CCM).
In cyanobacteria, CCMs have been well described; DIC uptake by three evolutionarily distinct HCO3− transporters creates an elevated concentration of cytoplasmic HCO3− (2). Cytoplasmic HCO3− enters the carboxysomes, where carbonic anhydrase converts some to CO2, which is then fixed by carboxysomal ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) (16). CO2 escaping the carboxysomes may be trapped by reconversion to HCO3− by membrane-associated complexes (2). CCMs in other autotrophs are incompletely described. Carboxysomes have been well studied, particularly in the gammaproteobacterium Halothiobacillus neapolitanus, and these, as well as carboxysomes from other autotrophic microorganisms, function similarly to those present in cyanobacteria. H. neapolitanus carboxysomes contain RubisCO and carbonic anhydrase (17), and mutants carrying disrupted carboxysomal genes lose the ability to grow under low-DIC conditions (18, 19). The HCO3− transporters necessary to generate elevated intracellular HCO3− concentrations to facilitate carboxysome function have not yet been identified in any chemolithoautotrophic microorganisms.
In T. crunogena, the genes encoding the carboxysomal components of the CCM (shell proteins, carboxysomal RubisCO, and carbonic anhydrase) are carried in its genome (10), and these genes are upregulated under low-DIC conditions (20). In contrast, the DIC uptake mechanism is not apparent from the genome sequence. Orthologs to cyanobacterial genes encoding HCO3− transporters are absent (10, 21), and upregulation of genes likely to encode membrane transporters was not apparent when assayed by oligonucleotide microarrays (20). The T. crunogena genome encodes a periplasmic carbonic anhydrase, which does not appear to play a role in DIC uptake under low-DIC conditions. This carbonic anhydrase is not upregulated by low-DIC conditions, and inhibiting it with acetazolamide does not hinder cells' ability to create high intracellular DIC concentrations (22). Intracellular DIC accumulation by T. crunogena cells is sensitive to CCCP (carbonyl cyanide-m-chlorophenylhydrazone) and DCCD (N,N-dicyclohexylcarbodiimide) and therefore likely driven by either proton potential, ATP, or both (21).
In an effort to characterize DIC uptake mechanisms and other CCM components beyond the cyanobacteria, the proteomes of T. crunogena cultivated under low- and high-DIC conditions were compared. To identify genes necessary for growth under low-DIC conditions, cells were subjected to random and site-directed mutagenesis, and the responses in transcript abundance to DIC concentrations during growth were reexamined. Mutants that were unable to grow under low-DIC conditions were phenotypically characterized with respect to carboxysome presence and DIC uptake.
RESULTS
DIC-responsive proteins identified by proteomics.
Fourteen proteins were more abundant in wild-type T. crunogena grown under DIC-limited conditions, with log2 abundance ratios of >2 in both replicates of the proteome and q values of <0.05 (Table 1; see also Table S2 in the supplemental material). These include carboxysome shell proteins as well as carboxysomal enzymes RubisCO and carbonic anhydrase (encoded by Tcr_0838 to Tcr_0842), as anticipated given the role of carboxysomes in CCMs (as described above). Other proteins upregulated under DIC limitation are encoded nearby, some of which have been implicated in carboxysome function in other organisms (proteins corresponding to Tcr_0847, Tcr_0850, Tcr_0852, and Tcr_0854). Upregulated proteins encoded elsewhere on the chromosome included a prophage protein (encoded by Tcr_0682), two porins (encoded by Tcr_1315 and Tcr_2159), and two others (encoded by Tcr_2157 and Tcr_2158) that are homologous to proteins involved in sulfur metabolism (23).
TABLE 1.
Proteins more abundant when cells were cultivated under DIC limitation
| Fractiona | Log2 ratiob | No. of peptides | P value | q value | Locus tag, protein description |
|---|---|---|---|---|---|
| M1 | 4.6 | 41 | 5.53E−11 | 1.46E−09 | Tcr_0682, hypothetical protein (prophage) |
| M2 | 2.6 | 30 | 4.58E−10 | 6.45E−09 | |
| S1 | 7.0 | 185 | 3.38E−24 | 1.14E−22 | Tcr_0838, carboxysomal form I RubisCO CbbL |
| S2 | 4.9 | 134 | 2.56E−13 | 8.23E−12 | |
| S1 | 6.4 | 9 | 4.64E−04 | 1.34E−03 | Tcr_0839, carboxysomal form I RubisCO CbbS |
| S2 | 7.0 | 6 | 4.45E−03 | 1.29E−02 | |
| M1 | 4.4 | 53 | 1.62E−13 | 8.07E−12 | Tcr_0840, carboxysome shell protein CsoS2 |
| M2 | 6.2 | 72 | 5.69E−19 | 3.34E−17 | |
| M1 | 4.5 | 15 | 2.38E−05 | 2.31E−04 | Tcr_0841, carboxysome shell protein CsoSCA |
| M2 | 4.1 | 19 | 1.82E−07 | 1.73E−06 | |
| S1 | 5.5 | 11 | 3.29E−03 | 6.13E−03 | Tcr_0842, carboxysome shell protein CsoS4A |
| S2 | 6.2 | 7 | 1.77E−04 | 1.14E−03 | |
| M1 | 3.6 | 12 | 4.16E−05 | 3.59E−04 | Tcr_0847, bacterioferritin family protein |
| M2 | 5.4 | 16 | 5.20E−06 | 3.98E−05 | |
| S1 | 4.3 | 17 | 6.21E−06 | 3.23E−05 | Tcr_0850, Ham1-like protein |
| S2 | 6.5 | 8 | 1.44E−05 | 1.33E−04 | |
| S1 | 2.8 | 11 | 8.77E−03 | 1.30E−02 | Tcr_0852, LysR family transcriptional regulator |
| S2 | 2.9 | 14 | 6.01E−04 | 2.92E−03 | |
| M1 | 2.3 | 31 | 3.78E−05 | 3.34E−04 | |
| M2 | 2.7 | 30 | 5.08E−15 | 1.49E−13 | |
| M1 | 5.4 | 22 | 8.23E−09 | 1.56E−07 | Tcr_0854, conserved hypothetical protein, DUF2309 |
| M2 | 5.5 | 37 | 5.84E−13 | 1.14E−11 | |
| M1 | 3.8 | 31 | 3.57E−13 | 1.58E−11 | Tcr_1315, porin |
| M2 | 4.4 | 46 | 1.31E−20 | 9.20E−19 | |
| M1 | 3.0 | 51 | 1.18E−21 | 2.35E−19 | Tcr_2157, sulfur relay, DsrE/F-like protein |
| M2 | 2.4 | 66 | 7.31E−17 | 2.86E−15 | |
| S1 | 2.7 | 42 | 8.91E−17 | 1.88E−15 | |
| S2 | 2.0 | 33 | 5.23E−06 | 5.33E−05 | |
| M1 | 4.0 | 34 | 1.11E−11 | 4.43E−10 | Tcr_2158, sulfur relay, DsrE/F-like protein |
| M2 | 3.1 | 35 | 1.57E−08 | 1.79E−07 | |
| S1 | 3.7 | 56 | 7.81E−18 | 1.89E−16 | |
| S2 | 3.3 | 42 | 6.43E−10 | 1.49E−08 | |
| M1 | 3.0 | 264 | 1.79E−51 | 7.10E−49 | Tcr_2159, porin |
| M2 | 2.4 | 345 | 8.99E−96 | 3.16E−93 | |
| S1 | 2.6 | 81 | 2.89E−24 | 1.09E−22 | |
| S2 | 2.0 | 61 | 5.36E−29 | 1.12E−26 |
M1 and M2 are combined membrane-associated fractions from low- and high-DIC cultures (i.e., M1 consisted of a membrane-associated fraction from cells cultivated in one low-DIC chemostat and a membrane-associated fraction from cells cultivated in a high-DIC chemostat; M2 is a biological replicate of M1); S1 and S2 are corresponding soluble fractions.
Log2 ratio was calculated as the log2 of the ratio of abundances of quantified peptides for cells cultivated under low-DIC conditions/high-DIC conditions.
Fourteen proteins were also upregulated in cells cultivated under high-DIC, NH3-limited conditions (Table 2). Many of these proteins reflected NH3 scarcity in the growth medium: cyanate lyase (encoded by Tcr_0040), urea uptake and metabolism proteins (encoded by Tcr_1790 and Tcr_1792), and P-II proteins (encoded by Tcr_1339 and Tcr_1499). Noncarboxysomal RubisCO enzymes were also upregulated (form I RubisCO and activation protein CbbQ, encoded by Tcr_0427 to Tcr_0429, and form II RubisCO, encoded by Tcr_0424).
TABLE 2.
Proteins more abundant when cells were cultivated under NH3 limitation
| Fractiona | Log2 ratiob | No. of peptides | P value | q value | Locus tag, protein description |
|---|---|---|---|---|---|
| S1 | 3.3 | 21 | 8.49E−04 | 2.13E−03 | Tcr_0040, cyanate lyase |
| S2 | 3.6 | 18 | 3.63E−08 | 6.90E−07 | |
| S1 | 3.2 | 10 | 2.13E−04 | 6.61E−04 | Tcr_0220, aminotransferase |
| S2 | 2.1 | 9 | 9.20E−04 | 4.09E−03 | |
| M1 | 3.5 | 5 | 2.47E−02 | 4.65E−02 | Tcr_0344, glutamate 5-kinase |
| M2 | 4.6 | 6 | 4.99E−04 | 2.19E−03 | |
| S1 | 3.6 | 22 | 2.02E−07 | 1.62E−06 | Tcr_0423, RubisCO activation protein CbbQ |
| S2 | 3.9 | 20 | 1.32E−07 | 2.30E−06 | |
| M1 | 3.3 | 28 | 2.34E−10 | 5.81E−09 | Tcr_0424, form II RubisCO CbbM |
| M2 | 4.4 | 49 | 1.58E−13 | 3.27E−12 | |
| S1 | 3.4 | 168 | 2.02E−38 | 2.28E−36 | |
| S2 | 4.8 | 149 | 6.46E−36 | 2.70E−33 | |
| M1 | 4.4 | 33 | 5.88E−04 | 3.29E−03 | Tcr_0427, form I RubisCO CbbL |
| M2 | 4.5 | 35 | 4.87E−09 | 5.91E−08 | |
| S1 | 4.3 | 174 | 2.32E−41 | 3.93E−39 | |
| S2 | 4.6 | 108 | 1.23E−20 | 1.03E−18 | |
| M1 | 4.2 | 35 | 5.30E−16 | 4.21E−14 | Tcr_0428, form I RubisCO CbbS |
| M2 | 5.0 | 41 | 6.62E−21 | 5.82E−19 | |
| S1 | 4.4 | 97 | 9.82E−36 | 6.65E−34 | |
| S2 | 4.7 | 80 | 1.51E−22 | 1.58E−20 | |
| M1 | 2.1 | 75 | 5.17E−14 | 2.93E−12 | Tcr_0429, RubisCO activation protein CbbQ |
| M2 | 2.1 | 85 | 9.97E−16 | 3.51E−14 | |
| S1 | 2.6 | 7 | 5.84E−03 | 9.36E−03 | Tcr_0772, HAD superfamily hydrolase |
| S2 | 2.5 | 5 | 3.89E−03 | 1.15E−02 | |
| M1 | 4.2 | 5 | 2.45E−02 | 4.65E−02 | Tcr_1339, nitrogen regulatory protein P-II (GlnB, GlnK) |
| M2 | 3.4 | 3 | 2.04E−03 | 7.05E−03 | |
| S1 | 6.4 | 9 | 2.13E−02 | 2.38E−02 | |
| S2 | 5.7 | 5 | 4.89E−03 | 1.38E−02 | |
| M1 | 7.0 | 40 | 1.43E−09 | 3.16E−08 | Tcr_1499, nitrogen regulatory protein P-II (GlnB, GlnK) |
| M2 | 5.9 | 55 | 7.84E−26 | 9.19E−24 | |
| S1 | 7.0 | 53 | 3.30E−12 | 4.85E−11 | |
| S2 | 5.5 | 40 | 2.82E−11 | 7.86E−10 | |
| S1 | 3.9 | 5 | 1.08E−03 | 2.46E−03 | Tcr_1790, urea carboxylase-associated protein 2 |
| S2 | 3.4 | 6 | 1.27E−05 | 1.20E−04 | |
| S1 | 3.1 | 23 | 3.77E−07 | 2.97E−06 | Tcr_1792, urea carboxylase/allophanate hydrolase |
| S2 | 3.2 | 21 | 3.85E−05 | 3.16E−04 | |
| M1 | 2.6 | 46 | 2.53E−09 | 5.29E−08 | Tcr_2004, methyl-accepting chemotaxis protein, Pas/Pac sensor |
| M2 | 3.3 | 41 | 1.34E−06 | 1.15E−05 | |
| S1 | 2.7 | 66 | 6.05E−19 | 1.58E−17 | |
| S2 | 3.6 | 31 | 3.07E−05 | 2.67E−04 |
M1 and M2 are combined membrane-associated fractions from low- and high-DIC cultures (i.e., M1 consisted of a membrane-associated fraction from cells cultivated in one low-DIC chemostat and a membrane-associated fraction from cells cultivated in a high-DIC chemostat; M2 is a biological replicate of M1); S1 and S2 are corresponding soluble fractions.
Log2 ratio was calculated as the log2 of the ratio of abundances of quantified peptides for cells cultivated under high-DIC (i.e., low-NH3) conditions/low-DIC (i.e., high-NH3) conditions.
The protein encoded by Tcr_0853, which was not detected by the quantitative proteomics approach, was detected by mass-spectrometric analysis of SDS-PAGE-separated membrane extracts from cells cultivated under DIC-limited conditions. This protein was identified by 3 unique peptides (false discovery rate, <1%) (see Fig. S1 in the supplemental material).
Mutants unable to grow under low-DIC conditions.
Random mutagenesis resulted in over 6,900 strains that were screened for diminished ability to grow under low-DIC conditions. Ten strains from the library of randomly generated mutants were unable to grow under low-DIC conditions; for 9 of these, Tn5-RL27 had inserted into the carboxysome operon. The 10th strain had an insertion in Tcr_0854, at nucleotide 2006 (Tcr_0854-2006) (see Table S3 in the supplemental material).
Site-directed mutant strains were successfully constructed; two were used as controls for phenotype experiments (Tcr_0668-1614-WF and csoS3-399-2XO mutants [Table S3]). Site-directed mutant Tcr_0854-737-MF strain was constructed to compare to the Tcr_0854-2006 strain. The Tcr_0853-710-WF strain was also constructed because Tcr_0853 encodes a protein with 13 predicted transmembrane helices (TMHMM) and a signal peptide (SignalP) and is in an apparent operon with Tcr_0854 immediately downstream from the carboxysome operon. Merodiploid site-directed mutant strains of T. crunogena were also successfully constructed for the other noncarboxysomal genes whose protein products are more abundant under low-DIC conditions.
Mutant growth, carboxysome, and DIC uptake phenotypes.
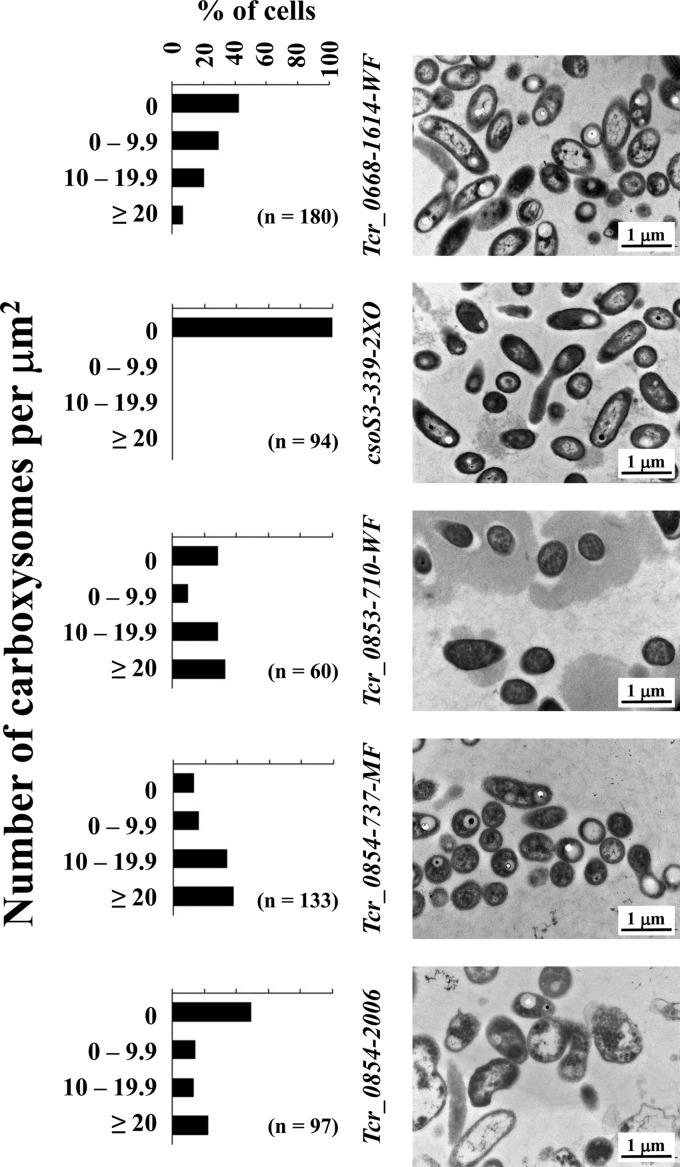
For the site-directed mutants, mutations in csoS3, Tcr_0853, and Tcr_0854 resulted in strains that could not grow under low-DIC conditions (Table S3). In order to elucidate whether this phenotype was due to defective carboxysomes, strains were examined via transmission electron microscopy. When cultivated under high-DIC conditions, carboxysomes were absent in all strains (data not shown). When cultivated under low-DIC conditions, all strains had frequencies of carboxysomes that were statistically significantly different from those of the Tcr_0668-1614-WF strain (chi-square test; P < 0.05). Carboxysomes were essentially absent from mutants with lesions within the carboxysome operon (csoS3-399-2XO) (Fig. 1). For strains with mutations in Tcr_0853 and Tcr_0854, carboxysome morphology was identical to that of wild-type and Tcr_0668-1614-WF strains. Carboxysomes in these cells were particularly abundant, with the numbers of cells whose carboxysome counts fell in the highest category (>20) exceeding what was observed for the Tcr_0668-1614-WF strain (Fig. 1).
FIG 1.
Carboxysome abundance in mutant strains of T. crunogena cultivated under low-DIC conditions. Carboxysomes are apparent as round, regularly sized electron-dense inclusions within cells. Histograms depict percentages of cells with a given number of carboxysomes per square micrometer of cytoplasm transected in the electron micrographs; n, number of cells examined.
The rates and magnitudes of DIC accumulation and fixation differed markedly among mutant strains. The Tcr_0668-1614-WF strain was capable of accumulating intracellular DIC and fixing carbon similarly to wild-type T. crunogena (15), confirming that Tcr_0668 does not play a role in the T. crunogena CCM and that plasmid and transposon insertion into the chromosome do not interfere with DIC uptake and fixation. Rates of accumulation in the Tcr_0854-2006 mutant were much lower (Fig. 2). An estimate for the rate of DIC uptake for both strains was calculated by adding the concentration of intracellular DIC to the concentration of fixed carbon at each time point and calculating a rate over the 30-s time course of the assay (106 ± 22 mM/min for the Tcr_0668-1614-WF mutant; 3.2 ± 0.6 for the Tcr_0854-2006 mutant). These estimates are conservative, since they do not include DIC loss from the cell either from diffusion (DIC) or excretion (fixed carbon).
FIG 2.
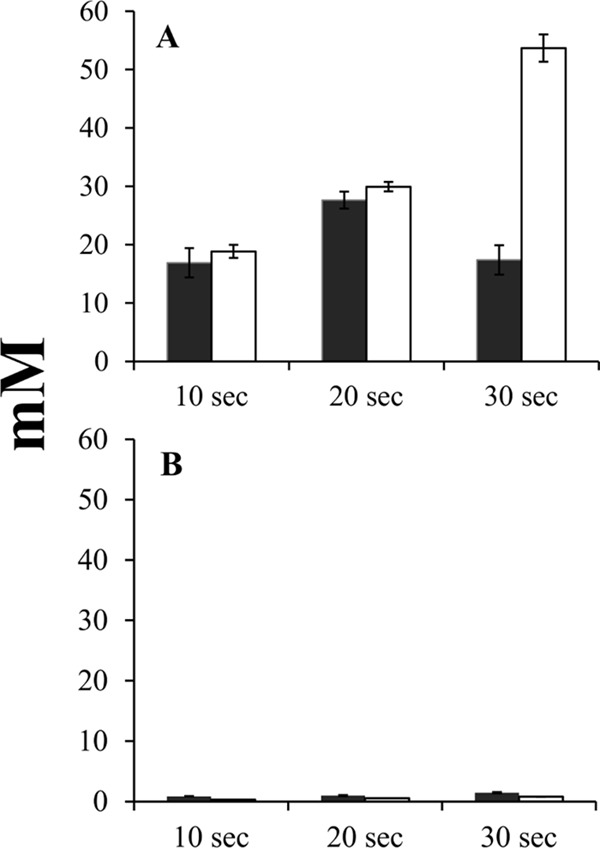
Time course of intracellular DIC accumulation (dark bars) and fixation (white bars) by mutant strains of T. crunogena. y axis, intracellular concentration of DIC or acid-stable (fixed) carbon. (A) Tcr_0668-1614-WF mutant strain; (B) Tcr_0854-2006 mutant strain. Mutant strains are described in Table S3 in the supplemental material. Error bars represent the standard errors.
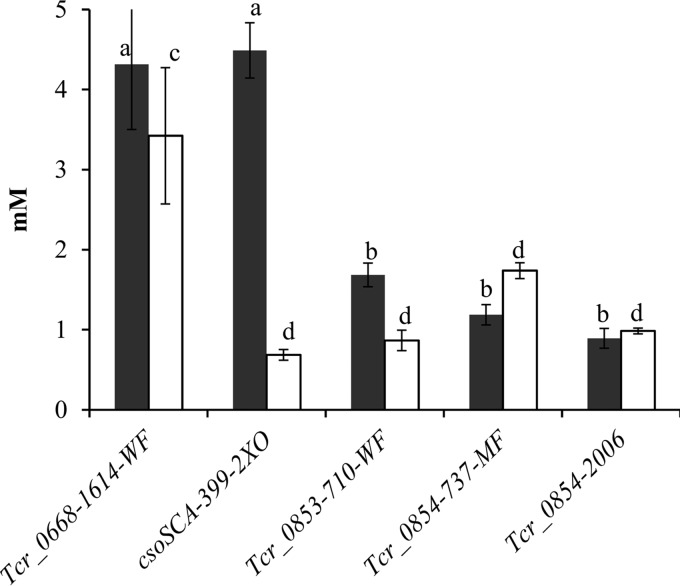
All mutants with interruptions in Tcr_0853 and Tcr_0854 had diminished abilities to accumulate and fix intracellular DIC under low-DIC conditions (Fig. 3) (P < 0.05). The csoS3-399-2XO mutant was capable of generating intracellular DIC to concentrations similar to those seen in the Tcr_0668-1614-WF mutant but had a substantially lower level of carbon fixation (Fig. 3) (Tukey's test; P < 0.05), which is consistent with diminished carboxysomal carbonic anhydrase activity (19). RubisCO activities were high in all strains when cultivated under low- and high-DIC conditions (see Fig. S2 in the supplemental material).
FIG 3.
Intracellular DIC accumulation (dark bars) and fixation (white bars) by mutant strains of T. crunogena. y axis, intracellular concentration of DIC or acid-stable (fixed) carbon. Mutant strains (described in Table S3) were incubated for 60 s. Error bars represent the standard errors, and letters indicate values that differ from the others significantly (Tukey's test; P < 0.05).
Transcription of Tcr_0853 and Tcr_0854 in wild-type and mutant strains.
In wild-type cells, transcript abundances for Tcr_0853, Tcr_0854, and csoS3 were markedly higher under DIC-limited conditions in chemostats (Table 3). This was also the case when wild-type cells were exposed to low-DIC conditions for 2 h (Table 4).
TABLE 3.
Fold change in Tcr_0853, Tcr_0854, and csoS3 transcript abundance in wild-type T. crunogena cells cultivated under DIC and NH3 limitation in chemostats
| Locus tag | Fold increasea | ΔΔCT ± SD |
|---|---|---|
| Tcr_0853 | 263 | −8.0 ± 1.8 |
| Tcr_0854 | 340 | −8.4 ± 1.6 |
| csoS3 | 859 | −9.7 ± 0.9 |
Fold increase in transcript abundance: DIC limited/NH3 limited = 2−ΔΔCT. RNA was extracted from wild-type cells grown in three DIC-limited chemostats and three NH3-limited chemostats.
TABLE 4.
Fold change in Tcr_0853, Tcr_0854, and csoS3 transcript abundance after 2 h of cultivation under low-DIC conditions
| Locus tag | Strain | Fold increasea | ΔΔCT ± SDb | Amplicon positionc |
|---|---|---|---|---|
| Tcr_0853 | Wild type | 643 | −9.3 ± 0.5 | 1283–1415 |
| Tcr_0668-1614-WF | 1,351 | −10.4 | ||
| csoS3-399-2XO | 1,097 | −10.1 | ||
| Tcr_0853-710-WF | 137 | −7.1 | ||
| Tcr_0854-737-MF | 1,097 | −10.1 | ||
| Tcr_0854-2006 mutant | 4,389 | −12.1 | ||
| Tcr_0854 | Wild type | 712 | −9.5 ± 0.1 | 1200–1339 |
| Tcr_0668-1614-WF | 832 | −9.7 | ||
| csoS3-399-2XO | 549 | −9.1 | ||
| Tcr_0853-710-WF | 5 | −2.3* | ||
| Tcr_0854-737-MF | 2 | −1.1* | ||
| Tcr_0854-2006 mutant | 1,176 | −10.2 | ||
| csoS3 | Wild type | 2,740 | −11.4 ± 0.8 | 588–736 |
| Tcr_0668-1614-WF | 10,809 | −13.4 | ||
| csoS3-399-2XO | 37 | −5.2* | ||
| Tcr_0853-710-WF | 6,208 | −12.6 | ||
| Tcr_0854-737-MF | 5,405 | −12.4 | ||
| Tcr_0854-2006 mutant | 14,263 | −13.8 |
Fold increase in transcript abundance is calculated as low DIC/high DIC (= 2−ΔΔCT).
For the wild type, standard deviations are based on two cultures that were cultivated under high-DIC conditions before exposure to low-DIC conditions. For the others, a single culture was cultivated (n = 1). Values with asterisks were detected as outliers from the others (P < 0.05) using Grubb's test.
Position, in nucleotides, relative to the first nucleotide of the start codon of the gene.
Likewise, strains carrying mutated copies of Tcr_0853, Tcr_0854, and to a lesser extent csoS3 had elevated levels of transcripts of csoS3 when shifted to low-DIC conditions for 2 h (Table 4). Based on Grubb's test (described in Materials and Methods), mutant Tcr_0853-710-WF and Tcr_0854-737-MF strains had smaller increases in transcript abundance from Tcr_0854 than the others (Table 4), consistent with a polar mutation in Tcr_0853-710-WF and insertion of pLD55 upstream (at nucleotide 710) of the reverse transcription-quantitative (qRT)-PCR amplicon position (nucleotides 1200 to 1339) in Tcr_0854-737-MF strain. The ΔΔCT value (where CT is threshold cycle) for Tcr_0854 in Tcr_0854-2006 strain was not affected, probably because insertion of Tn5-RL27 (at nucleotide 2006) was downstream of the amplicon position. Interestingly, in the Tcr_0853-710-WF mutant strain, the abundance of Tcr_0853 transcripts was lower than the others, though not to statistical significance (Table 4). In this strain, the region of Tcr_0853 targeted by qRT-PCR was downstream from the transposon insertion site in Tcr_0853-710-WF strain, and in this merodiploid strain, the native promoter was adjacent to the wild-type copy of Tcr_0853, which likely facilitated continued transcription of this gene.
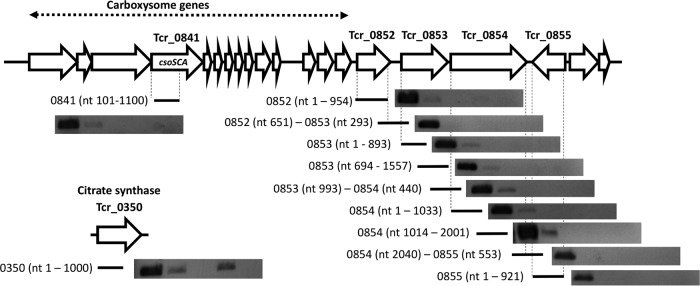
The similarity in fold increases in transcript abundance from Tcr_0843 and Tcr_0854 in wild-type cells (Tables 3 and 4), as well as their juxtaposition (Tcr_0854 has a start codon that overlaps the stop codon of Tcr_0853), is consistent with cotranscription, as are RT-PCR products targeting a potential polycistronic mRNA transcribed from these genes. For cells grown under low-DIC conditions, RT-PCR products were visible for transcripts from Tcr_0852, Tcr_0853, and Tcr_0854 but not from Tcr_0855 (Fig. 4). When using primers designed to produce amplicons spanning sequential genes, RT-PCR products were visible only when spanning Tcr_0853 and Tcr_0854, suggesting that these genes are cotranscribed. For cells grown under high-DIC conditions, RT-PCR products were not visible for Tcr_0852 to Tcr_0855, which is consistent with their transcripts being less abundant than in low-DIC cells. Citrate synthase transcripts were, however, successfully amplified from both low- and high-DIC cells, suggesting that RNA purified from high-DIC cells was of sufficient quality for transcripts to be detected (Fig. 4).
FIG 4.
Cotranscription of Tcr_0853 and Tcr_0854 in wild-type T. crunogena cells cultivated in chemostats under low- and high-DIC conditions. Agarose gel-electrophoresed, ethidium bromide-stained, RT-PCR amplicons are shown for each RT-PCR target, in five lanes, from left to right: genomic DNA, cells cultivated under low-DIC conditions, cells cultivated under low-DIC conditions (reverse transcriptase-free control), cells cultivated under high-DIC conditions, and cells cultivated under high-DIC conditions (reverse transcriptase-free control). Transcription of Tcr_0841 (csoS3) was monitored to verify CCM induction; Tcr_0350 was targeted to verify that the mRNA was of sufficient quality to act as an RT-PCR template.
Phylogenetic distribution of Tcr_0853 and Tcr_0854 homologs.
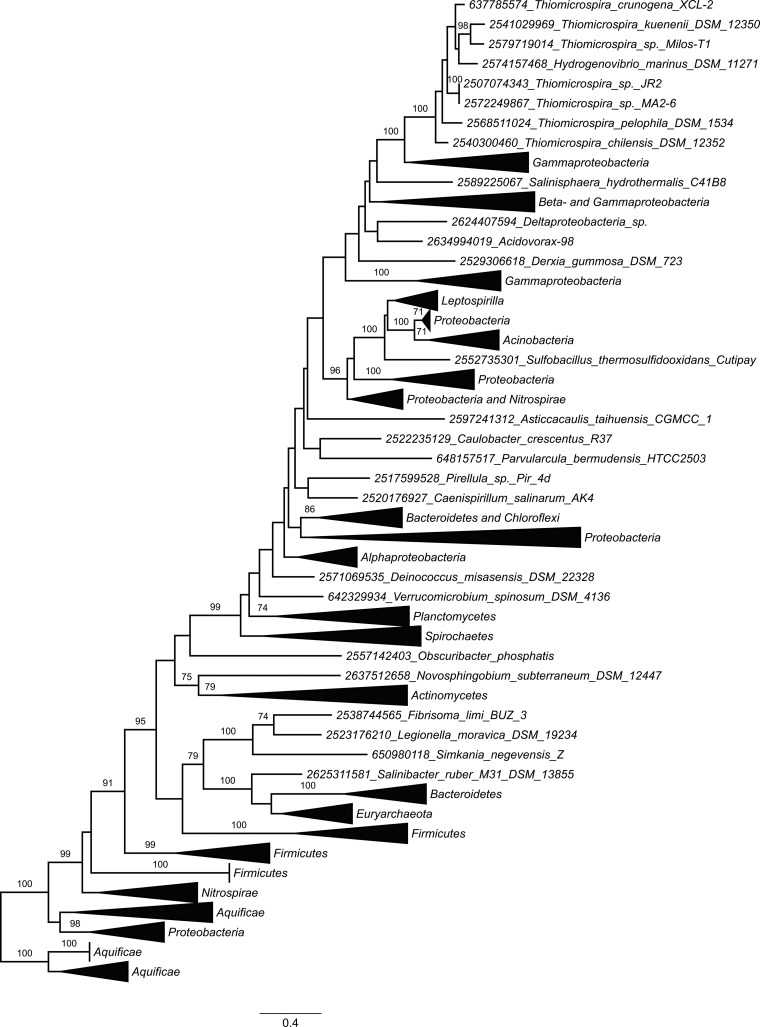
Genes homologous to Tcr_0853 and Tcr_0854 were found to be widespread, present in sequenced genomes from 14 phyla of Bacteria and the phylum Euryarchaeota (class Halobacteria) of Archaea. The phyla Proteobacteria, Firmicutes, Actinobacteria, and Euryarchaeota were particularly well represented (Fig. 5; see also Table S4 in the supplemental material). The phylogenetic tree constructed from the concatenated alignment of Tcr_0853 and Tcr_0854 homologs (Fig. 5) was congruent with trees generated from each gene individually (data not shown). Most of the homologs from the genus Thiomicrospira fell into a single well-supported clade nested within a larger clade dominated by genes from other gammaproteobacteria (Fig. 5).
FIG 5.
Maximum likelihood phylogenetic analysis of concatenated alignments of Tcr_0853 and Tcr_0854 homologs. Bootstrap values from 100 resamplings of the alignment are indicated when they exceeded 70%. Numbers preceding taxon names are gene object ID numbers from the Integrated Microbial Genomes database (http://img.jgi.doe.gov/), corresponding to the Tcr_0854 homolog of each pair. Collapsed clades are labeled by the dominant phyla or classes present in the clade.
DISCUSSION
Low-DIC conditions stimulated the expression of carboxysomes, as anticipated, and upregulated genes that may play a role in DIC uptake (porins, transport proteins) (Table 1; see also Table S2 in the supplemental material). In most cases, these proteins corresponded to genes with elevated transcript abundances under low-DIC conditions (Table 5) (20). Proteins corresponding to three abundant transcripts (Tcr_0843, 0845, 0853) could not be detected in the proteome (Table 5); either their expression was posttranscriptionally regulated, or they were present but not quantified due to analytical limitations. This appears to have been the case for the protein encoded by Tcr_0853, as it was detected by subsequent SDS-PAGE-based analysis (Fig. S1). In other cases (for Tcr_0692, Tcr_2157, and Tcr_2159), transcripts slightly less abundant under low-DIC conditions corresponded to more-abundant proteins, which may indicate posttranscriptional regulation.
TABLE 5.
Proteins and transcripts more abundant in cells cultivated under DIC limitation
| Locus tag, protein description | Ratios in proteomea | Ratios in transcriptomeb |
|---|---|---|
| Tcr_0682, hypothetical protein (prophage) | 6, 24 | 0.9, 1.1, 1.2 |
| Tcr_0838, carboxysomal form I RubisCO CbbL | 30, 128 | 4.5, 5.2 |
| Tcr_0839, carboxysomal form I RubisCO CbbS | 84, 128 | 9.2, 10.7 |
| Tcr_0840, carboxysome shell protein CsoS2 | 21, 74 | 2.9, 12.9, 14.3 |
| Tcr_0841, carboxysome shell protein CsoSCA | 17, 23 | 1.5, 3.2, 3.7 |
| Tcr_0842, carboxysome shell protein CsoS4A | 45, 74 | 3.2, 4.2, 5.1 |
| Tcr_0843, carboxysome peptide B | ND | 1.7, 2.4, 2.7 |
| Tcr_0844, carboxysome shell protein CsoS1 | 2, 2 | 6.1, 12.8 |
| Tcr_0845, carboxysome shell protein CsoS1 | ND | 2.7, 19.6 |
| Tcr_0846, carboxysome shell protein CsoS1C | S1, 45; M2, 23 | 12.6, 14.6 |
| Tcr_0847, bacterioferritin family protein | 12, 42 | 4.5, 11.1 |
| Tcr_0848, α-carboxysome RubisCO assembly factor | 13, 28c | 3.6, 5.5 |
| Tcr_0850, Ham1-like protein | 20, 91 | 1.2, 1.4, 1.7 |
| Tcr_0852, LysR family transcriptional regulator | S, 7, 7; M, 5, 6 | 0.9, 1.2, 1.4 |
| Tcr_0853, NADH subunit 5/MnhA homolog | ND | 1.1, 1.3, 1.6 |
| Tcr_0854, conserved hypothetical protein, DUF2309 | 42, 45 | 1.2, 1.4 |
| Tcr_1019, hypothetical protein | S2, 6 | 3.0, 3.0 |
| Tcr_1315, porin | 14, 21 | 1.3, 2.4, 3.3 |
| Tcr_2157, sulfur relay, DsrE/F-like protein | S, 4, 6; M, 5, 8 | 0.8, 0.8 |
| Tcr_2158, sulfur relay, DsrE/F-like protein | S, 10, 13; M, 9, 16 | 0.8, 0.9 |
| Tcr_2159, porin | S, 4, 6; M, 5, 8 | 0.8, 0.9, 0.9 |
Ratios of abundances of quantified peptides for cells cultivated under low-DIC conditions/high-DIC conditions. When relevant, the cell fractions are indicated (S, soluble; M, membrane; S1, M1, and M2 indicate biological replicates of these fractions). ND, not determined.
Ratios of transcripts for cells cultivated under low-DIC conditions/high-DIC conditions, from reference 20.
Not included in Table 1 because q values exceeded 0.05.
Interrupting csoS3 resulted in a requirement for high DIC for growth, as has been observed in H. neapolitanus (19), likely due to a disruption of carboxysome function, although in H. neapolitanus, cells with mutated csoS3 synthesize carboxysomes. Carboxysome absence in the T. crunogena csoS3 mutant is puzzling and could suggest a polar effect on the expression of carboxysome shell genes downstream from the mutated gene.
The low-DIC-induced protein encoded by Tcr_0854 clearly plays a role in DIC uptake. Transposon-mediated interruption of this gene at either of two positions (nucleotide 737 or 2006) or the gene Tcr_0853, immediately upstream, resulted in a decrease of DIC uptake rates (Fig. 2) and loss of ability to generate elevated concentrations of intracellular DIC, as well as a diminished ability to fix CO2 (Fig. 3), resulting in an inability to grow under low-DIC conditions (Table S3). In these mutants, RubisCO activities were similar to those in the Tcr_0668-1614-WF mutant (Fig. S2), as was carboxysome morphology (Fig. 1), indicating that the effect may be independent of the functioning of these microcompartments. Interestingly, carboxysome numbers in the cells were enhanced (Fig. 1), perhaps in an attempt to compensate for the lower concentrations of intracellular DIC (Fig. 3).
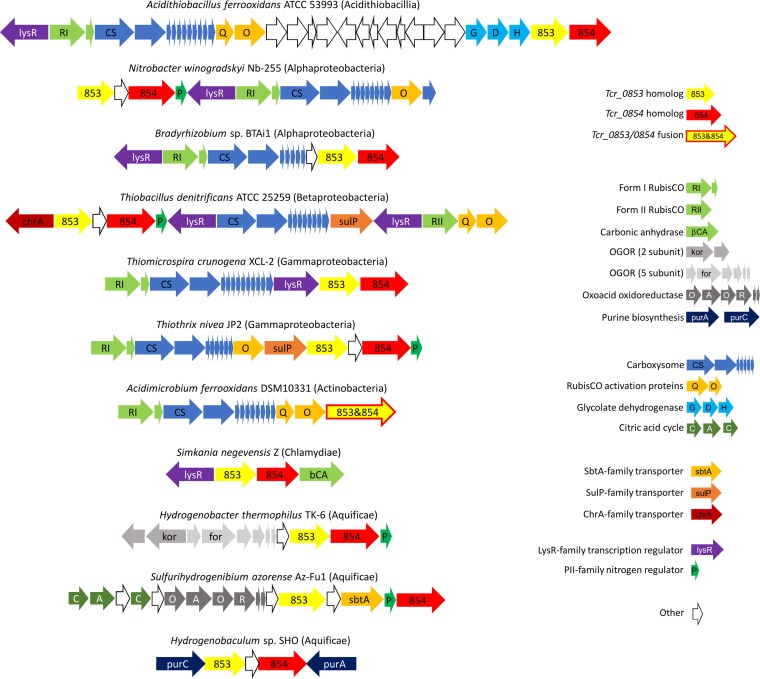
The protein encoded by Tcr_0854 belongs to PFam10070 (http://pfam.xfam.org/family/PF10070), which consists of a family of conserved proteins whose function is unknown. Many aspects of adjacent gene Tcr_0853 suggest a potential function. Tcr_0854 was cotranscribed with Tcr_0853 (Fig. 4), and both had greatly enhanced transcript abundances when cells were grown under low-DIC conditions (Table 3), suggesting a potential role in a CCM. Given that the Tcr_0853-encoded protein was predicted to have 13 transmembrane helices, it is possible that Tcr_0853 and Tcr_0854 act together to transport DIC. Interaction between the proteins encoded by these genes is also supported by the collocation of their homologs on chromosomes in many organisms. A total of 5,377 finished archaeal and bacterial genomes in IMG (Integrated Microbial Genomes database; http://img.jgi.doe.gov/) were queried via the “Cassette Search” feature, to identify organisms in which homologs are collocated in candidate operons (24). Of 664 taxa with a homolog to Tcr_0854, 650 had Tcr_0853 and Tcr_0854 homologs cooccurring in candidate operons. Indeed, in some taxa, a single fused gene in which the 5′ end is homologous to Tcr_0853 and the 3′ end is homologous to Tcr_0854 was found to be present (Fig. 6).
FIG 6.
Collocation of Tcr_0853 (yellow) and Tcr_0854 (red) homologs to genes encoding enzymes that consume dissolved inorganic carbon and associated pathways or cellular structures. For Acidimicrobium ferrooxidans, the Tcr_0853 and Tcr_0854 homologs exist as a single fused gene. OGOR is oxoglutarate:ferredoxin oxidoreductase, whose substrate (oxoglutarate) has been empirically determined (60). The alpha keto organic acid substrate for oxoacid oxidoreductase has not been characterized. Taxon names include class or phylum in parentheses.
The protein encoded by Tcr_0853 belongs to PFam00361 (http://pfam.xfam.org/family/PF00361), comprised of membrane-spanning proton transporters and their homologs. Included in this family are the proton-translocating subunit from both the NADH dehydrogenase complex (synonyms include NuoL, ND5, and NdhF [25]) and a subunit from Mrp-type Na+/H+ antiporters (26). Also included are the transmembrane subunits from the CO2 uptake systems present in some cyanobacteria (27). Genes encoding the other subunits of the Mrp-type Na+/H+ antiporter as well as the cyanobacterial CO2 uptake systems (CupA/ChpY or CupB/ChpZ) are absent from the T. crunogena genome, which suggests a different role for Tcr_0853.
One model for Tcr_0853-Tcr_0854 is that together they act as a proton-DIC symporter. This model is consistent with inhibitor studies of DIC uptake (21). Collapsing the proton potential with the protonophore CCCP resulted in low intracellular DIC concentrations, as would be expected if a proton-DIC symporter was responsible for generating elevated concentrations of intracellular DIC. DCCD addition also diminished intracellular DIC concentrations, and this was taken to indicate that DIC uptake was linked to the intracellular ATP pool, since this compound inhibits ATP synthase (21). DCCD can also interact with NADH dehydrogenase, where it has been shown to bind to the hydrophobic subunits of this complex (28). This has been well established for NuoH (29), but incubating the complex with [14C]DCCD results in labeling of multiple hydrophobic subunits (28). It is possible that Tcr_0853 also binds DCCD and its DIC uptake activity is inhibited. It is also possible that Tcr_0854 and perhaps Tcr_0853 stimulate DIC uptake via other mechanisms. They may act as an Na+/H+ antiporter that could create an Na+ gradient to power DIC uptake via a separate Na+/HCO3− symporter.
Genome data suggest that Tcr_0853 and Tcr_0854 homologs play a role in DIC uptake in several phyla. The genomes of many of the organisms carrying homologs to Tcr_0853 and Tcr_0854 also carry genes encoding steps in autotrophic carbon fixation pathways (Fig. 5 and 6; Table S4). Among them are many Proteobacteria that carry form I or form II RubisCO genes (active in the Calvin-Benson-Bassham cycle [30]), Chloroflexi with genes encoding malonyl coenzyme A (malonyl-CoA) reductase, propionyl-CoA synthase, and propionyl-CoA carboxylase, necessary for the hydroxypropionate cycle (31–33), and many members of Nitrospirae and Aquificae with ATP-dependent citrate lyase genes or citryl-CoA synthetase and citryl-CoA lyase, key enzymes of the reductive citric acid cycle (rCAC) (34). Further, Tcr_0853 and Tcr_0854 homologs are often juxtaposed to genes encoding carboxylases (RubisCO; oxoacid:acceptor oxidoreductase [rCAC carboxylases]) and other enzymes that consume DIC (carbonic anhydrase) or enzymes from biosynthetic pathways that consume DIC (purine synthesis [Fig. 6]).
Other genes found nearby include those needed to respond to low-DIC conditions, such as those encoding the subunits of glycolate dehydrogenase, used to metabolize the glycolate generated by the RubisCO oxidase reaction (35). In many organisms, Tcr_0853 and Tcr_0854 homologs are found near genes encoding carboxysomes. Sometimes genes encoding predicted permeases are nearby as well (Fig. 6). This collocation and the genes' widespread occurrence in autotrophs suggest that these genes might play a role in DIC uptake in a diverse group of organisms, including autotrophic representatives throughout the Proteobacteria, and much farther afield, in the phyla Firmicutes, Actinobacteria, Aquificae, Chloroflexi, and Nitrospirae.
Identifying a potential DIC transporter, particularly one that is present in many autotrophs in six phyla, will be quite helpful in understanding the activities of these organisms in situ. Given that some of these organisms are responsible for acid mine drainage (Acidithiobacillus spp. and Leptospirillum spp.) while others (Thiomicrospira spp.) are common and sometimes dominant at hydrothermal vents (36), it will be of interest to determine the degree to which active transport of DIC facilitates their growth in nature.
DIC transporters from cyanobacteria are currently being considered for bioengineering autotrophic organisms to produce compounds of industrial interest (37). Adding transporters from autotrophs beyond the cyanobacteria would expand the versatility of these bioengineered organisms, given the growth of these organisms in a very broad range of environmental conditions (e.g., pH) and the differences in transporter structure, which may make them more robust for use in industrial processes.
MATERIALS AND METHODS
Growth medium.
T. crunogena XCL-2 (10, 38) cultures were grown in chemostats (New Brunswick Scientific BioFlo 110) in TASW medium, an artificial saltwater medium, with 430 mM NaCl, 7.6 mM (NH4)2SO4, 6.1 mM MgSO4, 3.1 mM K2HPO4, 2.0 mM CaCl2, and 1 ml per liter trace element solution SL-8 (39). Modifications of the artificial seawater medium originally used to cultivate T. crunogena (14) included increasing the thiosulfate (Na2S2O3) to 40 mM and adding 100 mM Na HEPES (pH 8; (15). Chemostat dilution rates were 0.1 h−1.
Cells were cultivated under either DIC or NH3 limitation, as previous work had demonstrated that DIC limitation induced the expression of a CCM in this organism while NH3 limitation did not (15, 20, 22). To cultivate cells under DIC limitation, TASW was prepared with 2 mM NaHCO3 and the growth chamber was pulsed with O2 to maintain oxygen tensions at 5 to 20% atmospheric saturation using the BioFlo dO2 controller. For NH3 limitation, the concentration of NaHCO3 in TASW was raised to 50 mM, and (NH4)2SO4 was lowered to 0.4 mM. Under NH3 limitation, the growth chamber was pulsed with 5% CO2–95% O2 to maintain oxygen tensions at 5 to 20%. These conditions have been shown to result in 0.08 to 0.1 mM DIC in the growth chamber for DIC-limited cells and undetectable ammonia concentrations for NH3-limited cells (15).
For mutagenesis, T. crunogena was grown under DIC limitation in TASW with the NaCl content decreased to 20% (65 mM; referred to as “20% TASW”) in order to facilitate mating with Escherichia coli, which was grown in TYE medium (40) supplemented with kanamycin (25 mg/liter). TCTYE medium, used for mating T. crunogena with E. coli, consisted of TASW with the NaCl lowered as described above and supplemented with tryptone (10 g/liter), yeast extract (5 g/liter), and agar (15 g/liter).
Influence of DIC on the proteome.
T. crunogena was cultivated in four chemostats: two were DIC limited (low DIC), and two were NH3 limited (high DIC). The NH3-limited chemostats contained 0.8 mM 15NH3Cl. Once stable steady-state conditions had been attained and the growth chamber (1 liter) had been replenished with 5 volumes (5 liters) of fresh medium from the reservoirs, cells were harvested via centrifugation (5,000 × g, 15 min, 4°C) and stored frozen at −80°C.
Each cell pellet was fractionated into soluble and membrane fractions. Pellets were thawed and resuspended in 5 ml 50 mM Tris (pH 8), 10 mM MgSO4, and 1 mM dithiothreitol (DTT) and subjected to four freeze-thaw cycles. They were then sonicated with glass beads (5 × 30 s on ice, 1-min resting period between sonications) and centrifuged (6,000 × g, 30 min, 4°C) to remove intact cells. The pellets were resubjected to freeze-thaw, sonication, and centrifugation to maximize protein extraction. Supernatants from both cycles of treatment were combined and centrifuged to pellet membranes (75,000 × g, 30 min, 4°C). Membrane pellets and supernatants from this second centrifugation step were carried forward for subsequent analysis. Protein concentrations for supernatant and membrane fractions were quantified (RC DC protein assay [Bio-Rad, Inc.] using bovine serum albumin as the standard). Soluble fractions from unlabeled low-DIC cultures and 15N-labeled high-DIC cultures were combined at a protein ratio of 1:1, resulting in two mixed soluble fractions (since two low-DIC and two high-DIC chemostats were sampled). The same procedure was undertaken for membrane fractions.
The four samples with mixed unlabeled and 15N-labeled fractions were measured using a quantitative proteomics approach (41). Briefly, after denaturation in 6 M guanidine and 10 mM DTT, proteins were digested into peptides using sequencing grade trypsin (Promega Life Sciences). The peptide samples were analyzed using the 12-step, split-phase MudPIT (42) method. An LTQ-Orbitrap Elite instrument (Thermo Scientific) was used for data-dependent tandem mass spectrometry (MS/MS) analysis with each high-resolution full scan followed by 12 low-resolution MS/MS scans. The MS/MS data were searched against a concatenated forward-reverse protein database constructed from the T. crunogena genome (10) in unlabeled and 15N-labeled amino acids using Sequest (43) and DTASelect (44). Selected ion chromatogram extraction and peptide abundance ratio estimation were performed with ProRata (45). The P values for the abundance changes of proteins were calculated by Student's t test on the abundance changes of the constituent peptides of a protein and then were corrected to q values using the qvalue R package (46). The confidence intervals of protein abundance ratios were calculated by a profile likelihood algorithm in ProRata (46, 47).
The protein corresponding to Tcr_0853 was not detectable in protein profiles for low- and high-DIC cells despite its relevance to DIC uptake (see Results and Discussion). To determine whether low-DIC cells synthesize this protein, membrane preparations were subjected to SDS-PAGE (48). Coomassie blue-stained gel fragments were excised from the molecular mass region corresponding to approximately 50 to 75 kDa and processed as described previously (49) before being digested with trypsin/Lys-C overnight at 37°C. Peptides were dried in a vacuum concentrator (Labconco) and resuspended in 0.1% formic acid for liquid chromatography (LC)-MS/MS analysis. Experiments were performed in triplicate.
Peptides were separated using a C18 reversed-phase high-performance liquid chromatography (HPLC) column (New Objective) on an EASY-nLC1000 (Thermo Fisher Scientific) with a 60-min gradient (4 to 40% acetonitrile with 0.1% formic acid) and analyzed on a linear ion trap-Orbitrap instrument (Orbitrap XL; Thermo Fisher Scientific) using data-dependent acquisition, whereby the top 10 most-abundant ions were selected for MS/MS analysis in the linear ion trap.
Raw data files were processed in MaxQuant (version 1.5.0.30; www.maxquant.org) and searched against the UniprotKB T. crunogena protein sequence database. Search parameters included constant modification of cysteine by carbamidomethylation and the variable modification methionine oxidation. Proteins were identified using the filtering criteria of 1% protein and peptide false-discovery rate.
Mutagenesis.
Random mutagenesis was utilized to identify genes required for growth under low-DIC conditions. Wild-type T. crunogena cells were mated with E. coli BW20767 carrying plasmid pRL-27, which encodes transposon Tn5-RL27 (40). T. crunogena was cultivated in 20% TASW in DIC-limited chemostats. Cells were harvested via centrifugation (14,000 × g, 5 min, 4°C) and resuspended at an optical density at 600 nm (OD600) of 20 in 20% TASW medium. E. coli BW20767 was grown to an OD600 of 0.7, washed twice in 20% TASW, and resuspended as described for T. crunogena. To mate the cells, 50 μl of a 1:1 T. crunogena-E. coli suspension was spotted onto solid TCTYE medium. Plates were incubated overnight at 32°C under a 5% CO2 headspace. Growth was scraped from the surface of the solid medium, suspended, and washed three times with TASW, spread onto recovery plates (TASW medium plus 25 mg/ml kanamycin), and incubated under a 5% CO2 headspace.
A high-throughput assay was used to screen mutant strains for loss of ability to grow under low-DIC conditions. Phenol red was added to TASW (P-TASW); since T. crunogena produces sulfuric acid when using thiosulfate as an electron donor, P-TASW changes from red (pH 8) to yellow (pH <6) during growth. Random knockout colonies were inoculated into 96-well plates (200 μl P-TASW/well) and maintained under 5% CO2 headspace at room temperature for approximately 2 weeks. Strains were then transferred into two new 96-well plates containing either high- or low-DIC P-TASW. High-DIC 96-well plates were incubated under a 5% CO2 headspace, and the low-DIC plates were kept at ambient air. Strains that grew well under high-DIC conditions but were inhibited under low-DIC conditions were selected for further investigation.
It was anticipated that mutants whose carboxysomal genes were interrupted would be unable to grow under low-DIC conditions, as has been observed in H. neapolitanus (19). To remove these mutants and focus on novel CCM genes, each strain was screened with four PCRs, with four sets of primers, which together covered a substantial portion of the carboxysome operon (Tcr_0837-Tcr_0848) (see Table S1 in the supplemental material). Increased band size indicated an interruption within the operon. Strains that had wild-type carboxysome genes but were CO2 sensitive were selected for further scrutiny. To determine which gene had been interrupted in these mutant strains, arbitrarily primed PCR was used, followed by sequencing of the PCR products (50).
Site-directed mutagenesis (51) was used to interrupt Tcr_0841 (csoS3), the gene encoding carboxysomal carbonic anhydrase CsoSCA, anticipating that this would prevent growth under low-DIC conditions (19) and function as a positive control when screening for this phenotype. A strain that could function as a negative control (capable of growth under low-DIC conditions) was generated by targeting Tcr_0668, which is predicted to encode a conserved hypothetical protein located within a prophage. Transcript abundance from this gene is insensitive to the concentration of DIC during growth (20), suggesting that the protein product is unlikely to be part of a CCM. Based on results from random mutagenesis experiments (see Results), Tcr_0853 and Tcr_0854 were also targeted. Genes encoding proteins that were more abundant in cells cultivated under DIC limitation were also interrupted, to determine whether they might play a role in the T. crunogena CCM.
Target genes were PCR amplified from T. crunogena genomic DNA using primers directed to each gene (Table S1), digested, ligated into pcDNA3.1 (Invitrogen, Inc.), and introduced into chemically competent E. coli Top10 cells (Invitrogen, Inc.). After propagation, pcDNA3.1 plasmids carrying the target T. crunogena genes were purified, subjected to EZ-Tn5-transposon mutagenesis (EZ-Tn5<kan-2> kit; Epicentre, Inc.), and transformed into chemically competent E. coli Top10 cells (Invitrogen, Inc.). Once these plasmids were propagated and purified, mutated genes were amplified from them, restricted, and ligated into E. coli mating plasmid pLD55 (51). This DNA was introduced into chemically competent E. coli BW20767 (51). To introduce mutated target genes into T. crunogena, these E. coli BW20767 cells were then mated with T. crunogena as described above for random mutagenesis. RecA-mediated recombination between the mutated gene on the plasmid and the wild-type copy on the chromosome resulted in merodiploid mutant strains. These merodiploid strains were next cultivated in the presence of fusaric acid, favoring cells in which a second RecA-mediated crossover event had occurred, removing the plasmid and the wild-type copy of the gene (51).
Mutant phenotype characterization.
Mutant strains were screened for inability to grow under low-DIC conditions by cultivating them in 40-ml cultures of low-DIC (2 mM) and high-DIC (50 mM) TASW supplemented with kanamycin (25 mg/liter). Growth was monitored spectrophotometrically (OD600) until the cells reached stationary phase. Cultures of csoS3-399-2XO and Tcr_0668-1614-WF mutants were grown in parallel, to serve as a negative control for growth under low-DIC conditions (csoS3-399-2XO strain) and positive control for growth under low- and high-DIC conditions (Tcr_0668-1614-WF strain).
To determine whether mutants that were unable to grow under low-DIC conditions had aberrant carboxysomes, cells were examined via transmission electron microscopy. To prepare the cells, each strain was cultivated under high-DIC conditions in kanamycin-containing TASW. Once turbid, a 1-ml portion of each culture was preserved in 2.5% glutaraldehyde and refrigerated. A second 1-ml portion of each high-DIC culture was added to 25 ml low-DIC medium (2 mM DIC, 25 mg/liter kanamycin) and agitated overnight. The following morning, cells were pelleted via centrifugation (14,000 × g, 5 min, 4°C), resuspended in TASW, and preserved as described above for the high-DIC samples. These cells were prepared for electron microscopy, and the numbers of carboxysomes per square micrometer of cell cross-sectional area were calculated as described in reference 20. Carboxysome frequencies were binned into four categories (0, >0 to 9.99, 10 to 19.99, and >20 carboxysomes per μm2); differences between strains were tested for statistical significance with a chi-square test.
To measure differences in DIC uptake and fixation by mutant strains, cells were cultivated in chemostats under NH3-limited (50 mM DIC) conditions, harvested via centrifugation (5,000 × g, 15 min, 4°C), and resuspended in 1 liter low-DIC (2 mM DIC) medium, with pH and O2 maintained by the BioFlo 110 pH/dO2 module. For 2 h, cultures were monitored for continued consumption of O2, to verify that they remained viable, even if they were incapable of growth under these conditions. When treated this way, wild-type cells induced a CCM (carboxysome synthesis, ability to generate elevated intracellular DIC concentrations [20]). Cultures were then harvested, washed, and resuspended in DIC-free TASW. Cell suspensions were supplemented with 0.1 mg/ml bovine carbonic anhydrase and gently sparged with 1% O2-balance N2 for 5 min to remove residual DIC. DIC uptake and fixation were measured via incubations with 14C-DIC and silicone oil centrifugation as described in reference 15. RubisCO activities were also measured radiometrically (52).
Transcription of Tcr_0853 and Tcr_0854.
qRT-PCR was used to determine whether Tcr_0853 and Tcr_0854 transcript abundance was sensitive to the concentration of DIC present during growth. Wild-type cells were cultivated in DIC-limited and NH3-limited chemostats. After five vessel volumes had passed through the growth chamber, 30-ml portions were centrifuged (14,000 × g, 5 min, 4°C); cell pellets were flash-frozen with liquid nitrogen and stored at −80°C.
To determine whether the 2-h exposures to low-DIC conditions used above (DIC uptake and fixation experiments) affected the transcription of these genes in wild-type and mutant strains, 30-ml portions were removed from the high-DIC cultures as well as from the low-DIC cultures after 2 h, flash-frozen, and stored at −80°C.
Total RNA was extracted from samples with the RiboPure-Bacteria kit (Ambion, Inc.) and analyzed via gel electrophoresis to verify RNA quality (sharp 16S and 23S bands). RNA was quantified via Nanodrop 1000 (Nanodrop Technologies), and the QuantiTect SYBR green RT-PCR kit (Qiagen, Inc.) was used in an Applied Biosystems Step One real-time PCR system, with primers directed to Tcr_0853 and Tcr_0854 as well as csoS3 (positive control for CCM induction) and the citrate synthase gene (Tcr_0530; calibrator for the 2−ΔΔCT method) (Table S1). Relative amounts of transcripts were calculated using the 2−ΔΔCT method. For this method, fold change was calculated as 2−ΔΔCT, where ΔΔCT = ΔCT,q − ΔCT,cb, ΔCT,q = (CT,target − CT,citrate synthase) for the RNA from the DIC-limited cells and ΔCT,cb = (CT,target − CT,citrate synthase) for the RNA from the NH3-limited cells (20, 53).
To test for statistically significant differences in transcript abundance among strains cultivated for 2 h under low-DIC conditions, Grubb's test was used to detect outliers (e.g., strains whose ΔΔCT values differed significantly from the others). For finding outliers in ΔΔCT values for Tcr_0853 transcripts, ΔΔCT values from wild-type cells and cells with mutations in Tcr_0668 or csoS3 were pooled, and ΔΔCT values from Tcr_0853-710-WF, Tcr_0854-737-MF, and Tcr_0854-2006 mutants were individually added to determine whether any of them were sufficiently different from the pooled values to be detected as outliers. This process was repeated for Tcr_0854 transcripts. For csoS3 transcripts, ΔΔCT values from all types of cells were pooled, as mutations in Tcr_0853 and Tcr_0854 were not anticipated to affect csoS3 transcription.
To determine whether Tcr_0853 and Tcr_0854 were cotranscribed, RNA was also subjected to RT-PCR (Qiagen One-Step RT-PCR kit), using primer sets that covered overlapping sequential ∼1,000 nucleotide (nt) portions of the region spanning Tcr_0852 to Tcr_0855 (Table S1), and reverse transcriptase-free controls to check for DNA contamination. RT-PCRs with primers directed to Tcr_0841, encoding carboxysomal carbonic anhydrase, were used to verify that cells were responding to low-DIC conditions (22), and reactions with primers targeting Tcr_0350 (Table S1), encoding citrate synthase, were used as a positive control for RNA integrity, as this gene has been found to be transcribed similarly under low- and high-DIC conditions (20). RT-PCR amplicons were examined after subjecting the reaction products to gel electrophoresis (Tris-borate-EDTA [TBE], 1% agarose) and staining with ethidium bromide (48).
Phylogenetic distribution of Tcr_0853 and Tcr_0854 homologs.
Homologs to Tcr_0853 and Tcr_0854 were collected from the Integrated Microbial Genomes database (https://img.jgi.doe.gov/ [24]) using BLASTp (54) with query sequences translated from Tcr_0853 and Tcr_0854 nucleotide sequences. Gene pairs were winnowed down to 261 by selecting a single strain from each species and selecting taxa that represented the full phylogenetic diversity of the homologs. Tcr_0853 and Tcr_0854 homologs were aligned separately using MUSCLE (55) and then concatenated into a single alignment, which was subjected to GBLOCKS using stringent criteria (56). Phylogenetic trees were constructed using maximum likelihood (ML) analysis of amino acid sequences in a sequence alignment of 261 taxa. The best-fit model of evolution, estimated using “smart model selection” (SMS) in PhyML 3.0 (57), was determined to be the LG + G + I amino acid replacement model (58) with 4 categories using a discrete Gamma distribution (G = 0.739) and a proportion of Invariant sites (I = 0.062). Results were assessed using 100 bootstrap replicates. A consensus tree was visualized using FigTree (version 1.4.3) (59).
Supplementary Material
ACKNOWLEDGMENTS
USF MCB4404L includes Nourhan Abdelrahim, Fiorella Acosta, Mohammad Alak, Stefanie Albert, Lauren Allison, Abigail Anderson, Bashar Al-Turk, Kirsten Antonen, Alexander Aquino, Tiffany Arnold, Jervand Astrjan, Kylie Baggett, Erika Barajas, Betty Barisic, Lucas Bihary, Francesca Blazekovic, Jacob Byers, Camila Cabrera, Logan Cahoon, Gerson Calienes, Timothy Campbell, Gary Camper, George Cappos, Jakob Centlivre, Tess Chase, Jessica Cheer, Steven Cindric, Jacob Cohen, Montana Cole, Diana Contreras, Noella Cortinas, Alaina Covert, Stephen Cross, Emily Cutolo, Joseph Dalessio, Emilio De Narvaez, Susan Deng, Jomary Dox, Marcin Dragan, Cameron Durlacher, Deandra Dwyer, Alexandria Echevarria, Richard Eldridge, Raechel Ellis, Ricardo Escobar, Andrea Espina, Leonardo Estevez, Hiba Fatima, Quiana Fisher, Danielle Gagne, Christina Georgeades, Meghan Giroir, Fabiola Giron, Jonathon Grasham, Heather Groner, James Hanshew, Ashlee Hazle, Ian Heller, Ami Hewlett, Swaraj Hublikar, Bao Huynh, Raza Jabbar, Mirjana Janjus, Cody Johnson, Ryan Johnson, Amanda Justus, Michael Kemp, Brandi Kirby, Matthew Kondoff, Brittany Kurtright, Kristina Lamens, Lauren Lawres, Lauren Lefave, Christopher Leiker, Katherine Lewis, Jarrod Lindenmuth, Orlando Lugo, James Martin, Evan McClenthan, Michael McMeekin, Yael Medley, Swapnil Modi, Ekaterina Morozova, Brianna Neumann, Son Nguyen, Tracy Nguyen, Gael Nicolas, Jonathon Nino Charari, Karel Ortiz-Tavarez, Orville Pemberton, Jessica Paoletti, Robert Perkins, Annie Phillips, Cody Porter, Shaun Raquipo, Tony Rosquete, Shanetta Roundtree, Melissa Scheufler, Sasha Schuler, Robel Seifu, Maria Skopis, Kimberly Singleton, Nicole Stafford, Daniel Stehli, Valentina Stevanovich, Joseph Stewart, Roman Stickles, Sara Subar, Mrunal Tailor, Kathryn Trammel, Joshua Traina, Emily Trebour, Jared Tur, Ashley Ubaldini, Kathy Walls, Hannah Wapshott, Kenneth Warner, Raechel White, John Williams, Sarah Woods, and Nikolaos Zervos.
We are grateful to Abigail Shipp and Keegan Sabo for their assistance with the experiments and to the anonymous reviewers whose suggestions substantially improved the manuscript.
The National Science Foundation (NSF) provided funding to Kathleen M. Scott under grant numbers NSF-MCB-0643713 and NSF-IOS-1257532.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00871-16.
REFERENCES
- 1.Berg I. 2011. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price GD. 2011. Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth Res 109:47–57. doi: 10.1007/s11120-010-9608-y. [DOI] [PubMed] [Google Scholar]
- 3.Raven JA, Beardall J. 2016. The ins and outs of CO2. J Exp Bot 67:1–13. doi: 10.1093/jxb/erv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price GD, Badger MR, Woodger FJ, Long BM. 2009. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59:1441–1461. [DOI] [PubMed] [Google Scholar]
- 5.Brinkhoff T, Muyzer G. 1997. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl Environ Microbiol 63:3789–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinkhoff T, Sievert SM, Kuever J, Muyzer G. 1999. Distribution and diversity of sulfur-oxidizing Thiomicrospira spp. at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece). Appl Environ Microbiol 65:3843–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen M, Perner M. 2016. Hydrogenase gene distribution and H2 consumption ability within the Thiomicrospira lineage. Front Microbiol 7:99. doi: 10.3389/fmicb.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuenen JG, Robertson LA, Tuovinen OH. 1981. The genera Thiobacillus, Thiomicrospira, and Thiosphaera, p 2638–2657. In Starr MP. (ed), The prokaryotes. Springer-Verlag, New York, NY. [Google Scholar]
- 9.Takai K, Suzuki M, Nakagawa S, Miyazaki M, Suzuki Y, Inagaki F, Horikoshi K. 2006. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov and emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol 56:1725–1733. doi: 10.1099/ijs.0.64255-0. [DOI] [PubMed] [Google Scholar]
- 10.Scott KM, Sievert SM, Abril FN, Ball LA, Barrett CJ, Blake RA, Boller AJ, Chain PS, Clark JA, Davis CR, Detter C, Do KF, Dobrinski KP, Faza BI, Fitzpatrick KA, Freyermuth SK, Harmer TL, Hauser LJ, Hügler M, Kerfeld CA, Klotz MG, Kong WW, Land M, Lapidus A, Larimer FW, Longo DL, Lucas S, Malfatti SA, Massey SE, Martin DD, McCuddin Z, Meyer F, Moore JL, Ocampo LH, Paul JH, Paulsen IT, Reep DK, Ren Q, Ross RL, Sato PY, Thomas P, Tinkham LE, Zeruth GT. 2006. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena. PLoS Biol 4:e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson KS, Childress JJ, Hessler RR, Sakamoto-Arnold CM, Beehler CL. 1988. Chemical and biological interactions in the Rose Garden hydrothermal vent field, Galapagos spreading center. Deep Sea Res 35:1723–1744. doi: 10.1016/0198-0149(88)90046-5. [DOI] [Google Scholar]
- 12.Goffredi SK, Childress JJ, Desaulniers NT, Lee RW, Lallier FH, Hammond D. 1997. Inorganic carbon acquisition by the hydrothermal vent tubeworm Riftia pachyptila depends upon high external P-CO2 and upon proton-equivalent ion transport by the worm. J Exp Biol 200:883–896. [DOI] [PubMed] [Google Scholar]
- 13.Johnson KS, Childress JJ, Beehler CL. 1988. Short term temperature variability in the Rose Garden hydrothermal vent field. Deep Sea Res 35:1711–1722. doi: 10.1016/0198-0149(88)90045-3. [DOI] [Google Scholar]
- 14.Jannasch H, Wirsen C, Nelson D, Robertson L. 1985. Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol 35:422–424. doi: 10.1099/00207713-35-4-422. [DOI] [Google Scholar]
- 15.Dobrinski KP, Longo DL, Scott KM. 2005. A hydrothermal vent chemolithoautotroph with a carbon concentrating mechanism. J Bacteriol 187:5761–5766. doi: 10.1128/JB.187.16.5761-5766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerfeld CA, Melnicki MR. 2016. Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol 31:66–75. doi: 10.1016/j.pbi.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Kerfeld CA, Heinhorst S, Cannon GC. 2010. Bacterial microcompartments. Annu Rev Microbiol 64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- 18.English RS, Jin S, Shively JM. 1995. Use of electroporation to generate a Thiobacillus neapolitanus carboxysome mutant. Appl Environ Microbiol 61:3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dou Z, Heinhorst S, Williams E, Murin E, Shively J, Cannon G. 2008. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem 283:10377–10384. doi: 10.1074/jbc.M709285200. [DOI] [PubMed] [Google Scholar]
- 20.Dobrinski KP, Enkemann SA, Yoder SJ, Haller E, Scott KM. 2012. Transcription response of the sulfur chemolithoautotroph Thiomicrospira crunogena to dissolved inorganic carbon limitation. J Bacteriol 194:2074–2081. doi: 10.1128/JB.06504-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menning KJ, Menon BB, Fox G, USF MCB4404L 2012, Scott KM. 2016. Dissolved inorganic carbon uptake in Thiomicrospira crunogena XCL-2 is Δp- and ATP-sensitive and enhances RubisCO-mediated carbon fixation. Arch Microbiol 198:149–159. doi: 10.1007/s00203-015-1172-6. [DOI] [PubMed] [Google Scholar]
- 22.Dobrinski KP, Boller AJ, Scott KM. 2010. Expression and function of four carbonic anhydrase homologs in the deep-sea chemolithoautotroph Thiomicrospira crunogena. Appl Environ Microbiol 76:3561–3567. doi: 10.1128/AEM.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahl C. 2015. Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life 67:268–274. doi: 10.1002/iub.1371. [DOI] [PubMed] [Google Scholar]
- 24.Markowitz VM, Chen IA, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich T, Scheide D. 2000. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett 479:1–5. doi: 10.1016/S0014-5793(00)01867-6. [DOI] [PubMed] [Google Scholar]
- 26.Morino M, Suzuki T, Ito M, Krulwich T. 2014. Purification and functional reconstitution of a seven-subunit Mrp-type Na+/H+ antiporter. J Bacteriol 196:28–35. doi: 10.1128/JB.01029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A, Ogawa T. 2001. Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc Natl Acad Sci U S A 98:11789–11794. doi: 10.1073/pnas.191258298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vuokila PT, Hassinen IE. 1988. NN′-dicyclohexylcarbodi-imide-sensitivity of bovine heart mitochondrial NADH: ubiquinone oxidoreductase. Inhibition of activity and binding to subunits. Biochem J 249:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yagi T, Hatefi Y. 1988. Identification of the dicyclohexylcarbodiimide-binding subunit of NADH-ubiquinone oxidoreductase (Complex I). J Biol Chem 263:16150–16155. [PubMed] [Google Scholar]
- 30.Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. 2008. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc B Biol Sci 363:2629–2640. doi: 10.1098/rstb.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strauss G, Fuchs G. 1993. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium, Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur J Biochem 215:633–643. doi: 10.1111/j.1432-1033.1993.tb18074.x. [DOI] [PubMed] [Google Scholar]
- 32.Klatt CG, Bryant DA, Ward DM. 2007. Comparative genomics provides evidence for the 3-hydroxypropionate autotrophic pathway in filamentous anoxygenic phototrophic bacteria and in hot spring microbial mats. Environ Microbiol 9:2067–2078. doi: 10.1111/j.1462-2920.2007.01323.x. [DOI] [PubMed] [Google Scholar]
- 33.Tang K, Barry K, Chertkov O, Dalin E, Han CS, Hauser LJ, Honchak BM, Karbach LE, Land ML, Lapidus A, Larimer FW, Mikhailova N, Pitluck S, Pierson BK, Blankenship RE. 2011. Complete genome sequence of the filamentous anoxygenic phototrophic bacterium Chloroflexus aurantiacus. BMC Genomics 12:334. doi: 10.1186/1471-2164-12-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beh M, Strauss G, Huber R, Stetter KO, Fuchs G. 1993. Enzymes of the reductive citric acid cycle in the autotrophic eubacterium Aquifex pyrophilus and in the archaebacterium Thermoproteus neutrophilus. Arch Microbiol 160:306–311. doi: 10.1007/BF00292082. [DOI] [Google Scholar]
- 35.Eisenhut M, Schubert H, Ibelings BW, Bauwe H, Matthijs HC, Hagemann M. 2007. Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC. 6803. Plant Physiol 144:1946–1959. doi: 10.1104/pp.107.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brazelton WJ, Baross JA. 2010. Metagenomic comparison of two Thiomicrospira lineages inhabiting contrasting deep-sea hydrothermal environments. PLoS One 5:e13530. doi: 10.1371/journal.pone.0013530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaudana SB, Zarzycki J, Moparthi VK, Kerfeld CA. 2015. Bioinformatic analysis of the distribution of inorganic carbon transporters and prospective targets for bioengineering to increase Ci uptake by cyanobacteria. Photosynth Res 126:99–109. doi: 10.1007/s11120-014-0059-8. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad A, Barry JP, Nelson DC. 1999. Phylogenetic affinity of a wide, vacuolate, nitrate-accumulating Beggiatoa sp. from Monterey Canyon, California, with Thioploca spp. Appl Environ Microbiol 65:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biebl H, Pfennig N. 1978. Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate reducing bacteria. Arch Microbiol 117:9–16. doi: 10.1007/BF00689344. [DOI] [Google Scholar]
- 40.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 41.Pan C, Oda Y, Lankford PK, Zhang B, Samatova NF, Pelletier DA, Harwood CS, Hettich RL. 2008. Characterization of anaerobic catabolism of p-coumarate in Rhodopseudomonas palustris by integrating transcriptomics and quantitative proteomics. Mol Cell Proteomics 7:938–948. doi: 10.1074/mcp.M700147-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates Iii JR. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom 219:245–251. doi: 10.1016/S1387-3806(02)00563-8. [DOI] [Google Scholar]
- 43.Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 44.Tabb DL, McDonald WH, Yates JR III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan C, Kora G, Tabb DL, Pelletier DA, McDonald WH, Hurst GB, Hettich RL, Samatova NF. 2006. Robust estimation of peptide abundance ratios and rigorous scoring of their variability and bias in quantitative shotgun proteomics. Anal Chem 78:7110–7120. doi: 10.1021/ac0606554. [DOI] [PubMed] [Google Scholar]
- 46.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan C, Kora G, McDonald WH, Tabb DL, VerBerkmoes NC, Hurst GB, Pelletier DA, Samatova NF, Hettich RL. 2006. ProRata: a quantitative proteomics program for accurate protein abundance ratio estimation with confidence interval evaluation. Anal Chem 78:7121–7131. doi: 10.1021/ac060654b. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 50.Das S, Noe JC, Paik S, Kitten T. 2005. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J Microbiol Methods 63:89–94. doi: 10.1016/j.mimet.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 51.Metcalf WM, Jiang W, Daniels LL, Kim S, Haldimann A, Wanner BL. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 52.Schwedock J, Harmer TL, Scott KM, Hektor HJ, Seitz AP, Fontana MC, Distel DL, Cavanaugh CM. 2004. Characterization and expression of genes from the RubisCO gene cluster of the chemoautotrophic symbiont of Solemya velum: cbbLSQO. Arch Microbiol 182:18–29. doi: 10.1007/s00203-004-0689-x. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 57.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 58.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 59.Rambaut A. 2016. FigTree: tree figure drawing tool version 1.4.3. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, Scotland. [Google Scholar]
- 60.Yamamoto M, Arai H, Ishii M, Igarashi Y. 2003. Characterization of two different 2-oxoglutarate:ferredoxin oxidoreductases from Hydrogenobacter thermophilus TK-6. Biochem Biophys Res Commun 312:1297–1302. doi: 10.1016/j.bbrc.2003.11.078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.