ABSTRACT
Peptidoglycan is a vital component of nearly all cell wall-bearing bacteria and is a valuable target for antibacterial therapy. However, despite decades of work, there remain important gaps in understanding how this macromolecule is synthesized and molded into a three-dimensional structure that imparts specific morphologies to individual cells. Here, we investigated the particularly enigmatic area of how peptidoglycan is synthesized and shaped during the first stages of creating cell shape de novo, that is, in the absence of a preexisting template. We found that when lysozyme-induced (LI) spheroplasts of Escherichia coli were allowed to resynthesize peptidoglycan, the cells divided first and then elongated to recreate a normal rod-shaped morphology. Penicillin binding protein 1B (PBP1B) was critical for the first stage of this recovery process. PBP1B synthesized peptidoglycan de novo, and this synthesis required that PBP1B interact with the outer membrane lipoprotein LpoB. Surprisingly, when LpoB was localized improperly to the inner membrane, recovering spheroplasts synthesized peptidoglycan and divided but then propagated as amorphous spheroidal cells, suggesting that the regeneration of a normal rod shape depends on a particular spatial interaction. Similarly, spheroplasts carrying a PBP1B variant lacking transpeptidase activity or those in which PBP1A was overproduced could synthesize new peptidoglycan and divide but then grew as oddly shaped spheroids. We conclude that de novo cell wall synthesis requires the glycosyltransferase activity of PBP1B but that PBP1B transpeptidase activity is needed to assemble cell walls with wild-type morphology.
IMPORTANCE Bacterial cell wall peptidoglycan is synthesized and modified by penicillin binding proteins (PBPs), which are targeted by about half of all currently prescribed antibiotics, including penicillin and its derivatives. Because antibiotic resistance is rising, it has become increasingly urgent that we fill the gaps in our knowledge about how PBPs create and assemble this protective wall. We report here that PBP1B plays an essential role in synthesizing peptidoglycan in the absence of a preexisting template: its glycosyltransferase activity is responsible for de novo synthesis, while its transpeptidase activity is required to construct cell walls of a specific shape. These results highlight the importance of this enzyme and distinguish its biological roles from those of other PBPs and peptidoglycan synthases.
KEYWORDS: PBP1B, PBP1A, spheroplast, bacterial morphology, peptidoglycan
INTRODUCTION
For rod-shaped Escherichia coli to grow and multiply, new cell wall must be incorporated into the existing structure, followed by symmetrical division so that each daughter cell retains the size and shape of the mother cell (1, 2). These two processes, elongation and division, share several enzymes, proteins, and substrates but are distinguished from one another by the presence of unique components and by the fact that the MreB protein guides cell elongation and the FtsZ protein initiates and guides cell division (3–7). In both cases, the peptidoglycan (PG) component of the wall is synthesized by one or more bifunctional class A penicillin binding proteins (PBPs), which polymerize and cross-link the glycan chains via glycosyltransferase (GTase) and transpeptidase (TPase) activities, respectively (1). Alternately, one or more SEDS family proteins may supply the GTase polymerization activity, while one of the class B PBPs acts as the TPase (8, 9). In E. coli, PBPs 1A and 1B are the major bifunctional class A PG synthases, and one or the other must be present for cell viability (10–13). The fact that only one of these PBPs is required for E. coli to grow with a normal rod shape indicates that either enzyme can drive both elongation and division (4, 10, 14), despite intimations that PBP1A may prefer the elongation complex (5) and that PBP1B may associate more strongly with the division complex (14–17).
Although PBPs 1A and 1B were once considered redundant and interchangeable, mutants lacking one or the other exhibit different phenotypes, indicating that the two enzymes have different abilities and biological roles. For example, unlike cells lacking PBP1A, mutants lacking PBP1B are more sensitive to d-methionine and some β-lactams (11, 18, 19), survive less well during stationary phase (20), grow poorly in the absence of NaCl (12), and form biofilms less efficiently (21). They are also more sensitive to sodium citrate, EDTA, vancomycin, and the lytic effects of overproducing the dd-carboxypeptidase PBP5 (B. M. Meberg, S. Kannan, and K. D. Young, unpublished data). Also notable is the fact that mutants lacking PBP1B lyse either when the elongation-specific protein PBP2 is inactivated or when the division-specific PBP3 is inactivated, whereas under the same conditions, mutants lacking PBP1A continue to grow as spherical or filamentous cells, respectively (22–24). Thus, the two class A PBPs are not functionally identical, even though they can substitute for one another during cell elongation and division.
One of the ways in which PBPs 1A and 1B differ is in their effect on lysozyme-induced (LI) spheroplasts. Spheroplasts and L forms are cells that have lost their peptidoglycan wall either temporarily (spheroplasts and unstable L forms) or permanently (stable L forms) (25, 26). These forms can be cultivated under the proper osmotic conditions, and the first group can sometimes regenerate their cell walls and return to a wild-type morphology (26–30). This behavior raises the following important but unresolved question: how do spheroplasts and L forms rebuild their original morphology in the absence of a preexisting template? It appears that a large part of the answer lies in the nature and activity of PBPs 1A and 1B: E. coli LI spheroplasts that lack PBP1B cannot reconstruct their original rod shape but instead continue to expand until they lyse, whereas similar spheroplasts lacking PBP1A recover and recreate a wild-type shape (28). Here, we show that PBP1B plays a key role in the earliest stages of the shape recovery process. In the absence of PBP1B, spheroplasts containing wild-type amounts of PBP1A cannot synthesize new PG de novo and the cells do not divide. Surprisingly, E. coli LI spheroplasts grow and multiply if they are supplied with a PBP1B having no TPase activity, but the resulting progeny are amorphous or spheroidal. Thus, the GTase activity of PBP1B is essential for cell division during spheroplast recovery, but reconstruction of a wild-type rod shape requires a functional TPase domain. Sufficient overproduction of PBP1A can also induce LI spheroplasts to divide, but the progeny are similarly amorphous and spheroidal, suggesting that the two PBPs have different cross-linking abilities. In short, only PBP1B can recreate normally shaped cells de novo in the absence of a preexisting cell wall template, further distinguishing the biological role of PBP1B from that of other cell wall-synthesizing enzymes.
RESULTS
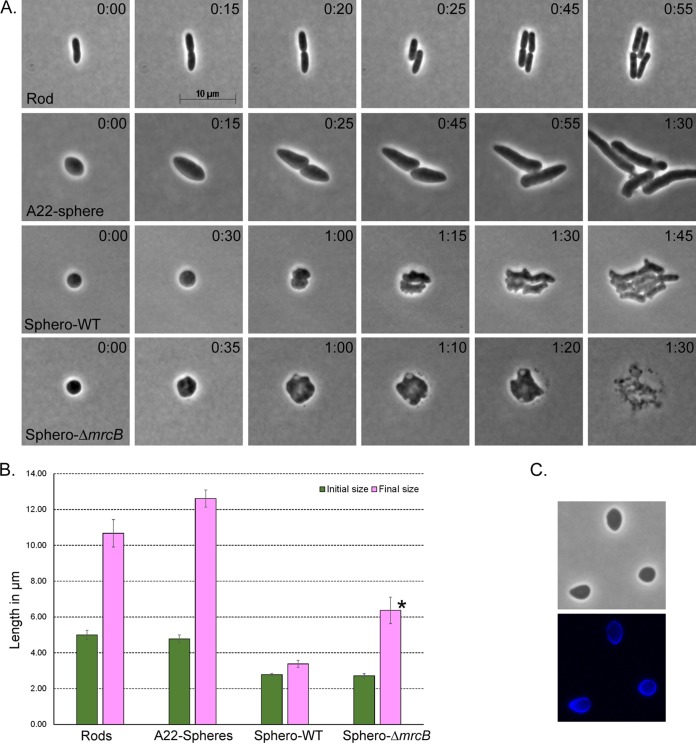
Cell division initiates cell shape restoration in LI spheroplasts.
To determine how E. coli lysozyme-induced (LI) spheroplasts begin to rebuild their original wild-type morphology, we first compared their growth behavior to that of normal rod-shaped cells and to that of spherical cells that retained a peptidoglycan (PG) wall. Wild-type cells grow by elongating to approximately double their original length before dividing in half, which we confirmed to be true for our strain (Fig. 1A, rod, and B, rods). This well-known cycle suggests that elongation is the principal means by which cells recreate their original shape. To compare the behavior of spherical cells with and without PG, we first grew cells in the presence of compound A22 [S-(3,4-dichlorobenzyl)isothiourea], which inhibits MreB and prevents cell wall elongation so that E. coli loses its rod shape and becomes spherical or ovoid (31, 32). Such A22-induced cells have an intact, preexisting PG cell wall, which was clearly visible after labeling with the fluorescent d-alanine derivative coumarin-carbonyl-amino–d-alanine (HADA) (Fig. 1C). When A22 was removed so that the cells could recommence growth, these spherical or ovoid cells first elongated to almost three times their original diameter before dividing in half (Fig. 1A, A22-sphere, and B, A22-Spheres). Thus, when PG was present, spherical or nearly spherical cells elongated before dividing.
FIG 1.
Shape reconstruction in spheroplasts begins with cell division. (A) Time-lapse microscopic examination of the normal cell cycle of a rod (n = 4), A22-induced spheres (n = 11), and lysozyme-induced spheroplasts from wild-type (WT) E. coli (Sphero-WT; n = 5) or a PBP1B-deficient mutant (Sphero-ΔmrcB; n = 4). The time after plating (in hours:minutes) is displayed in the upper right corner of each panel. (B) Graphical comparison of cell sizes during the growth cycle of rods and the recovery phase of spherical cells. The initial size is the cell length immediately after the spheroplasts were plated onto the agar pad; the final size is the maximum cell length prior to division. Error bars represent standard deviations. *, PBP1B-deficient spheroplasts (Sphero-ΔmrcB) never divided but instead enlarged to about twice their original diameter before lysing. (C) A22-induced spheres were labeled with the fluorescent d-alanine derivative HADA. (Top) phase image; (bottom) fluorescent image.
In contrast, E. coli LI spheroplasts did not elongate in the first stage of the shape recovery process. Instead, the initial event was always that spheroplasts enlarged slightly and then divided, producing two imperfect ovoids (Fig. 1A, Sphero-WT, times 0:30 and 1:00). The preliminary increase in diameter before division was very slight (Fig. 1B, Sphero-WT). Only after the initial division did the cells begin to elongate (Fig. 1A, Sphero-WT, time 1:15) before reestablishing the normal elongation-division cycle (Fig. 1A, Sphero-WT, times 1:30 and 1:45), consistent with previous observations (28). Thus, in the presence of a preexisting PG template, cells elongated before dividing, whereas in the absence of a preexisting template, cells divided before elongating.
PBP1B synthesizes new peptidoglycan in spheroplasts.
Spheroplasts lacking PBP1B fail to recover a normal rod shape and instead enlarge until they lyse (28). We reconfirmed this result (Fig. 1A, Sphero-ΔmrcB) and found that spheroplasts expanded to about twice their diameter before lysing (Fig. 1B, Sphero-ΔmrcB). However, these cells retained the PG synthase PBP1A, so it was not clear if PBP1B was required for cell wall synthesis or whether it played some other role in spheroplast recovery.
To clarify the role of PBP1B, we labeled PG in recovering LI spheroplasts with the fluorescent d-alanine derivative HADA (33) (Fig. 2). In wild-type E. coli cells, PG was clearly visible in rod-shaped cells immediately prior to treatment with lysozyme (Fig. 2A and B, before). This preformed PG was degraded when cells were converted into spheroplasts (Fig. 2A and B, time 0:00), consistent with the findings of previous work showing that PG disappears from newly formed spheroplasts (28). As the wild-type spheroplasts began to recover, newly synthesized PG became visible 20 min into the recovery phase (Fig. 2A and B). At this early stage, newly synthesized PG was not distributed uniformly around the cells but instead appeared as streaks or patches (Fig. 2A and B). However, at 40 min into the recovery period, new PG was concentrated most strongly near the midpoint of each cell, consistent with the initiation and maturation of division sites (Fig. 2A and B). After 1 h, new PG was spread fairly uniformly around the circumference of daughter cells (Fig. 2A and B). Thus, new PG synthesis in recovering spheroplasts was not uniform but began as patches or streaks, moved rapidly to potential division sites, and then spread throughout the cell as each spheroplast became more rod shaped.
FIG 2.
Synthesis of new peptidoglycan in recovering spheroplasts. E. coli MG1655 and its isogenic ΔmrcB mutant lacking PBP1B were labeled with the fluorescent d-alanine derivative HADA. Cells were washed and examined immediately before lysozyme treatment and immediately after the creation of LI spheroplasts (0:00). Newly synthesized PG was visualized by fluorescence microscopy. The time after plating onto spheroplast recovery medium (in hours:minutes) is displayed. The images are representative. (A and B) E. coli MG1655 (n = 15); (C and D) E. coli DR7 (ΔmrcB) (n = 11).
Just like wild-type cells, E. coli cells lacking PBP1B (E. coli ΔmrcB cells) synthesized PG before lysozyme treatment (Fig. 2C and D, before), indicating that this mutant could synthesize PG to maintain a normal rod shape, but this PG disappeared from spheroplasts (Fig. 2C and D, time 0:00). As observed previously (28), these PBP1B-deficient spheroplasts did not divide but grew into large spheroids (Fig. 2C). In sharp contrast to the findings for wild-type cells, though, new PG was never observed in spheroplasts lacking PBP1B, even after 1 h or more into the recovery period (Fig. 1D and not shown). Thus, PBP1B was required for the de novo synthesis of PG in spheroplasts.
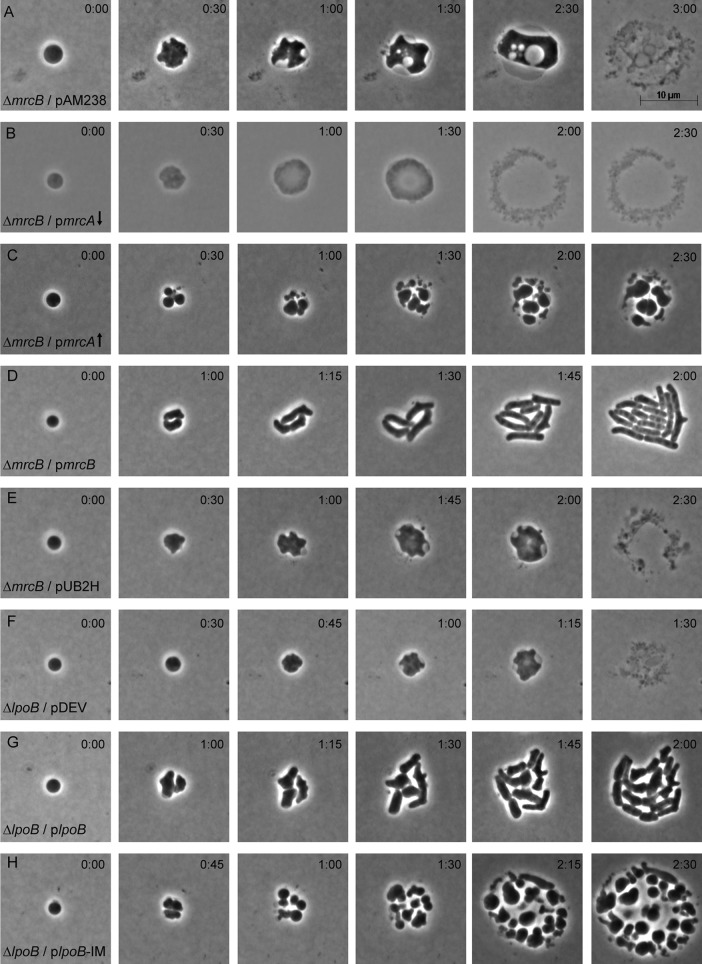
The preceding results indicated that normal amounts of PBP1A, the alternate E. coli PG synthase, did not synthesize PG in recovering spheroplasts. It was possible, though, that additional PBP1A would provide enough synthetic capacity to restore recovery to spheroplasts lacking PBP1B. Initially, we found that the overproduction of PBP1A from a low-copy-number vector (∼5 copies/cell) did not rescue such spheroplasts (Fig. 3B). However, the production of PBP1A from a medium-copy-number vector (∼25 copies/cell) enabled spheroplasts to divide (Fig. 3C). Surprisingly, instead of generating cells with a normal rod morphology, these spheroplasts gave rise to roughly spheroidal progeny, with some exhibiting short amorphous projections (Fig. 3C). These semicoccoidal cells continued to divide to yield yet more spheroidal progeny, resulting in microcolonies composed of loosely dispersed cells with heterogeneous sizes and shapes (Fig. 3C; see also Fig. S1 in the supplemental material). The simultaneous cooverproduction of PBP1A with its activator, LpoA, also allowed spheroplasts to divide but did not rescue the cell shape (not shown). Overall, the results strongly suggest that the PG synthase activity of PBP1B but not that of PBP1A is essential for the regeneration of a wild-type rod morphology.
FIG 3.
LpoB activation of PBP1B is required for spheroplast recovery. Spheroplasts lacking PBP1B (ΔmrcB) (A to E) or LpoB (ΔlpoB) (F to H) were grown on osmotically protected sucrose recovery medium, and the recovery process was monitored by time-lapse phase-contrast microscopy. The time after plating (in hours:minutes) is displayed in the upper right corner of each panel. The images are representative. (A) E. coli DR7V1(pAM238), vector-only control (n = 7); (B) E. coli DR7A(pSK17), wild-type PBP1A overexpressed from a low-copy-number vector (↓), 100 μM IPTG (n = 7); (C) E. coli MAJ595(pMAJ60), wild-type PBP1A overexpressed from a medium-copy-number vector (↑), 25 μM IPTG (n = 45); (D) E. coli DR7C(pSK12), PBP1B complementation with basal expression, no IPTG (n = 9); (E) E. coli DR7U(pUB2H), PBP1B-ΔUB2H overexpressed (n = 8); (F) E. coli DR8V(pDEV), vector-only control (n = 14); (G) E. coli DR8C(pLpoB), wild-type LpoB overexpressed (n = 5); (H) E. coli DR8CIM(pLpoB-IM), inner membrane-bound LpoB variant overexpressed (n = 15).
We next asked if native levels of PBP1A failed to restore spheroplast recovery because of two other complications, i.e., because PG synthesis was impeded by the accumulation of toxic undecaprenyl-diphosphate (Und-PP)-linked colanic acid (CA) intermediates (34) or by the depletion of Und-PP-linked peptidoglycan precursors (35, 36). In E. coli spheroplasts, the Rcs stress response is strongly induced and synthesizes a large amount of CA (34). This reaction may have redirected so much undecaprenyl-phosphate (Und-P) into the CA pathway that too few Und-PP-PG precursors were available for PBP1A. To rule out this possibility, we deleted wcaJ from an E. coli mutant lacking PBP1B, thus inhibiting the first step in CA biosynthesis and ensuring that no Und-PP intermediates could be drawn away from the PG synthetic pathway (34). However, spheroplasts lacking both PBP1B and CA did not recover (not shown), indicating that PBP1A activity was not inhibited by this pathway artifact. In addition, we overproduced the UppS protein, which increases the pool of Und-P (35, 36), but this also failed to rescue the spheroplast recovery of cells lacking PBP1B and CA (not shown). These results suggest that PBP1A activity was not limited by a buildup of lethal Und-P intermediates or by a dearth of Und-PP-linked precursors but was more likely caused by a difference in the function of PBP1A from that of PBP1B.
LpoB-mediated activation of PBP1B is required for spheroplast recovery.
PBP1B is anchored to the inner membrane by a transmembrane helix and part of its GTase domain, with the active sites of its GTase and TPase domains being in the periplasm (37). These GTase and TPase domains are separated by the UB2H segment that interacts with the outer membrane lipoprotein LpoB, which activates PBP1B (38, 39). In the absence of a functional Tol-Pal system, Typas et al. speculated that a PBP1B-LpoB transmembrane complex might promote outer membrane constriction during division (38), though Markovski et al. later argued that this was not the case (40). In either case, since spheroplasts lacking either PBP1B or LpoB fail to divide (28), it was possible that this complex drives constriction during spheroplast recovery. Because removal of the UB2H domain prevents the PBP1B-LpoB association (37–39), we used this abbreviated construct to determine if UB2H is required for spheroplast recovery. Spheroplasts lacking PBP1B but carrying the empty vector pAM238 did not divide but instead enlarged into large spheroids with periplasmic vacuoles and eventually lysed (Fig. 3A), as observed previously (28). When PBP1B was supplied in trans, the spheroplasts grew, divided, and gradually recovered their original rod shape (Fig. 3D). However, recovery did not occur if the spheroplasts were provided with PBP1B lacking the UB2H domain (Fig. 3E), strongly suggesting that recovery depended on PBP1B being able to associate with LpoB.
The preceding results could be explained in one of two ways: either PBP1B and LpoB must interact physically so as to help synthesize PG during cell division or the PBP1B-LpoB complex is required to invaginate the outer membrane. To distinguish between these alternatives, we first assayed spheroplast recovery in cells producing the LpoB-independent PBP1B variant with the E313D mutation [PBP1B(E313D)] (40). Spheroplasts producing PBP1B(E313D) and LpoB grew, divided, and recovered a normal rod shape (Fig. S2A). Surprisingly, cells producing PBP1B(E313D) but lacking LpoB did not divide but lysed instead (Fig. S2B). As expected, when LpoB was supplied in trans, these spheroplasts grew, divided, and gradually recovered a normal rod shape (Fig. S2C). Thus, the PBP1B-LpoB interaction is required for shape recovery, even though this PBP1B variant may be enzymatically active in the absence of LpoB.
We next examined spheroplasts in which PBP1B interacted with a variant of LpoB that is retained in the inner membrane (IM) instead of being transported to the outer membrane (the LpoB-IM variant) (38). Spheroplasts lacking LpoB but carrying the empty vector pDEV failed to divide and instead grew into spheroids and lysed (Fig. 3F), while spheroplasts expressing wild-type LpoB in trans divided and returned to normal rod shapes (Fig. 3G). In contrast, when LpoB-IM was supplied in trans, the mutant spheroplasts divided (Fig. 3H), indicating that a transperiplasmic interaction was not essential for invagination of these wall-less cells. However, these spheroplasts gave rise to misshapen, roughly spheroidal progeny that formed loosely packed microcolonies (Fig. 3H), characteristics equivalent to those observed when PBP1A was overproduced in cells lacking PBP1B (Fig. 3C). Although a few cells produced tube-shaped projections after 5 to 6 h (Fig. 3H, times 2:15 and 2:30), none of these regained a normal rod-like morphology during the observed time course (e.g., see Fig. S3). As an aside, we also established that the dispersed microcolony phenotype was caused by the production of extracellular CA, because CA-negative spheroplasts carrying LpoB-IM formed tightly packed microcolonies devoid of this translucent material (Fig. S3, upper right inset).
The preceding results indicated that a transperiplasmic PBP1B-LpoB complex was not required for spheroplast invagination during cell division but that the complex was required so that the ensuing progeny could return to a normal rod shape. Although the exact mechanism remains unknown, the simplest explanation is that PBP1B-LpoB helps reconstruct a wild-type morphology by incorporating or cross-linking PG into the cell wall in a specific pattern during septation.
PBP1B glycosyltransferase activity is required for spheroplast recovery.
PBP1B may contribute to spheroplast recovery either because the enzyme synthesizes PG via its GTase domain or because it cross-links this new PG via its TPase domain. These domains can be inactivated independently; e.g., the E233Q mutation (here denoted GT*) inactivates the GTase domain of PBP1B, and the S510A mutation (here denoted TP*) inactivates the TPase domain (41, 42). Note, however, that the E233Q mutation also eliminates the TPase activity of PBP1b, so that this mutant lacks both GTase and TPase activities (14). Nonetheless, we were able to use these PBP1B variants to determine which catalytic function was important for spheroplast recovery.
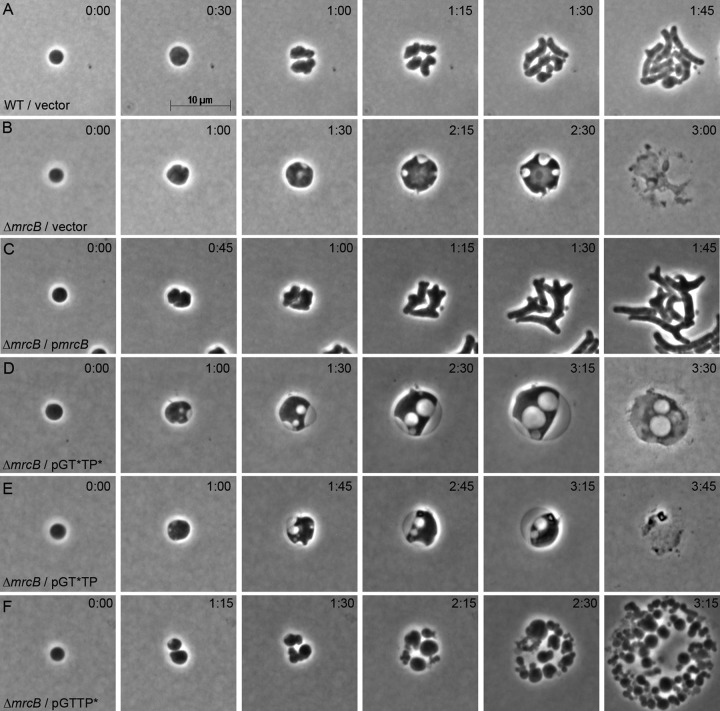
Wild-type spheroplasts carrying the control vector pJFK118EH recovered normally (Fig. 4A), whereas spheroplasts lacking PBP1B and carrying the vector did not recover (Fig. 4B). As expected, spheroplasts expressing PBP1B in trans were rescued and recovered normally (Fig. 4C). The PBP1B variant in which neither catalytic domain was active (the GT*TP* variant) failed to rescue the recovery defect of PBP1B-null spheroplasts (Fig. 4D), consistent with PBP1B being required for recovery. Furthermore, because the PBP1B-LpoB complex was present but inactive, this result also indicated that the two proteins did not simply physically link the IM and outer membrane so that the septum could invaginate. Similarly, the PBP1B GTase-inactive variant (the GT*TP variant) also failed to rescue PBP1B-null spheroplasts (Fig. 4E), again emphasizing the importance of the GTase domain for PG synthesis and supporting the view that PBP1B-LpoB does not play a simple physical role during septation.
FIG 4.
Contributions of PBP1B GTase and TPase domains to spheroplast recovery. Spheroplasts carrying wild-type PBP1B (A) or lacking PBP1B (ΔmrcB) (B to E) were grown on osmotically protected sucrose recovery medium, and the recovery process was monitored by time-lapse phase-contrast microscopy. Individual strains carried the plasmid vector (pJFK118EH) (A and B) or plasmids expressing PBP1B variants in which one or both catalytic domains were inactivated (inactive domains are designated GT* or TP*) (D to F). The time after plating (in hours:minutes) is displayed in the upper right corner of each panel. The images are representative. (A) E. coli MG1655 pJFK118EH, vector-only control (n = 10); (B) E. coli DR7V2(pJFK118EH), vector-only control (n = 6); (C) E. coli DR7C1, PBP1B complementation with basal expression, no IPTG (n = 6); (D) E. coli DR7N(pGT*TP*), PBP1B-GT*TP* basal expression, no IPTG (n = 23); (E) E. coli DR7TP(pGT*TP), PBP1B-GT*TP basal expression, no IPTG (n = 8); (F) E. coli DR7GT(pGTTP*), PBP1B-GTTP* basal expression, no IPTG (n = 16).
Surprisingly, in cells lacking wild-type PBP1B, the PBP1B variant with an inactive TPase domain but carrying an active GTase domain (the GTTP* variant) rescued the cell division defect but not the morphological defect (Fig. 4F). Instead, these progeny cells continued to divide relatively normally but grew as heterogeneous spheroidal cells that did not recover a wild-type rod shape (Fig. 4F and S4). Intriguingly, microcolonies of PBP-null spheroplasts carrying the PBP1B variant (the GTTP* variant) appeared strikingly similar to those that overexpressed PBP1A (compare the cells in Fig. 3C and S1 to those in Fig. S4) and to LpoB-deficient spheroplasts in which LpoB-IM was produced (compare the cells in Fig. 3H and S3 to those in Fig. S4). Thus, it was possible that the TPase domain of PB1B was not activated when LpoB was tethered to the inner membrane, which would explain why this construct behaved like the GTTP* mutant. Regardless of the exact mechanism, the glycosyltransferase activity of PBP1B was required for division during spheroplast recovery, whereas transpeptidation was required to recover a wild-type rod shape.
PBP1B-associated GTase activity synthesizes new peptidoglycan in spheroplasts.
The growth and division of spheroplasts expressing a PBP1B variant with GTase activity only implied but did not prove that this variant enzyme synthesized PG during invagination. To confirm this supposition, we labeled newly synthesized PG during spheroplast recovery by labeling cells with HADA. Normal rod-shaped cells lacking PBP1B but supplied with PBP1B (GTTP* variant cells) synthesized PG (Fig. 5, before), and these cells lost the PG signal after being converted into spheroplasts (Fig. 5, time 0:00). Note that in the latter case some rods escaped lysozyme treatment and retained their PG (Fig. 5B, time 0:00). When they were allowed to recover, the spheroplasts synthesized new PG, though the cells did not recover a normal rod shape (Fig. 5, times 1:00 and 2:00). This behavior was in sharp contrast to the behavior of spheroplasts lacking any PBP1B, which failed to synthesize new PG (Fig. 2D). Thus, the GTase activity of PBP1B was essential for synthesizing PG during spheroplast division, though this new PG did not by itself allow the cells to recover the wild-type morphology.
FIG 5.
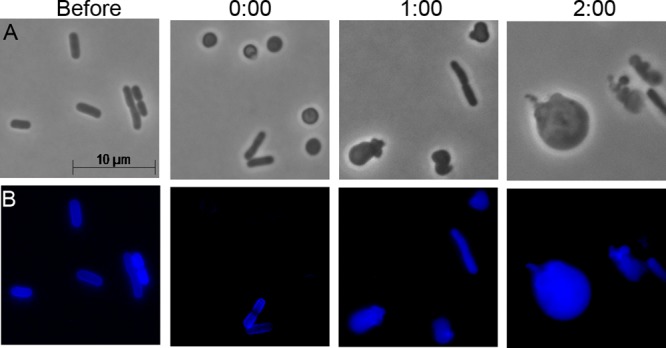
A PBP1B variant with only glycosyltransferase activity synthesizes new peptidoglycan during spheroplast recovery. E. coli DR7GT (ΔmrcB strain carrying plasmid pGTTP*) was labeled with the fluorescent d-alanine derivative HADA. Cells were washed and examined immediately before lysozyme treatment and immediately after the creation of LI spheroplasts (0:00). Newly synthesized PG in recovering spheroplasts was visualized after 1 and 2 h of growth in liquid spheroplast recovery medium (i.e., images on agar are not time-lapse observations). The recovery time (in hours:minutes) is displayed above the panels. The images are representative (n = 19).
DISCUSSION
We strongly agree with Billings et al. that “de novo cell-wall synthesis … [is] a powerful tool to study how cell shape is programmed in bacteria” (43). Such experiments shed important mechanistic light on the components and reactions required to create different cellular morphologies. Here, we show that PBP1B is the major synthase required for initiating de novo PG synthesis and for generating wild-type cell shape in recovering E. coli spheroplasts. Under these conditions the glycosyltransferase activity of PBP1B is required to initiate PG synthesis and drive cell division, and the final rod-shaped morphology depends on the transpeptidation carried out by this enzyme. These results argue that PBPs with similar enzymatic abilities are not redundant but instead play distinct roles in creating cellular morphology in the absence of a preexisting cell wall.
Cell division precedes cell elongation.
As we have shown here and elsewhere, spheroplast recovery begins with division and is followed by cell elongation that eventually recreates the normal rod shape of E. coli (28). Although this hierarchy of events seems straightforward, spheroplasts generated by another method exhibit a slightly different mode of recovery. For example, Joseleau-Petit established a model system in which E. coli spheroplasts are created by incubating cells in the presence of cefsulodin (44), thereby creating what we term “cefsulodin-induced (CI) spheroplasts,” as opposed to the lysozyme-induced (LI) spheroplasts that we used here. After cefsulodin is removed, CI spheroplasts first develop tubular protrusions composed of newly incorporated PG, and these elongating cells eventually recover a normal rod shape within a few generations (43, 45). CI spheroplasts that do not recover do not extrude these protrusions and so remain spheroidal, suggesting that in this system elongation initiates shape recovery (43, 45).
Why is the order of events different between CI spheroplasts and LI spheroplasts, and what does this tell us about the requirements for de novo cell wall synthesis and shape recovery? The major difference is the status of the wall in the two types of spheroplasts. CI spheroplasts are billed as having no intact cell wall (45), which is technically correct, in the sense that the PG in these cells cannot be cross-linked normally because cefsulodin inactivates the TPase activities of PBPs 1A and 1B (43). However, the GTase activities of these enzymes are still functional (14, 43, 44), and in fact, cefsulodin enhances the GTase activity of PBP1a in vitro so that the enzyme synthesizes glycan chains that are 1.7 times longer than normal (5). Also, the residual PG in CI spheroplasts continues to be cross-linked by an alternate l,d-transpeptidase route, meaning that these cells contain at least some cell wall fragments, though they cannot protect against osmotic lysis (44). In sum, in CI spheroplasts, PBPs 1A and 1B have functional GTase activity (but not TPase activity) and these cells contain a small amount of abnormally cross-linked PG. In contrast, in LI spheroplasts, PBPs 1A and 1B are fully functional and these cells contain no measurable amount of PG (28).
The sequence of recovery events in these two types of spheroplasts can be reconciled as follows. In CI spheroplasts, which retain PG remnants and glycan strands, the GTase activities of PBPs 1A and/or 1B can use this residual material as a preexisting template to which new glycan chains may be added. Therefore, cell wall elongation can begin immediately, followed by normal cell division of the rod-like protrusions. On the other hand, in LI spheroplasts there is little or no preexisting PG, which means that a PG template must be synthesized before elongation can begin. We find that this is accomplished solely or primarily by the GTase activity of PBP1B, which drives cell division first, and only after this is the new template extended and reshaped by the PBPs. Thus, PBP1B activity during cell division probably represents the initial event in shape recovery in both LI and CI spheroplasts. Corroborating evidence for this interpretation is that CI spheroplasts grow very poorly in the absence of PBP1B, which led Joseleau-Petit et al. to conclude that such cells require the GTase activity of PBP1B for efficient growth or division, even while they are in the spheroidal state (44). Only a slight amount of PG is visible around each spheroidal cell, but the concentration of PG is quite prominent at division sites during constriction (43, 45). This material likely represents poorly cross-linked PG created by the residual GTase activity of PBP1B. Thus, PBP1B GTase activity probably accompanies or drives division in propagating CI spheroplasts. When cefsulodin is removed to allow recovery, these cells already have a template that can be elongated, which coincides nicely with the results reported here. PBP1B, then, is required for CI spheroplast division before recovery begins, whereas in LI spheroplasts, PBP1B must synthesize PG and initiate division prior to subsequent shape recovery.
What happens to wall-less cells that cannot initiate cell division during the transition stage? Can they recover a normal morphology? This question has been addressed indirectly by Mercier et al., who created E. coli L-form-like cells by adding fosfomycin, an inhibitor that prevents the synthesis of PG precursors and glycan chains (46). When fosfomycin is removed, these spherical fosfomycin-induced (FI) spheroplasts divide and recover their normal rod shape, although the details of this transition have not been documented (29, 46). What is clear is that efficient recovery requires FtsZ, which orchestrates normal cell division in most bacteria. When FtsZ is removed from such FI spheroplasts and the cells are placed in recovery medium lacking fosfomycin, most cells lyse, but a few recover and grow as elongated rods before they die (46). This argues that PBP1B (or some other PG synthase) can synthesize PG de novo in recovering FI spheroplasts and can, in a few cells, recreate the rod-shaped morphology in the absence of FtsZ-driven cell division. This sequence of events implies that in the absence of cell division, a normal morphology can be recovered, though at a much reduced frequency, consistent with PBP1B having a principal role in restoring cell shape.
Surprisingly, using the fosfomycin system, Mercier et al. isolated a small number of suppressor strains that grew as highly amorphous spheroidal cells that nevertheless synthesized a cell wall. Of the three suppressors examined, all contained mutations that disabled (in one way or another) PBP1B or LpoB, a lipoprotein that activates PBP1B (46). The existence of such strains would seem to contradict our conclusion that PBP1B is required for de novo PG synthesis. However, in two of these strains, lpoB transcription was impaired to an unknown degree (E. coli sup-1) or the protein was truncated at about half its normal length (E. coli sup-7) (46). Wild-type PBP1B would retain measurable activity if some LpoB remained active in these cells and might do so in any case because LpoB enhances PBP1B activity by up to 8-fold (39, 47, 48). This means that PBP1B in these two strains may initiate de novo cell wall synthesis but be unable to recreate a rod morphology. In the third strain (E. coli sup-5) PBP1B was truncated but not eliminated, leaving its GTase domain virtually intact (46). In fact, amino acid residues missing from the truncated protein are not well conserved (37), so this variant could easily retain GTase activity and be capable of polymerizing glycan chains. Thus, the GTase-proficient PBP1B in the sup-5 strain may synthesize peptidoglycan de novo but be unable to recreate rod-shaped cells because it lacks TPase activity, in agreement with what we report here. Of course, the LI spheroplast versus FI spheroplast (L-form) systems are not exactly equivalent, so more work will be required to fill in the details and differences.
Finally, the above-described considerations may explain why Mercier et al. could not recreate a ΔftsZ suppressor strain simply by removing PBP1B (as in their supplementary Fig. 4) (46), because this would eliminate an essential GTase activity. Similarly, the expression of wild-type PBP1B (or LpoB) would be lethal for the suppressor strains that arose spontaneously, because the combined GTase/TPase activities of PBP1B would recreate a normal rod shape, thus leading to filamentation and death (as in Fig. 4 of Mercier et al. [46]). In sum, then, the results obtained with all model systems argue that PBP1B plays a prime role in the genesis of bacterial shape. Here, we have delineated that role more clearly.
Why PBP1B and not PBP1A? The transpeptidation possibility.
For over 30 years it has been clear that PBP1A can support E. coli growth and division in the absence of PBP1B (12, 16, 17). At first, this simple fact suggested that the two enzymes were interchangeable and perhaps redundant (11). Why, then, under normal circumstances, can PBP1B but not PBP1A synthesize PG de novo? Although the two enzymes are similar, compared to cells lacking PBP1A, those without PBP1B grow at a decreased rate, have reduced PG surface density (19), are hypersensitive to beta-lactam antibiotics (49, 50), lyse with increased readiness during exponential and stationary growth (51; S. Kannan, M. A. Jorgenson, and K. D. Young, unpublished data), and cannot support spheroplast recovery (28). The answer may be rooted in enzymatic differences. For example, in an in vitro membrane assay, PBP1B is approximately 10-fold more resistant to penicillin than PBP1A (52) and PBP1B synthesizes more peptidoglycan than PBP1A in ether-permeabilized cells (53). Thus, the most straightforward possibility is that PBP1B is simply a more efficient enzyme, which may explain why PBP1A cannot support spheroplast recovery, unless the latter protein is overproduced in sufficient quantity.
However, general kinetic differences between the enzymes either are irrelevant or are, at best, only a partial explanation, since the overproduction of PBP1A in PBP1B-deficient spheroplasts restores growth but not the ability to regenerate rod-shaped cells. Because a PBP1B variant lacking TPase activity behaves like PBP1A in this regard, we suggest that the two wild-type enzymes differ in the way that they cross-link PG and that this difference determines whether spheroplasts can recover a wild-type morphology. The conclusion that transpeptidation may be required for recreating a normal morphology is consistent with the findings of previous work showing that altered PG cross-linking affects cell shape. For example, the removal of multiple PBPs from E. coli, including those that affect cross-linking, results in abnormally shaped cells (24, 54). Similarly, non-PBP proteins with endopeptidase activity determine the normal spiral shape of Helicobacter pylori (55, 56). Indirect evidence and modeling studies also suggest a connection between PG cross-linking and rod shape in E. coli (57, 58). Curiously, the opposite situation has recently been observed in pathogenic strains of Chlamydia, in which an MreB-associated GTase synthesizes PG but in which cell division is driven by PG cross-linking (59). Of course, possibilities other than a defining role for transpeptidation can be imagined, and further work will be needed to illuminate the exact mechanism by which PBP1B directs the synthesis of rod-shaped E. coli.
Relationship to functions of SEDS proteins.
Recently, members of the SEDS protein family were implicated as newly identified alternate GTases that may be fundamentally important PG polymerases (8, 9). These novel observations predict that the SEDS members RodA and FtsW may act in concert with two class B PBPs (in E. coli, PBP2 and PBP3, respectively) to synthesize PG, in addition to or perhaps independently of PBPs 1A and 1B (8, 9). The results that we report here do not directly address how this new SEDS-plus-class B PBP paradigm might fit into the creation of cells with a defined morphology. However, indirectly, our current results do place constraints on how these enzymes function in de novo shape determination. First, regarding the SEDS proteins, it is clear that in the absence of PBP1B neither RodA nor FtsW plus the RodA- and FtsW-associated proteins can synthesize sufficient PG to regenerate a normal cylindrical cell wall in E. coli. Thus, PBP1B plays a central role in creating cell shape, at least in cells making wild-type levels of all other proteins. Second, the results strongly imply that the TPase activities of PBPs 1A, 1B, 2, and 3 are differentiated in some way. The data indicate that the TPase activity of PBP1A cannot substitute for that of PBP1B, at least not under normal conditions. The fact that the TPase activity of PBP1B is required to create normally shaped cells is somewhat surprising because the TPase activities of the two class B PBPs have long been considered the primary determinants of whether E. coli elongates to form a rod shape (PBP2) or whether the cells divide (PBP3) (7). However, even though both of these class B PBPs are present in recovering spheroplasts, neither is able to synthesize a cylindrical cell wall in the absence of the TPase supplied by PBP1B. Biochemically, it has been difficult to establish the extent of TPase activity associated with these two class B PBPs. The TPase activity of PBP2 was detected only recently (5), and though PBP3 exhibits some TPase activity toward artificial substrates (60, 61), no such activity has been measured in the presence of natural substrates (14). Nonetheless, regardless of their enzymatic capabilities, our present data indicate that the combined TPase activities of PBPs 1A, 2, and 3 by themselves cannot initiate de novo rod-like growth in E. coli. In short, then, in vivo there must be structural or functional differences between PBP1B and both the PBP1A machinery and the SEDS-driven machinery. We conclude that under normal growth conditions the GTase activity of PBP1B is essential for de novo PG polymerization and its TPase activity plays a pivotal role in constructing uniformly rod-shaped E. coli cells. Under these circumstances, other enzymes cannot substitute for PBP1B.
Summary.
As Billings et al. aptly summarize, the ability to resynthesize a rod-shaped cell wall de novo “suggests that wild-type morphology is strongly programmed by the cell-wall synthesis machinery” (43). Here, we identify PBP1B to be an integral part of that machinery and establish that the two enzymatic activities of PBP1B play different roles in creating bacterial cell shape. Specifically, one of the earliest steps in building a bacterial wall de novo requires the GTase of PBP1B, while the TPase activity of this enzyme helps ensure that the cells recover a wild-type rod shape. Future work on the overall process should expand our understanding of these early stages of de novo cell wall synthesis, define how these activities are integrated with subsequent enzymatic steps to create cells of defined shapes, and help explain why most bacteria produce multiple PG synthases.
MATERIALS AND METHODS
Bacterial strains, plasmids, DNA manipulation, and media.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2, respectively. E. coli DH5α was used as the intermediate cloning strain, and all experiments were performed in the strain MG1655 genetic background. Routine cultures were grown in Luria-Bertani (LB) medium, and when appropriate, ampicillin (100 μg/ml), kanamycin (50 μg/ml), or spectinomycin (50 μg/ml) was added. The spheroplast recovery assay was conducted in sucrose recovery medium (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose, 0.23 M sucrose, pH 7.0) (28). Standard DNA, PCR, and molecular biological techniques were utilized for cloning and plasmid construction (62), and the sequence of each plasmid and clone was verified by DNA sequencing (UAMS DNA Sequencing Core Facility).
TABLE 1.
E. coli strains
| Strain | Relevant features | Source or reference |
|---|---|---|
| DH5α | ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 | Lab collection |
| MG1655 | F− λ− ilvG rfb-50 rph-1 | Lab collection |
| CAG60377 | BW25113 LpoB-IM | 38 |
| DR7 | MG1655 ΔmrcB::frt | 28 |
| DR7C | DR7(pSK12) | 28 |
| DR7A | DR7(pSK17) | This work |
| DR7U | DR7(pUB2H) | This work |
| DR7V1 | DR7(pAM238) | This work |
| DR7V2 | DR7(pJFK118EH) | This work |
| DR7C1 | DR7(pPBP1B) | This work |
| DR7N | DR7(pGT*TP*) | This work |
| DR7TP | DR7(pGT*TP) | This work |
| DR7GT | DR7(pGTTP*) | This work |
| DR8 | MG1655 ΔlpoB::frt | 28 |
| DR8C | DR8(pLpoB) | 28 |
| DR8V | DR8(pDEV) | This work |
| DR8CIM | DR8(pLpoB-IM) | This work |
| MAJ594 | DR7(pDSW361) | This work |
| MAJ595 | DR7(pMAJ60) | This work |
| MAJ596 | DR7(pMAJ63) | This work |
| MM43 | MG1655 mrcB (E313D) yadC::Tn10 | 40 |
| MM49 | MG1655 mrcB (E313D) ΔlpoB::frt yadC::Tn10 | 40 |
| MM49C | MM49(pLpoB) | This work |
TABLE 2.
Plasmids
| Plasmida | Relevant features | Source or reference |
|---|---|---|
| pDEV | colE1 lacIq Plac kan | 28 |
| pAM238 | pSC101 Plac spec | 64 |
| pDSW361 | pDSW361 is a Kanr derivative of pDSW204; pBR lacIq Ptrc99A kan | 63 |
| pJFK118EH | pMB1 lacIq Ptac kan | 42 |
| pSK12 | pAM238-Plac::mrcB | S. Kannan |
| pUB2H | pAM238-Plac::mrcB Δ(340–573)b | This work |
| pSK17 | pAM238-Plac::mrcA | S. Kannan |
| pMAJ60 | pDSW361-Ptrc99A::mrcA | This work |
| pMAJ63 | pDSW361-Ptrc99A::mrcA-lpoA | This work |
| pLpoB | pDEV-Plac::lpoB | 28 |
| pLpoB-IM | pDEV-Plac::lpoB (T62C, G36C, G65A) | This work |
| pPBP1B | pJFK118EH-Ptac::mrcB | 42 |
| pGT*TP* | pJFK118EH-Ptac::mrcB (G697C, T1528G) | 42 |
| pGT*TP | pJFK118EH-Ptac::mrcB (G697C) | 42 |
| pGTTP* | pJFK118EH-Ptac::mrcB (T1528G) | 42 |
Inactive domains of PBP1B are indicated with asterisks: GT*, glycosyltransferase with the E233Q mutation; TP*, transpeptidase with the S510A mutation.
Δ(340–573), deletion of nucleotides 340 to 573.
Plasmid constructions.
The plasmids used for the rescue of ΔmrcB cells were constructed in pDEV (28) or pDSW361 (63). To construct pLpoB-IM, primers LpoB-EcoRI-F (CGCGAATTCGTATTAACTTTATAAGGAGGAAAAACATAGTACAAAAAGTAGTCGCTACGC) and LpoB-HindIII-R (CGCAAGCTTTTATGTCGTCGAAACGGCACCT) were used to PCR amplify lpoB from the chromosome of E. coli CAG60377 (38) and cloned into the EcoRI and HindIII restriction sites (the underlined sequences) of plasmid pDEV. To construct pUB2H, inverse PCR was used to remove the UB2H region from pSK12. First, primers UB2H-1-F (TTCGGTTTCTTCCGTCTGTATCCGC) and UB2H-1-R (AAGATGTACCATTCGGCCATAAACT) were used to PCR amplify pSK12 to delete the region from G340 to G573 of mrcB that encodes the UB2H domain. The resulting DNA fragment was treated with DpnI, gel purified, and ligated to itself to generate pUB2H. To construct pMAJ60, mrcA was amplified from E. coli MG1655 DNA with primers P135 (CAGGAATTCAAGTTCGTAAAGTATTTTTTGATCC) and P366 (TTGGAGCTCTCAGAACAATTCCTGTGCCTC). The 2,568-bp product was cut with EcoRI and SacI (restriction sites are underlined) and ligated to the corresponding restriction sites of pDSW361. pMAJ63 was constructed by amplifying lpoA with primers P367 (CAAGAGCTCAATTTCACACAGGAAACAGACCATGGAATTCGTACCCTCAACATTTTCTCG) and P368 (TTGGGATCCTTAACTGACGGGGACTACCTG). The 2,083-bp product was cut with SacI and BamHI (restriction sites are underlined) and ligated to pMAJ60.
Spheroplast recovery.
The spheroplast recovery assay was performed as described previously (28). Briefly, 0.1 ml of E. coli cells from an exponentially growing LB broth culture was harvested when the cells reached an optical density at 600 nm (OD600) of 0.2 and washed with phosphate-buffered saline (PBS; pH 8.0). To improve the spheroplasting efficiency, cells were plasmolyzed by resuspending them in 0.625 M sucrose plus lysozyme (20 μg/ml) for 10 min at 37°C. We later found that the efficiency of spheroplasting could be improved further without affecting recovery by increasing the lysozyme concentration to 150 μg/ml. A mild osmotic shock was applied by diluting these cells with 1.5 times the original volume of PBS (no sucrose) containing lysozyme (20 μg/ml) to reduce the sucrose concentration to 0.25 M, after which the cells were incubated at 37°C for an additional 10 min. To remove the lysozyme, the cells were pelleted by centrifugation at 500 × g for 15 min and then washed with sucrose recovery medium. The resulting cell pellet was resuspended in 5 μl of recovery broth, and 1 to 2 μl was transferred onto a sucrose recovery soft agar pad in a chambered slide. These slides were incubated on the stage of a Zeiss Axio Imager Z1 microscope that was enclosed in a 37°C incubation chamber. Time-lapse observations and photographs were obtained as described below.
Recovery of ΔmrcB spheroplasts.
ΔmrcB cells harboring plasmids were grown at 37°C, spheroplasted, and spotted onto sucrose recovery agar containing 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside; pSK17 and pLpoB), 25 μM IPTG (pMAJ60 and pMAJ63), or no IPTG (pSK12, pPBP1B, pGT*TP, pGTTP*, and pGT*TP*).
Peptidoglycan labeling.
E. coli PG was labeled with the fluorescent d-alanine derivative hydroxy coumarin-carbonyl-amino–d-alanine (HADA) (33), a gift from Erkin Kuru and Michael S. VanNieuwenhze (33). E. coli cells were incubated in LB medium at 37°C with shaking until they reached an OD600 of 0.2. Live spherical cells that still retained PG were created by adding compound A22 [S-(3,4-dichlorobenzyl)isothiourea; final concentration, 5 μg/ml], incubating for an additional 1 h at 37°C, and then labeling with 500 μM HADA for 20 min, after which the cells were fixed at room temperature for 15 min in 2.8% formaldehyde plus 0.04% glutaraldehyde, washed twice with PBS, pH 7.4, and prepared for microscopy.
The following procedure was used to visualize the peptidoglycan in rod-shaped cells before and after spheroplast formation, as well as the new peptidoglycan synthesis that occurred in spheroplasts during the cell wall recovery period. E. coli cells were incubated in LB medium at 37°C with shaking until the culture reached an OD600 of 0.2. This culture was divided into four parts. One part was used to monitor the quality of spheroplast formation. HADA (500 μM) was added to this portion, incubation was continued for an additional 20 min, and half of the cells were fixed and examined by microscopy to show that PG was present before spheroplasts were generated (these are referred to as “before samples”). The other half of these cells were converted into spheroplasts, fixed, and examined by microscopy to verify that peptidoglycan had been removed (these are referred to as “after samples”), as described previously (28). The unlabeled rod-shaped cells in the remaining three parts of the original culture were converted into spheroplasts to detect the synthesis of new peptidoglycan at different points during the recovery period. The spheroplasts were inoculated into three separate tubes of liquid sucrose recovery broth and incubated at 37°C in a stationary water bath. HADA was added immediately to the first tube, the mixture was incubated for 20 min, and the cells were fixed, washed, and prepared for microscopy (the 20-min time point). The second set of spheroplasts was incubated for 20 min at 37°C, HADA was added, the culture was incubated for an additional 20 min, and the cells were fixed, washed, and prepared for microscopy (the 40-min time point). The third set of spheroplasts was incubated for 40 min at 37°C, HADA was added, the culture was incubated for an additional 20 min, and the cells were fixed, washed, and prepared for microscopy (the 1-h time point). The level of HADA incorporation represented the amount of new peptidoglycan synthesized during these three recovery periods.
Microscopy.
Spheroplasts and intact cells were visualized by using a wide-field epifluorescent Zeiss Axio Imager Z1 microscope fitted with a ×100 differential interference contrast objective (numerical aperture, 1.45). Fluorescent images of HADA-labeled cells were captured by using 4′,6-diamidino-2-phenylindole (DAPI) filters (excitation, 358 nm; emission, 461 nm). Images were acquired with a Zeiss AxioCam MRm camera and were processed with AxioVision software and Adobe Photoshop.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matylda Zietek and Athanasios Typas for providing strain CAG60377 and Suresh Kannan for providing plasmids pSK12 and pSK17. We are very grateful to Erkin Kuru and Michael S. VanNieuwenhze for providing HADA. We thank William MacCain for helpful discussions and comments. We thank Allen Gies of the UAMS DNA Sequencing Core Facility for help with DNA sequencing.
The research reported in this publication was supported by award number R01-GM061019 from the National Institute of General Medical Sciences of the National Institutes of Health to Kevin D. Young.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00612-16.
REFERENCES
- 1.Höltje J-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62:181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapiro JA, Hsu C. 1989. Escherichia coli K-12 cell-cell interactions seen by time-lapse video. J Bacteriol 171:5963–5974. doi: 10.1128/jb.171.11.5963-5974.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szwedziak P, Löwe J. 2013. Do the divisome and elongasome share a common evolutionary past? Curr Opin Microbiol 16:745–751. doi: 10.1016/j.mib.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Egan AJ, Vollmer W. 2013. The physiology of bacterial cell division. Ann N Y Acad Sci 1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 5.Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, Zapun A, Breukink E, Nguyen-Distèche M, den Blaauwen T, Vollmer W. 2012. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol 85:179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- 6.Margolin W. 2009. Sculpting the bacterial cell. Curr Biol 19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA, Suh H, Marto JA, Garner EC, Bernhardt TG. 2016. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 1:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki H, Nishimura Y, Hirota Y. 1978. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci U S A 75:664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yousif SY, Broome-Smith JK, Spratt BG. 1985. Lysis of Escherichia coli by β-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol 131:2839–2845. [DOI] [PubMed] [Google Scholar]
- 12.Kato J-I, Suzuki H, Hirota Y. 1985. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet 200:272–277. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- 13.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 14.Egan AJF, Biboy J, Ivan't Veer I, Breukink E, Vollmer W. 2015. Activities and regulation of peptidoglycan synthases. Philos Trans R Soc Lond B Biol Sci 370:20150031. doi: 10.1098/rstb.2015.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexeeva S, Gadella TW Jr, Verheul J, Verhoeven GS, den Blaauwen T. 2010. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol Microbiol 77:384–398. doi: 10.1111/j.1365-2958.2010.07211.x. [DOI] [PubMed] [Google Scholar]
- 16.Muller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, Fraipont C, Terrak M, Nguyen-Disteche M, Vollmer W. 2007. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem 282:36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- 17.Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, von Rechenberg M, Nguyen-Disteche M, den Blaauwen T, Holtje JV, Vollmer W. 2006. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol 61:675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- 18.Caparrós M, Torrecuadrada JLM, de Pedro MA. 1991. Effect of d-amino acids on Escherichi coli strains with impaired penicillin-binding proteins. Res Microbiol 142:345–350. doi: 10.1016/0923-2508(91)90050-K. [DOI] [PubMed] [Google Scholar]
- 19.Caparrós M, Quintela JC, de Pedro MA. 1994. Variability of peptidoglycan surface density in Escherichia coli. FEMS Microbiol Lett 121:71–76. [DOI] [PubMed] [Google Scholar]
- 20.Pepper ED, Farrell MJ, Finkel SE. 2006. Role of penicillin-binding protein 1b in competitive stationary-phase survival of Escherichia coli. FEMS Microbiol Lett 263:61–67. doi: 10.1111/j.1574-6968.2006.00418.x. [DOI] [PubMed] [Google Scholar]
- 21.Kumar A, Sarkar SK, Ghosh D, Ghosh AS. 2012. Deletion of penicillin-binding protein 1b impairs biofilm formation and motility in Escherichia coli. Res Microbiol 163:254–257. doi: 10.1016/j.resmic.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 22.García del Portillo FG, de Pedro MA. 1990. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J Bacteriol 172:5863–5870. doi: 10.1128/jb.172.10.5863-5870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García del Portillo F, de Pedro MA. 1991. Penicillin-binding protein 2 is essential for the integrity of growing cells of Escherichia coli ponB strains. J Bacteriol 173:4530–4532. doi: 10.1128/jb.173.14.4530-4532.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181:3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith DG. 1969. Bacteria with their coats off: spheroplasts, protoplasts and L forms. Sci Prog 57:169–192. [PubMed] [Google Scholar]
- 26.Allan EJ, Hoischen C, Gumpert J. 2009. Bacterial L forms. Adv Appl Microbiol 68:1–39. doi: 10.1016/S0065-2164(09)01201-5. [DOI] [PubMed] [Google Scholar]
- 27.Domingue GJ Sr, Woody HB. 1997. Bacterial persistence and expression of disease. Clin Microbiol Rev 10:320–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranjit DK, Young KD. 2013. The Rcs stress response and accessory envelope proteins are required for de novo generation of cell shape in Escherichia coli. J Bacteriol 195:2452–2462. doi: 10.1128/JB.00160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercier R, Kawai Y, Errington J. 2014. General principles for the formation and proliferation of a wall-free (L form) state in bacteria. eLife 2014:3. doi: 10.7554/eLife.04629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawai Y, Mercier R, Errington J. 2014. Bacterial cell morphogenesis does not require a preexisting template structure. Curr Biol 24:863–867. doi: 10.1016/j.cub.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bendezú FO, de Boer PA. 2008. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwai N, Nagai K, Wachi M. 2002. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci Biotechnol Biochem 66:2658–2662. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- 33.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew Chem Int Ed Engl 51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranjit DK, Young KD. 2016. Colanic acid intermediates prevent de novo shape recovery of Escherichia coli spheroplasts, calling into question biological roles previously attributed to colanic acid. J Bacteriol 198:1230–1240. doi: 10.1128/JB.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jorgenson MA, Kannan S, Laubacher ME, Young KD. 2016. Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol Microbiol 100:1–14. doi: 10.1111/mmi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgenson MA, Young KD. 2016. Interrupting biosynthesis of O antigen or the lipopolysaccharide core produces morphological defects in Escherichia coli by sequestering undecaprenyl phosphate. J Bacteriol 198:3070–3079. doi: 10.1128/JB.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sung MT, Lai YT, Huang CY, Chou LY, Shih HW, Cheng WC, Wong CH, Ma C. 2009. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc Natl Acad Sci U S A 106:8824–8829. doi: 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan AJF, Jean NL, Koumoutsi A, Bougault CM, Biboy J, Sassine J, Solovyova AS, Breukink E, Typas A, Vollmer W, Simorre J-P. 2014. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc Natl Acad Sci U S A 111:8197–8202. doi: 10.1073/pnas.1400376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markovski M, Bohrhunter JL, Lupoli TJ, Uehara T, Walker S, Kahne DE, Bernhardt TG. 2016. Cofactor bypass variants reveal a conformational control mechanism governing cell wall polymerase activity. Proc Natl Acad Sci U S A 113:4788–4793. doi: 10.1073/pnas.1524538113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terrak M, Ghosh TK, van Heijenoort J, Van Beeumen J, Lampilas M, Aszodi J, Ayala JA, Ghuysen JM, Nguyen-Disteche M. 1999. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol Microbiol 34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- 42.Meisel U, Holtje JV, Vollmer W. 2003. Overproduction of inactive variants of the murein synthase PBP1B causes lysis in Escherichia coli. J Bacteriol 185:5342–5348. doi: 10.1128/JB.185.18.5342-5348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billings G, Ouzounov N, Ursell T, Desmarais SM, Shaevitz J, Gitai Z, Huang KC. 2014. De novo morphogenesis in L forms via geometric control of cell growth. Mol Microbiol 93:883–896. doi: 10.1111/mmi.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joseleau-Petit D, Liebart JC, Ayala JA, D'Ari R. 2007. Unstable Escherichia coli L forms revisited: growth requires peptidoglycan synthesis. J Bacteriol 189:6512–6520. doi: 10.1128/JB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cambré A, Zimmermann M, Sauer U, Vivijs B, Cenens W, Michiels CW, Aertsen A, Loessner MJ, Noben J-P, Ayala JA, Lavigne R, Briers Y. 2015. Metabolite profiling and peptidoglycan analysis of transient cell wall-deficient bacteria in a new Escherichia coli model system. Environ Microbiol 17:1586–1599. doi: 10.1111/1462-2920.12594. [DOI] [PubMed] [Google Scholar]
- 46.Mercier R, Kawai Y, Errington J. 2016. Wall proficient E. coli capable of sustained growth in the absence of the Z-ring division machine. Nat Microbiol 1:16091. doi: 10.1038/nmicrobiol.2016.91. [DOI] [PubMed] [Google Scholar]
- 47.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupoli TJ, Lebar MD, Markovski M, Bernhardt T, Kahne D, Walker S. 2013. Lipoprotein activators stimulate Escherichia coli penicillin-binding proteins by different mechanisms. J Am Chem Soc 136:52–55. doi: 10.1021/ja410813j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki H, Nishimura Y, Yasuda S, Nishimura A, Yamada M, Hirota Y. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol Gen Genet 167:1–9. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt LS, Botta G, Park JT. 1981. Effects of furazlocillin, a β-lactam antibiotic which binds selectively to penicillin-binding protein 3, on Escherichia coli mutants deficient in other penicillin-binding proteins. J Bacteriol 145:632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paradis-Bleau C, Kritikos G, Orlova K, Typas A, Bernhardt TG. 2014. A genome-wide screen for bacterial envelope biogenesis mutants identifies a novel factor involved in cell wall precursor metabolism. PLoS Genet 10:e1004056. doi: 10.1371/journal.pgen.1004056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramachandran V, Chandrakala B, Kumar VP, Usha V, Solapure SM, de Sousa SM. 2006. Screen for inhibitors of the coupled transglycosylase-transpeptidase of peptidoglycan biosynthesis in Escherichia coli. Antimicrob Agents Chemother 50:1425–1432. doi: 10.1128/AAC.50.4.1425-1432.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kraus W, Holtje J-V. 1987. Two distinct transpeptidation reactions during murein synthesis in Escherichia coli. J Bacteriol 169:3099–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meberg BM, Paulson AL, Priyadarshini R, Young KD. 2004. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J Bacteriol 186:8326–8336. doi: 10.1128/JB.186.24.8326-8336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonis M, Ecobichon C, Guadagnini S, Prevost MC, Boneca IG. 2010. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol 78:809–819. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- 56.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. 2010. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell 141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. 2008. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci U S A 105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furchtgott L, Wingreen NS, Huang KC. 2011. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Mol Microbiol 81:340–353. doi: 10.1111/j.1365-2958.2011.07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liechti G, Kuru E, Packiam M, Hsu YP, Tekkam S, Hall E, Rittichier JT, VanNieuwenhze M, Brun YV, Maurelli AT. 2016. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 12:e1005590. doi: 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishino F, Matsuhashi M. 1981. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem Biophys Res Commun 101:905–911. doi: 10.1016/0006-291X(81)91835-0. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen-Disteche M, Fraipont C, Buddelmeijer N, Nanninga N. 1998. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol Life Sci 54:309–316. doi: 10.1007/s000180050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 63.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gonzalez MD, Akbay EA, Boyd D, Beckwith J. 2010. Multiple interaction domains in FtsL, a protein component of the widely conserved bacterial FtsLBQ cell division complex. J Bacteriol 192:2757–2768. doi: 10.1128/JB.01609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.