Abstract
Purpose
Pterygium is claimed to be a benign proliferation triggered by prolonged exposure to ultraviolet radiation. The frequency of K-ras oncogene mutation, which is among the initial mutations in tumorigenesis, is evaluated in this study.
Patients and methods
In this prospective randomized clinical, trial pterygium tissues and normal conjunctiva tissue specimens are obtained from the superotemporal quadrant of patients who underwent primary pterygium excision with autograft transplantation. DNA extraction from tissues was performed using the QIAamp DNA FFPE tissue kit. A PCR reaction was performed to amplify sequences containing codons 12, 13, and 61 of the K-ras gene in DNA. These PCR products then underwent the ‘pyrosequencing' procedure for mutations at these codons.
Results
Pterygium and normal conjunctival tissue samples of 25 patients (10 females, 15 males) were evaluated in the study. The mean age of the patients was 54.54±13.13 years. Genetic analysis revealed no K-ras mutations in normal conjunctival tissues, whereas pterygium tissues of the same cases demonstrated mutation at codon 12 in one case and mutations at codon 61 in seven cases, which was statistically significant (P<0.05). The point missense mutations at codon 61 were glutamine to arginine (Glu61Arg CAA>CGA) in four cases and glutamine to leucine (Glu61Leu CAA>CTA) in three cases.
Conclusion
The significantly higher frequency of codon 61 mutation of the ras oncogene in primary and bilateral pterygium specimens compared with normal conjunctiva supports the tumoral origin of pterygium, and thus set the stage for research into a targeted therapy for pterygium with better outcomes than surgical excision.
Introduction
The original description of pterygium dates back to the time of Hippocrates; however, the pathogenesis of this benign fibrovascular proliferation advancing on the corneal surface is still not fully understood.1, 2 The lesion has a characteristic wing-like appearance and occurs more frequently on the nasal limbus than the temporal. It may cause irritation and redness due to tear film irregularity and visual impairment related to irregular astigmatism. Treatment is limited to surgical excision and there is a high recurrence rate, which has led researchers to seek potential prognostic and predictive factors to pave the way for novel, targeted treatment alternatives other than surgery.2, 3
Although several risk factors have been proposed, the higher incidence in tropical and subtropical regions supports the role of ultraviolet (UV) radiation in the formation of pterygium.4, 5 However, exactly how UV radiation triggers pterygium formation is a matter of debate. Some authors hypothesized that scattered light follows an alternative transcameral optical path and hits limbal stem cells from their inner surface. Other studies have provided evidence implicating antiapoptotic mechanisms, immunological mechanisms, cytokines, growth factors, extracellular matrix modulators following UVR injury. There is also compelling evidence toward uncontrolled cell proliferation as a mechanism, as pterygium requires cell migration, proliferation, and local angiogenesis.2, 6, 7, 8
Ultraviolet radiation is known to lead to tumor formation in other tissues in the human body. For example, in skin tumors UV light has been shown to affect tumor suppressor genes such as p53 and oncogenes such as K-ras.9, 10 The role of the p53 gene in pterygium pathogenesis had been evaluated in several studies.11 However, there is only one study by Detorakis et al12 investigating the frequency of K-ras mutation.
K-ras is the most frequently mutated oncogene in cancer.13, 14 It encodes low molecular weight G-proteins that are located on the internal surface of cell membranes and have GTPase activity.15, 16, 17 They function as molecular switches, regulating critical cellular processes including mitosis, cell differentiation, apoptosis, and gene expression.16, 17, 18 Mutational activation of K-Ras has been found in 22% of tumors. It has been shown to play an important role in colorectal cancer progression and in 90% of pancreatic adenocarcinomas because it causes the downstream cascade proteins to be permanently switched on.18 Approximately 40% of colorectal cancers present point mutations in the K-ras gene.19 These mutations are generally located at codons 12 and 13 in the majority of cases and less frequently at other codons, like codon 61 or 64.20 These point substitutions in codons 12 and 13 were confirmed to be negative predictors of response to targeted therapies with anti-epidermal growth factor receptor antibodies.21 Therefore, determining the K-ras mutational status of tumor samples prior to treatment has become an essential step toward getting optimal benefit from treatment.
The preliminary study of Detorakis et al,12 investigating the possible role of K-ras mutation in pterygium pathogenesis, evaluated K-ras mutation at codons 12 and 13 in pterygium samples and found a mutation rate of 10% at codon 12, especially evident in recurrent and young cases. In this study we endeavored to elucidate the mutation rate of K-ras oncogene at codon 61 in addition to codons 12 and 13. As a difference from the study of Detorakis et al12 which included both primary and recurrent cases, our study enrolled only primary pterygium tissues and compared it with the patients' own normal conjunctival tissue. Of the several mutation analysis methods available, the highly sensitive method of pyrosequencing is preferred because it allows direct sequencing by the synthesis of short fragments of DNA via a novel enzymatic cascade system.22
Materials and methods
Patients with pterygium who were scheduled for an excisional pterygium surgery with conjunctival autograft transplantation were enrolled in the study. The nature of the study was explained to all participants before obtaining written informed consent for the use of collected samples or any accompanying images in this research project. The research presented in this manuscript was performed in compliance with the Helsinki Declaration according to the study ethics proposal approved by the Ethics Board of Selçuk University.
Patients completed an information form including occupation, duration of outside activity, sunglass use, family history, duration of pterygium, and treatment history. Patients who had received any ocular medication or chemical agent or underwent previous surgery for any ocular disease including pterygium were excluded from the study.
Bilateral best-corrected visual acuity and biomicroscopic findings were recorded for each patient. Pterygium was classified as type 1 if it did not extend beyond the limbus, type 2 if it extended 2–4 mm over limbus but did not encroach onto the optic zone, and type 3 if it extended into the optic zone. In subjects with bilateral pterygium the eye with more advanced type has been preferred for excision.
Pterygium surgery with the conjunctival autograft transplantation technique was performed under local anesthesia, and 1 × 1 mm samples were taken from both the pterygium specimens and from the superotemporal bulbar conjunctiva after taking a conjunctival autograft. All fresh tissues were put in separate Eppendorf tubes filled with 0.9% NaCl and stored at −80 °C until DNA isolation.
Laboratory procedures
Frozen pterygium and normal conjunctival tissues were allowed to return to room temperature, then incubated overnight at 56 °C in proteinase K (Qiagen, Hilden, Germany) for lysis. Afterwards, DNA from the lysate was precipitated using AL buffer and ethanol. DNA purification was performed using the spin column procedure (QIAamp DNA FFPE Tissue Kit; Qiagen) and AW1, AW2, and ATL buffers were added sequentially according to the manufacturer's protocol. Finally, the DNA content was measured with a Nanodrop-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) at 260 and 280 nm. Samples with a DNA quantity less than 100 ng/ml were excluded and reanalyzed from the stored tissue samples.
Mutation in the K-ras gene at codons 12, 13, and 61 in the pterygium and normal tissues were detected with the pyrosequencing method using the PyroMark K-ras Kit (Qiagen, Catalog Nr: 971450). Prior to pyrosequencing, the corresponding mutation sequences at codons 12, 13, and 61 underwent PCR amplification using 10 ng/μl of genomic DNA with the HotStarTaq Plus Master Mix Kit and PyroMark K-ras Kit (Qiagen) including sequencing primers for codons 12, 13, and 61. The primary PCRs were generated in 20 μl reaction volumes, which contained 1 μl DNA, 1 μl PCR primers for codon 12/13 or codon 61, 12.5 μl HotStarTaq Plus Master Mix, 2.5 μl Coral Load, 3 μl Q solution, and 1 μl MgCl2. The primary PCR cycling conditions were 95 °C for 5 min for activation of HotStarTaq Plus DNA polymerase; denaturation at 92 °C for 20 s, annealing at 53 °C for 30 s, elongation at 72 °C for 20 s, and a final hold at 72 °C for 5 min. In addition to the patients' samples, PCR was also conducted on positive and negative controls.
The amplicons were immobilized on Streptavidine High Performance beads. Two microliters of Streptavidine Sepharose High Performance Beads were added to 40 μl binding buffer and mixed with 10 μl PCR product and 28 μl distilled water for 10 min at room temperature. The beads were captured on the filter probes using the Pyrosequencing Vacuum Prep Tool after the vacuum was applied and then washed with 70% ethanol for 5 s, denaturation solution (0.2 M NaOH) for 10 s, and washing buffer for 5 s. The vacuum was then stopped and the beads were released into a PyroMarkQ24 Plate containing 24.2 μl annealing buffer and 0.8 μl sequencing primer. Pyrosequencing reactions were then performed using the PyroMark Gold Q24 Reagents, PyroMark Q24 instrument (Qiagen), and PyroMark Q24 version 1.0.6.3 software.
Data were analyzed using the Statistical Package for the Social Sciences (SPSS version 13.0) for Windows. Differences between pterygium and control tissues were analyzed using χ2 and Fisher exact tests. All tests were performed at an error level of 5%.
Results
Primary pterygium tissue and normal conjunctival tissues as the control group were harvested from 25 subjects whose mean age was 54.54±13.13 years. Ten (40%) of the patients were women and 15 (60%) were men. The ratio of sunglasses use was 16% (4/25) and the mean duration of outdoors work was 4.08 (2–8) h/day. Family history for pterygium was not reported.
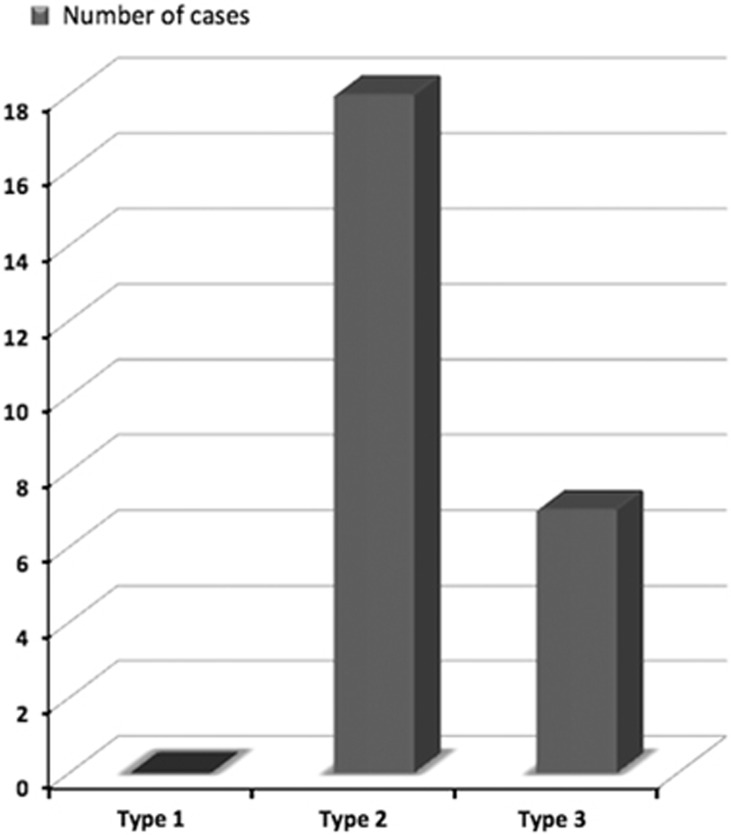
Pterygia were located nasally in all patients and exceeded the cornea by 2–4 mm (type 2) in 18 (72%) cases and by more than 4 mm (type 3) in 7 (28%) cases (Figure 1). Fifteen cases presented with bilateral pterygium. Pterygium grading in the fellow eyes were type 1 in eight subjects, type 2 in three subjects, and type 3 in four subjects.
Figure 1.
Distribution of cases according to pterygium types.
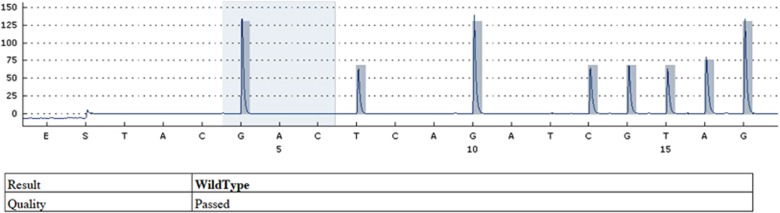
No K-ras mutations were found at codons 12, 13, and 61 in normal conjunctival tissues (Figure 2). In pterygium tissues of the same subjects, only one patient exhibited a missense mutation from GGT (glycine) to GTT (valine) at codon 12. No mutations were detected at codon 13, as in normal conjunctiva. Statistical analysis using Fisher's exact test demonstrated no significant difference between pterygium and normal tissue in respect to codons 12 and 13 (P=0.32, P=1) (Table 1).
Figure 2.
Sequencing graphic for codone 12 of the 25th case demonstrating normal tissue sample without any mutation (wild type).
Table 1. Frequency of mutation at codone 12, 13, and 61 in normal conjunctiva and pterygium tissues.
| Codone | Normal tissue (n=25) | Pterygium (n=25) |
|---|---|---|
| Codone 12 | 0 | 1 |
| Codone 13 | 0 | 0 |
| Codone 61 | 0 | 7 |
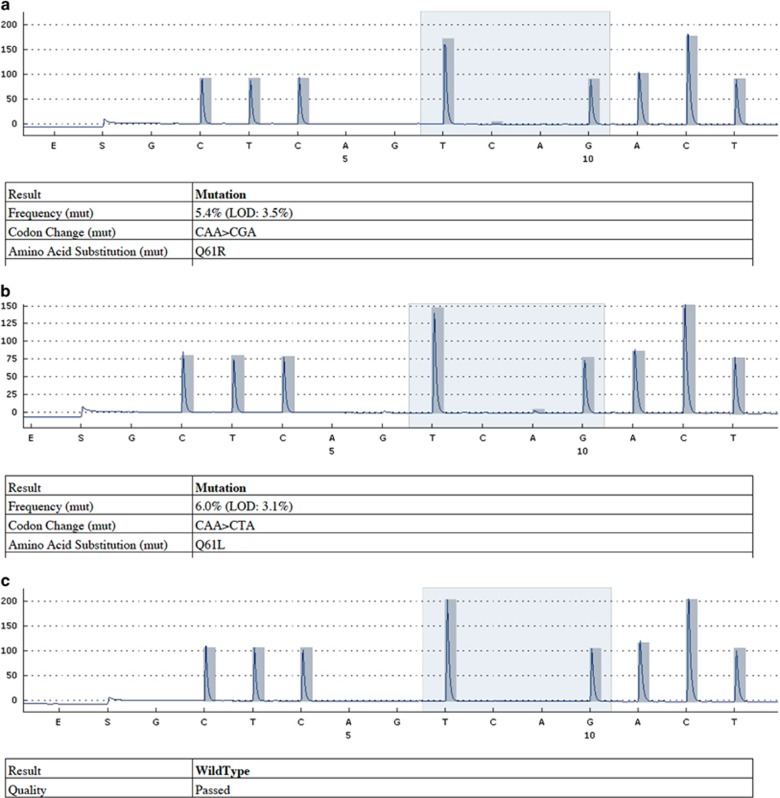
Pyrosequencing analysis of codon 61 revealed mutations at codon 61 in seven subjects (Table 2). All were missense mutations; the CAA (glutamine) was replaced with CGA (arginine) in four cases and CTA (leucine) in three cases (Figures 3a–c). The frequency of codon 61 mutation in pterygium tissue compared with normal tissue was found to be statistically significant according to Fisher's exact test (P=0.01).
Table 2. Outcome of mutation analysis at codone 61.
| Case no. |
Codone 61 |
|
|---|---|---|
| Normal tissue | Pterygium | |
| 1 | N | N |
| 2 | N | CAA>CGA |
| 3 | N | N |
| 4 | N | N |
| 5 | N | CAA>CGA |
| 6 | N | N |
| 7 | N | N |
| 8 | N | N |
| 9 | N | N |
| 10 | N | N |
| 11 | N | CAA>CTA |
| 12 | N | CAA>CGA |
| 13 | N | N |
| 14 | N | N |
| 15 | N | CAA>CTA |
| 16 | N | N |
| 17 | N | CAA>CGA |
| 18 | N | N |
| 19 | N | N |
| 20 | N | N |
| 21 | N | CAA>CTA |
| 22 | N | N |
| 23 | N | N |
| 24 | N | N |
| 25 | N | N |
Figure 3.
Sequencing graphic of codone 61. (a) CGA mutation of codone 61. (b) CTA mutation of codone 61. (c) Normal conjunctival tissue without mutation (wild type).
Subgroup analysis of the seven cases with codon 61 mutations (5 women, 2 men) yielded a mean age of 53.71±14.16 years. The mean age of the patients without codon 61 mutations was 54.88±13.12 years, but the age difference between patients with and without codon 61 mutation was insignificant (P=0.95). With respect to the clinical findings of the patients with codon 61 mutations, six had type 2 and one had type 3 pterygium, and all had bilateral pterygium. Pterygia in the fellow eyes were type 1 in six cases and type 2 in one case. Only one of the seven patients with mutation at codon 61 used sunglasses. The median duration of outdoor work was 3 (range 2–5) h/day in patients with codon 61 mutation and 4 (range, 1–8) h/day in patients without; this difference was not statistically significant (P=0.10).
Discussion
Although the clinical features and surgical treatment methods of pterygium have been well described, its pathogenesis has not been fully elucidated. It is characterized by hyperplasia and dysplasia of the conjunctiva and has several biological features in common with tumors, such as uncontrolled cell proliferation, corneal invasion, and recurrence after resection.23, 24, 25 Another source of evidence that pterygium should be considered a neoplasia rather than a degenerative disease is the better visual outcome obtained with application of 5-fluorouracil or mitomycin during surgery.26 The lack of metastasis, cell migration, local angiogenesis, and recurrence seen in cancer tissue may be ascribed to the benign nature of this tumor-like proliferation.27
Tan et al28 concluded that pterygium resulted from disruption of the normal process of apoptosis. The increased cellular proliferation and low level of apoptosis in pterygium tissue was further confirmed by Liang et al.29
Many recent studies also consider pterygium as a neoplastic condition. The MAPK signal pathway, which is activated in nearly all cancers, has been shown to also be activated in pterygium tissue.30, 31, 32 Loss of heterozygosity (LOH) and microsatellite instability (MI) are characteristics of transformed cells.33, 34, 35 Detorakis et al33 have found that LOH is common with pterygia most often at 9p (48%), followed by 17q (42%), but MI of DNA was uncommon. LOH was not detected in normal conjunctiva protected from UV radiation beneath the upper eyelid, proving that these genetic alterations are likely due to UV exposure.34, 36, 37
UV light exposure induces formation of pyrimidine dimers containing thymine and cytosine bases in DNA via photochemical reactions. These dimers are mutagenic if not repaired.38 Among various types of lesions formed in DNA after UV light exposure, cyclobutane pyrimidine dimers are the most mutagenic based on their frequency, slow repair, and distinct mutagenicity.39, 40 Studies on UV light-induced skin tumors have shown that one of the genes most sensitive to cyclobutain pyrimidine dimer formation is p53 and immunohistochemical studies further revealed overexpression of p53 in many types of UV radiation damaged cells.41, 42, 43
P53 is a key element in genomic stability and is involved in several aspects of cell cycle arrest, apoptosis, control of genome integrity, and DNA repair.44 P53 mutations are known to be the most common genetic markers in human neoplasias.45, 46 Several previous studies reported increased p53 levels in pterygium tissue, while no positive staining has been observed in normal conjunctiva.45, 46, 47 Further studies found that p53 expression levels in pterygia differ between epithelial layers, being higher in basal cells compared with more superficial layers. These findings support the proposed theory of transcameral exposure of limbal basal cells to solar light.6
Oncogenes are also known to be affected by UV radiation. Mutations in ras family proto-oncogenes are encountered in cultured cells of mouse skin tumors and cutaneous melanoma following exposure to near-UV radiation. Such mutations may convert these proto-oncogenes into active oncogenes.9, 10 Ras genes are the most frequent proto-oncogene in tumoral proliferations of the human body. They are encoded by three genes: H-ras, K-ras, and N-ras. Of these, K-ras is the most frequent isoform encountered in the COSMIC data set, but there are limited data about ras mutations detected at early stages of carcinogenesis in precancerous or benign tumors.13 It has been reported in benign skin tumors like keratoacanthoma and also in precancerous colon adenoma. Ninety-eight percent of human K-ras mutations are reported at codons 12 and 13. Codon 61 mutations are rarely seen in malignancies.13, 48 Mutation of ras proteins at codon 61 impairs the coordination of water molecules necessary for phosphate binding, whereas mutations at codons 12 and 13 inhibit the formation of van der Waals linkage. The glutamine residue at position 61 (Gln61) has a vital role in catalysis as it forms a hydrogen bond with a specific residue (Arg 789) of GAP p120 to allow the nucleophilic attack of a water molecule that is crucial for GTP hydrolysis. Mutation of this residue therefore results in impaired hydrolysis of GTP, which gives rise to prolonged activation of the ras–GTP complex, subsequently overstimulating the Ras/Raf/MEK/MAPK pathway and leading to uncontrolled cell proliferation.49
The preliminary study about the role of K-ras mutation in pterygium performed by Detorakis et al12 evaluated only codon 12 and 13 mutations and found a significant mutation rate of 10% at codon 12. They additionally noted a correlation between postoperative recurrence and young age, implying a relationship between clinical profile and locus of K-ras mutation. Contrastingly the mutation at codon 12 was detected in only one case in our study. This discrepancy may be attributed to the differences in the methods used for genetic analysis or to the criteria for patient selection. Detorakis et al12 used the restriction fragment length polymorphism method to determine the frequency of K-ras mutations. As a difference we preferred pyrosequencing method as it enables regional sequencing with a higher sensitivity in addition to screening for known mutations.50 The patient inclusion criteria demonstrated also some differences among the two studies. Detorakis et al12 enrolled both primary and recurrent ptergium cases whereas only primary pterygium cases are included in our study. Regarding this fact the lower frequency of codon 12 mutations in primary pterygium cases in our study compared with the study of Detorakis et al12 may be attributed to a possible correlation between pterygium recurrence and codon 12 mutation in the K-ras oncogene.
Another information our study contributes to that of Detorakis et al12 is the higher frequency of codon 61 mutations in these primary pterygium cases. Evaluation of 25 cases revealed codon 61 mutation in seven cases: Glu61Arg in four cases and Glu61Leu in three cases. Transition from glutamine to leucine as a result of transversion from A–T to T–A at codon 61 of the H-ras oncogene has been previously reported in the benign skin tumor keratoacanthoma, supporting a possible relationship between codon 61 mutation and benign degenerative diseases.48 As Detorakis et al12 did not evaluate codon 61 mutations, no comparison could be made with our study results.
Further evaluation of cases with K-ras codon 61 mutations demonstrated similar mean age and clinical stage compared with the cases without a mutation at codon 61 except for the fact that all cases with codon 61 mutation exhibited bilateral pterygium. This is a remarkable finding implying a genetic predisposition in bilateral pterygium cases. However, the higher number of bilateral cases (15 out of 25) in the study might have affected the outcome as well. The time spent outdoors was even shorter in subjects with codon 61 mutation; however, not using sunglasses might also explain the higher frequency of genetic mutation. The low reliability of patients' responses on questionnaires is another confounding factor that must be kept in mind when evaluating these data.
The significantly higher frequency of K-ras oncogene codon 61 mutation in primary pterygium, we detected in our preliminary analysis has not been reported previously in the literature. We found a significant difference in codon 61 mutation frequency between pterygium and normal conjunctival tissues of the same patient. For this study, normal conjunctiva tissues were obtained from the upper fornix, a location that is not exposed to UV radiation, and no mutations were found in these tissue samples. This finding suggests a relationship of pterygium with oncogenes and thus tumoral proliferation. There are also other studies in the literature supporting the relationship between K-ras codon 61 mutation and UV radiation. Törmänen et al51 demonstrated that UV radiation targets pyrimidine dimmers, resulting in a high bias toward ras Q61 mutations generation. This may be considered another supporting finding for the similarity of pterygium to a benign tumor. Lack of sunglasses use in patients with codon 61 mutation raises the possibility of a cumulative effect of UV in patients with a genetic mutation, a hypothesis that is further supported by the bilaterality of their pterygia.
Evaluation of recurrent cases and more detailed documentation of the risk factors, especially quantification of UV exposure, would improve the power of this study, and their absence can be accepted as this study's limitations. The higher number of bilateral cases in the study (15/25) should be respected also as a confounding factor in the study. Evaluation of fellow eye for the same mutation in bilateral cases and comparing the outcome with the same number of unilateral patients are warranted to confirm this finding.
Until the risk factors and mechanisms leading to pterygium formation are determined definitively, surgical excision remains the sole and best treatment option for pterygium. However, excision also entails certain risks and complications, including dellen formation and permanent astigmatism. Even the high recurrence rate following surgical excision highlights the fact that more effective treatment options targeting the underlying molecular pathways and inhibiting this tumor-like formation at the proximal stages are warranted. The significantly higher frequency of codon 61 mutation of the ras oncogene in primary and bilateral pterygium specimens compared with normal conjunctiva found in this study might support the tumoral origin of this conjunctival hyperplasia and be an initial step toward targeted therapies that will improve the prognosis of pterygium.
Acknowledgments
This study was granted by the Konya Necmettin Erbakan University Scientific Research Project Coordinatory (project number: 131318009).
Footnotes
The study was presented as a poster at the National Meeting of Turkish Ophthalmological Society, Antalya, Turkey, 4–8 November 2015.
The authors declare no conflict of interest.
References
- Duke-Elder S. System of ophthalmology. In: Diseases of the Outer Eye. Mosby: St Louis, MO, 1965; pp; 573–574. [Google Scholar]
- Detorakis ET, Spandidos DA. Pathogenetic mechanisms and treatment options for ophthalmic pterygium: trends and perspectives. Int J Mol Med 2009; 23(4): 439–447. [DOI] [PubMed] [Google Scholar]
- Ang LP, Chua JL, Tan DT. Current concepts and techniques in pterygium treatment. Curr Opin Ophthalmol 2007; 18(4): 308–313. [DOI] [PubMed] [Google Scholar]
- Coroneo MT. Pterygium as an indicator of ultraviolet insulation: a hypothesis. Br J Ophthalmol 1993; 77(11): 734–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran DJ, Hollows FC. Pterygium and ultraviolet radiation: a positive correlation. Br J Ophthalmol 1984; 68: 343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Girolamo N, Chui J, Coroneo MT, Wakefield D. Pathogenesis of pterygia: role of cytokines, growth factors, and matrix metalloproteinases. Prog Retin Eye Res 2004; 23(2): 195–228. [DOI] [PubMed] [Google Scholar]
- Bradley JC, Yang W, Bradley RH, Reid TW, Schwab IR. The science of pterygia. Br J Ophthalmol 2010; 94(7): 815–820. [DOI] [PubMed] [Google Scholar]
- Liu T, Yangwuyue L, Xie L, He X, Bai J. Progress in the pathogenesis of Pterygium. Curr Eye Res 2013; 38(12): 1191–1197. [DOI] [PubMed] [Google Scholar]
- Dushku N, Reid TW. Immunohistochemical evidence that human pterygia originate from an invasion of vimentin-expressing altered limbal epithelial basal cells. Curr Eye Res 1994; 13(7): 473–481. [DOI] [PubMed] [Google Scholar]
- Nishigori C, Wang S, Miyakoshi J, Sato M, Tsukada T, Yagi T et al. Mutations in ras genes in cells cultured from mouse skin tumors induced by ultraviolet irradiation. Proc Natl Acad Sci USA 1994; 91(15): 7189–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein O, Rosenthal G, Zirkin H, Monos T, Lifshitz T, Argov S. Overexpression of p53 tumor suppressor gene in pterygia. Eye 2002; 16: 619–2110. [DOI] [PubMed] [Google Scholar]
- Detorakis ET, Zafiropoulos A, Arvanitis DA, Spandios DA. Detection of point mutations at codone 12 of K-ras in ophthalmic pterygia. Eye 2005; 19(2): 210–214. [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. A comprehensive survey of ras mutations in cancer. Cancer Res 2012; 72(10): 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011; 39(database issue): D945–D950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A, Bazan V, Agnese V, Rodolico V, Gebbia N. Prognostic and predictive factors in colorectal cancer: Kirsten Ras in CRC (RASCAL) and TP53CRC collaborative studies. Ann Oncol 2005; 16(Suppl 4): iv44–iv49. [DOI] [PubMed] [Google Scholar]
- Bazan V, Agnese V, Corsale S, Calo V, Valerio MR, Latteri MA et al. Specific TP53and/or Ki-ras mutations as independent predictors of clinical outcome in sporadic colorectal adenocarcinomas: results of a 5-year Gruppo Oncologico dell'Italia Meridonale (GOIM) prospective study. Ann Oncol 2005; 16(Suppl 4): iv50–iv55. [DOI] [PubMed] [Google Scholar]
- Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM et al. K-ras oncogene mutations in sporadic colorectal cancer in the Netherlands Cohort Study. Carcinogenesis 2003; 24(4): 703–710. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002; 418(6901): 934. [DOI] [PubMed] [Google Scholar]
- Saif MW, Shah M. K-ras mutations in colorectal cancer: a practice changing discovery. Clin Adv Hematol Oncol 2009; 71: 45–53, 64. [PubMed] [Google Scholar]
- Li Z, Chen Y, Wang D, Wang G, He L, Suo J. Detection of KRAS mutations and their associations with clinicopathological features and survival in Chinese colorectal cancer patients. J Int Med Res 2012; 40(4): 1589–1598. [DOI] [PubMed] [Google Scholar]
- Morelli MP, Kopetz S. Hurdles and complexities of codon 13 KRAS mutations. J Clin Oncol 2012; 30(29): 3565–3567. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Poe AC, Limor J, Grady KK, Godman I, McCollum AM et al. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J Clin Microbiol 2006; 44(11): 3900–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kase S, Osaki M, Jin XH, Ohgami K, Yoshida K, Saito W et al. Increased expression of erythropoietin receptor in human pterygial tissues. Int J Mol Med 2007; 20(5): 699–702. [PubMed] [Google Scholar]
- Chowers I, Pe'er J, Zamir E, Livni N, Ilsar M, Frucht-Pery J. Proliferative activity and p53 expression in primary and recurrent pterygia. Ophthalmology 2001; 108(5): 985–988. [DOI] [PubMed] [Google Scholar]
- Tan DT, Liu YP, Sun L. Flow cytometry measurement of DNA content in primary and recurrent pterygia. Invest Ophthalmol Vis Sci 2000; 41: 1684–1686. [PubMed] [Google Scholar]
- Yağmur M. Pterjium cerrahisi. Türkiye Klinikleri J Surg Med Sci 2007; 3(1): 67–74. [Google Scholar]
- Karukonda SR, Thompson HW, Beuerman RW, Lam DS, Wilson R, Chew SJ et al. Cell cycle kinetics in pterygium at three latitudes. Br J Ophthalmol 1995; 79: 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DT, Tang WY, Liu YP, Goh HS, Smith DR. Apoptosis and apoptosis related gene expression in normal conjunctiva and pterygium. Br J Ophthalmol 2000; 84(2): 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Jiang Z, Ding B, Cheng P, Huang D, Tao L. Expression of cell proliferation and apoptosis biomarkers in pterygia and normal conjunctiva. Mol Vis 2011; 17: 1687–1693. [PMC free article] [PubMed] [Google Scholar]
- De Luca A, Maiello MR, D'Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/ AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets 2012; 16(Suppl 2): S17–S27. [DOI] [PubMed] [Google Scholar]
- del Barco Barrantes I, Nebreda AR. Roles of p38 MAPKs in invasion and metastasis. Biochem Soc Trans 2012; 40(1): 79–84. [DOI] [PubMed] [Google Scholar]
- Cui D, Pan Z, Zhang S, Zheng J, Huang Q, Wu K. Downregulation of c-Myc in pterygium and cultured pterygial cells. Clin Exp Ophthalmol 2011; 39(8): 784–792. [DOI] [PubMed] [Google Scholar]
- Detorakis ET, Sourvinos G, Tsamparlakis J, Spandidos DA. Evaluation of loss of heterozygosity and microsatellite instability in human pterygium: clinical correlations. Br J Ophthalmol 1998; 82(11): 1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detorakis ET, Drakonaki EE, Spandidos DA. Molecular genetic alterations and viral presence in ophthalmic pterygium (Review). Int J Mol Med 2000; 6(1): 35–41. [PubMed] [Google Scholar]
- Spandidos DA, Sourvinos G, Kiaris H, Tsamparlakis J. Microsatellite instability and loss of heterozygosity in human pterygia. Br J Ophthalmol 1997; 81(6): 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BG, Sahni D, Torres JC, Dushku N, Reid TW. MLH1 and MSH2 expression in pterygia. Cornea 2007; 26(4): 468–472. [DOI] [PubMed] [Google Scholar]
- Taylor HR, West S, Munoz B, Rosenthal FS, Bressler SB, Bressler NM. The long-term effects of visible light on the eye. Arch Ophthalmol 1992; 110(1): 99–104. [DOI] [PubMed] [Google Scholar]
- Goodsell DS. The molecular perspective: ultraviolet light and pyrimidine dimers. Oncologist 2001; 6: 298–299. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Lee CS, O'Connor TR, Yasui A, Pfeifer GP. The DNA damage spectrum produced by simulated sunlight. J Mol Biol 2000; 299(3): 681–693. [DOI] [PubMed] [Google Scholar]
- You YH, Lee DH, Yoon JH, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J Biol Chem 2001; 276(4): 44688–44694. [DOI] [PubMed] [Google Scholar]
- Kanjilal S, Pierceall WE, Cummings KK, Kripke ML, Ananthaswamy HN. High frequency of p53 mutations in ultraviolet radiation-induced murine skin tumors: evidence for strand bias and tumor heterogeneity. Cancer Res 1993; 53(13): 2961–2964. [PubMed] [Google Scholar]
- van Kranen HJ, de Gruijl FR, de Vries A, Sontag Y, Wester PW, Senden HC et al. Frequent p53 alterations but low incidence of ras mutations in UV-B-induced skin tumors of hairless mice. Carcinogenesis 1995; 16(5): 1141–1147. [DOI] [PubMed] [Google Scholar]
- Wood RD, Skopek TR, Hutchinson F. Changes in DNA base sequence induced by targeted mutagenesis of lambda phage by ultraviolet light. J Mol Biol 1984; 173(3): 273–291. [DOI] [PubMed] [Google Scholar]
- Perra MT, Maxia C, Corbu A, Minerba L, Demurtas P, Colombari R et al. Oxidative stress in pterygium: relationship between p53 and 8-hydroxydeoxyguanosine. Mol Vis 2006; 12: 1136–1142. [PubMed] [Google Scholar]
- Pfeifer GP. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem Photobiol 1997; 65(2): 270–283. [DOI] [PubMed] [Google Scholar]
- Frezza C, Martins CP. From tumor prevention to therapy: empowering p53 to fight back. Drug Resist Updat 2012; 15: 258–267. [DOI] [PubMed] [Google Scholar]
- Bargonetti J, Manfredi JJ. Multiple roles of the tumor suppressor p53. Curr Opin Oncol 2002; 14(1): 86–91. [DOI] [PubMed] [Google Scholar]
- Corominas M, Sloan SR, Leon J, Kamino H, Newcomb EW, Pellicer A. ras activation in human tumors and in animal model systems. Environ Health Perspect 1991; 93: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W. The Ras-RasGAP complex: structural basisfor GTPase activation and its loss in oncogenic Ras mutants. Science 1997; 277(5324): 333–338. [DOI] [PubMed] [Google Scholar]
- Zhiyong Z, Poe AC, Limor J, Grady KK, Goldman I, McCollum AM et al, Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum J Clin Microbiol 2006; 44(11): 3900–3910. [DOI] [PMC free article] [PubMed]
- Törmänen VT, Pfeifer GP. Mapping of UV photoproducts within ras proto-oncogenes in UV-irradiated cells: correlation with mutations in human skin cancer. Oncogene 1992; 7(9): 1729–1736. [PubMed] [Google Scholar]