Abstract
Purpose
Autosomal-dominant optic atrophy (ADOA), often associated with mutations in the OPA1 gene (chromosome 3q28-q29) is rarely reported in Asia. Our aim was to identify and describe this condition in an Asian population in Singapore.
Patients and methods
Preliminary cross-sectional study at the Singapore National Eye Centre, including patients with clinical suspicion of ADOA, who subsequently underwent genetic testing by direct sequencing of the OPA1 gene.
Results
Among 12 patients (10 families) with clinically suspected ADOA, 7 patients (5 families) from 3 different ethnic origins (Chinese, Indian, and Malay) carried a heterozygous pathogenic variant in the OPA1 gene. The OPA1 mutations were located on exons 8, 9, 11, and 17: c.869G>A (p.Arg290Glu), c.892A>G (p.Ser298Gly), c.1140G>A (splicing mutation), and c.1669C>T (p.Arg557*), respectively. One splicing mutation (c.871-1G>A) was identified in intron 8. We also identified a novel mutation causing optic atrophy and deafness (c.892A>G (p.Ser298Gly)). Among the phenotypic features, colour pupillometry disclosed a dissociation between low vision and preserved pupillary light reflex in ADOA.
Conclusion
We report the first cases of genetically confirmed OPA1-related ADOA from Singapore, including a novel mutation causing ‘ADOA plus' syndrome. Further epidemiological studies are needed in order to determine the prevalence of ADOA in South-East Asia.
Introduction
Autosomal-dominant optic atrophy (ADOA) is the most common inherited optic neuropathy, characterized by bilateral visual loss, typically occurring in early childhood, associated with central or cecocentral visual field defects, colour vision deficits, and temporal or diffuse pallor of the optic disc.1, 2 OPA1 mutations are responsible for about 60–80% of genetically confirmed ADOA cases.3, 4, 5 To date, the locus-specific database dedicated to OPA1 (http://opa1.mitodyn.org/), under our curation, has listed a total of 377 OPA1 gene variants, of which 65% are considered pathogenic.6, 7 The OPA1 gene, localized on 3q28-q29, has 30 coding exons including three alternative exons. It encodes a mitochondrial dynamin-related GTPase critical to mitochondrial function.3, 4 Despite the reported autosomal-dominant inheritance, there is incomplete penetrance of the condition8 and almost half of the OPA1 mutations have been detected in apparently sporadic cases with no demonstrable family history.9 As a result of the wide spectrum of mutations, there is considerable inter and intrafamilial variability in the ADOA phenotype ranging from minimal to significant visual loss to severe phenotypes affecting young adults with systemic associations such as sensorineural deafness, ataxia, external ophthalmoplegia, peripheral neuropathy, and myopathy, also known as the ADOA 'plus' phenotypes.10 Severe syndromes affecting young children, due to recessive OPA1 inheritance, were also recently reported.5, 11, 12
Although ADOA is an ubiquitous condition,13, 14, 15, 16, 17, 18 several studies have reported its possibly lower incidence in Asia.19, 20 Our aim was to investigate, for the first time, its occurrence in the multiethnic population of Singapore.
Materials and methods
Patients with unexplained optic neuropathies, clinically suggestive of ADOA, seen at Singapore National Eye Centre (January 2013 to August 2014) were genetically tested for OPA1 mutations. Inclusion criteria were: bilateral visual loss, dyschromatopsia, optic disc pallor or atrophy, and visual field loss, irrespective of the presence of ophthalmic family history. Other causes of optic neuropathies were ruled out in all included patients. The study was approved by our central institutional review board ethics committee.
A blood sample was taken from 12 included patients after informed consent. The 30 coding OPA1 exons and the exon–intron junctions were sequenced using Sanger technique. The OPA1 variants are described according to the isoform 1 (RefSeq: NM_015560.2). Phenotyping of genetically confirmed carriers included monocular chromatic pupillometry, described in detail elsewhere.21 In brief, one eye of each subject was exposed to blue (469 nm) and red (631 nm) light (order of light exposure randomized) of gradually increasing intensity from 6.8 to 13.8 log photons/cm2/s1 at the level of the cornea, over 2 min, preceded and followed by 1 min of darkness. The dark pupil diameter before light exposure was used to convert the rest of the pupillary constriction data into constriction ratio. The pupillary constriction during the light exposure was binned in bins of 0.5 log units between 7 and 14 log photons/cm2/s1, generating 14 data points per light exposure recording. Pupillometry results were compared with those of 54 healthy control subjects that underwent the same protocol.
Results
Patient 1
A 27-year-old Malay man, with a family history of poor vision, was evaluated for unexplained poor vision and recent, bilateral hearing loss. He admitted having poor vision since primary school as well as a family history of poor childhood vision (in a second-degree relative). Best corrected visual acuity (BCVA) was 6/45 OU and colour vision was 13/15 on Ishihara plates OU. Fundoscopy disclosed bilateral temporal disc pallor and normal appearance of the retina. MRI brain was normal. Genetic testing revealed a novel pathogenic heterozygous missense variant c.892A>G (p.Ser298Gly) in exon 9 of OPA1 gene.
Patient 2
A 28-year-old Indian male with a family history of unexplained, poor vision, was evaluated for longstanding visual loss. His BCVA was 6/24 RE and 6/30 LE, and fundoscopy disclosed pale, cupped discs. Work-up for toxic, nutritional, and compressive causes of optic atrophy (including MRI brain and anterior visual pathway) was negative. Genetic testing disclosed a pathogenic missense pathogenic variant c.869G>A (p.Arg290Glu) in exon 8 of OPA1.4
Patient 3
A 70-year-old male Chinese patient with no significant ophthalmic family history had unexplained visual loss since his youth. BCVA was 6/120 in both eyes, associated with bilaterally pale, cupped discs; the remainder of the neuro-ophthalmic examination was normal. A dedicated CT of brain, orbits, and pituitary fossa ruled out compression of the afferent pathways and the remainder of the evaluation did not disclose a nutritional, toxic, inflammatory, or infectious cause of this bilateral optic neuropathy. Genetic testing disclosed a pathogenic splicing variant in the intron 8 of the OPA1 gene (c.871-1G>A).22
Patient 4
A 24-year-old Chinese man with a family history of visual loss presented unexplained bilateral progressive visual loss since childhood. BCVA was 6/45 OU, and fundoscopy revealed bilateral temporal disc pallor. Genetic testing revealed a pathogenic heterozygous non sense variant in the exon 17 of the OPA1 gene (c.1669C>T (p.Arg557*)).22
Patients 5, 6, and 7
This family, originally from Myanmar, was composed of three affected male patients: two brothers (patients 5 and 6) and their father (patient 7). No other family members were affected. The proband, age 25, (patient 5), was independently, initially diagnosed with Leber's hereditary optic neuropathy (LHON) in a context of poor, unexplained visual loss (6/60 OU), and a mitochondrial point mutation (m.11253T>C (p.Ile165Thr)) in the MT-ND4 gene.23 At that time, no nuclear DNA testing was performed. His brother (patient 6), also suffered of unexplained visual loss, thought to be related to LHON. The LHON diagnosis was later challenged, when the father, age 61 (patient 7), was evaluated for longstanding visual loss (6/45 OU) and centrocaecal scotomas. Genetic testing in all three patients disclosed a pathogenic, heterozygous c.1140G>A splicing variant in exon 11 of OPA1 gene. This mutation is responsible for exon 11 skipping and has been previously reported to be pathogenic in ADOA.24
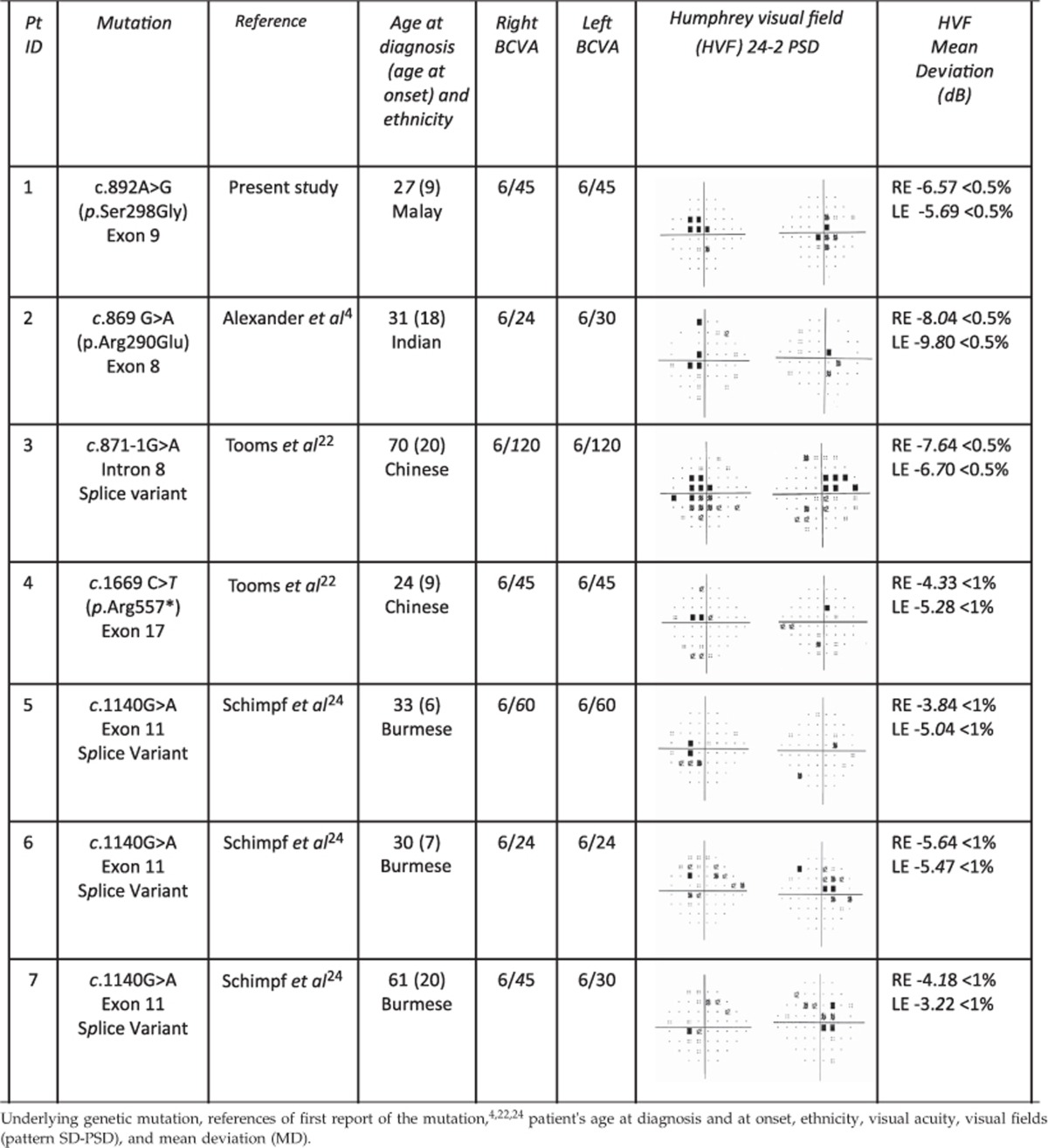
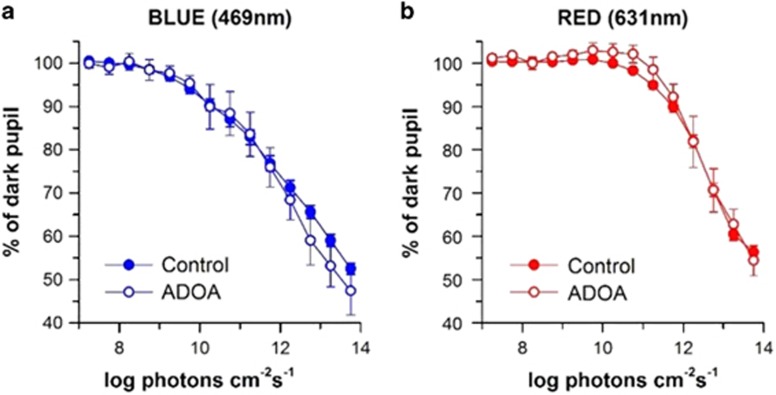
Humphrey visual fields were variably abnormal in all included patients (Table 1). Chromatic pupillometry was performed in five of the seven subjects with genetically confirmed ADOA and compared with those obtained in 54 healthy control subjects. Dose–response curves for pupillary constriction were generated for controls and patients with ADOA for both blue 469 nm light and red 631 nm light. The mean constriction amplitude of pupils, both to blue and red light, at each point of the curve was comparable in the ADOA group and in the healthy control group (Figure 1).
Table 1. Summary of included patients.
Figure 1.
Colour pupillometry in genetically confirmed ADOA patients. Dose–response curves for pupillary constriction are shown for controls (n=54, filled circles) and patients with ADOA (n=5, open circles), who were exposed to blue 469 nm light (a, left, blue trace) and red 631 nm light (b, right, red trace). There was no significant difference of the pupil responses in ADOA patients, compared with controls. Pupil diameter is expressed as a percentage of the dark pupil measured prior to each light exposure. The mean±SEM is shown in the graphs.
Discussion
This is the first report of genetically confirmed ADOA in Singapore and in South-East Asia. ADOA is rare in Asia, occurring in only 6.3% of the Chinese patients with hereditary optic neuropathies.20 This is in contrast to a European study, which reported 30% positivity for OPA1 mutations in 980 patients from France and Spain with suspected hereditary optic neuropathy.9 It is not clear though, whether ADOA is less prevalent in Asia, or merely less frequently detected, due to unfamiliarity with the condition. Indeed, milder phenotypes and limited access to genetic testing may explain, partly, the reduced prevalence of the disease in Asia. It has nevertheless been suggested that screening for OPA1 mutation is needed in Chinese populations suspected with hereditary optic neuropathies.25
Our patients encompassed all major ethnic groups in Singapore, including Chinese, Indian, and Malay, presenting with classical clinical findings. Phenotypically, our patients had similar findings with those disclosed in Caucasian ADOA populations, that is, early onset of a slowly progressive optic neuropathy. The pupillometry results were also similar to those reported in Caucasian cohorts.26 It has been postulated that the ipRGCs expressing melanopsin, mediating blue light responses, are relatively resistant to the intracellular metabolic dysfunction induced by the various genetic defects in mitochondrial optic neuropathies.27 In all five patients tested among our group, there was no significant difference in the pupil responses to red and blue light, when compared with healthy controls, suggesting resistance of ipRGCS in ADOA.
The detected mutations were classical, spanning across exons 8, 9, 11, and 17 and in intron 8.6 We also report an ADOA ‘plus' patient due to a novel missense mutation c.892A>G (p.Ser298Gly) in exon 9, not previously reported.6, 7, 28 This variant is predicted by the state of the art in silico methods to be disease causing with high confidence; (1) the substituted nucleic acid is highly conserved29 (phyloP score: 4.88) leading to the substitution of a serine amino acid, which is highly conserved in the eight species aligned30 in this study (Figure 2), (2) the main prediction tools for predicting damaging effects of missense mutations predict this mutation to affect the protein function with high confidence31, 32, 33 (Mutation Taster probability: 0.99; PolyPhen-2 probability: 1.00; SIFT score: 0.00, with sequences used for prediction diverse enough), (3) we submitted the first report of this variant in the reference OPA1 locus-specific database6, 7 (DB-ID: OPA1_00306), and this mutation is unknown in the catalog of genome variations34 and both in exome35, 36 or genome37 databases of healthy individuals.
Figure 2.
Evolutionary conservation analysis of the OPA1 p.(Ser298Gly) mutation (patient id #1). Eight species were aligned using the multiple sequence alignment tool CLUSTAL Omega30 (version 1.2.2) to analyze the evolutionary conservation of the OPA1 proteins. Sequences of Caenorhabditis elegans (CAA87771.3), Drosophila melanogaster (NP_610941.1), Danio rerio (NP_001007299.1), Gallus gallus (NP_001034398.1), Mus musculus (NP_001186106.1), Bos taurus (NP_001179890.1 2), Pan troglodytes (XP_003310226.1), and Homo sapiens (NP_056375.2) were retrieved from GenBank.
One family (patients 5, 6, and 7) was carrying a pathogenic, heterozygous OPA1 mutation, c.1140G>A. Interestingly, one of the affected male members in this family (patient 5) was also carrying the mitochondrial DNA variant m.11253T>C, detected by an initial, limited genetic work-up. The m.11253T>C variant is currently referenced both as a polymorphism and as a candidate LHON mutation in the Mitomap reference database, indicating that the pathogenic status of this variant is not definitive. It is, therefore, well possible that a synergistic effect might have played a role, between the OPA1 and the mtDNA variant, given also that patient 5 had a lower visual acuity than his affected father.
In conclusion, we report here the first seven cases of genetically confirmed ADOA, in a multiethnic population from Singapore. Although ADOA may be a rare condition in Asia, it should be part of the differential diagnosis in unexplained optic neuropathies, even in the absence of family history.

Acknowledgments
This work was supported by the National Medical Research Council, Singapore (Grant Nos: NMRC/CIRG/1401/2014(DM) and NMRC/NIG/1000/2009 (JJG)) and the Singapore National Eye Centre Health Research Endowment Fund, Singapore (Grant No: 1005/20/2013 (DM)). The funding organisations had no role in the design and conduct of the research.
Footnotes
The authors declare no conflict of interest.
References
- Lenaers G, Hamel C, Delettre CC, Amati-Bonneau P, Procaccio V, Bonneau D et al. Dominant optic atrophy. Orphanet J Rare Dis 2012; 7: 46–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milea D, Amati-Bonneau P, Reynier P, Bonneau D. Genetically determined optic neuropathies. Curr Opin Neurol 2010; 23(1): 24–28. [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin J-M, Gigarel N, Lorenzo C, Belenguer P et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet 2000; 26(2): 207–210. [DOI] [PubMed] [Google Scholar]
- Alexander C, Votruba M, Pesch UEA, Thiselton DL, Mayer S, Moore A et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet 2000; 26(2): 211–215. [DOI] [PubMed] [Google Scholar]
- Chao de la Barca JM, Prunier-Mirebeau D, Amati-Bonneau P, Ferré M, Sarzi E, Bris C et al. OPA1-related disorders: diversity of clinical expression, modes of inheritance and pathophysiology. Neurobiol Dis 2016; 90: 20–26. [DOI] [PubMed] [Google Scholar]
- Ferré M, Amati-Bonneau P, Tourmen Y, Malthièry Y, Reynier P. eOPA1: an online database for OPA1 mutations. Hum Mutat 2005; 25(5): 423–428. [DOI] [PubMed] [Google Scholar]
- Ferré M, Caignard A, Milea D, Leruez S, Cassereau J, Chevrollier A et al. Improved locus-specific database for OPA1 mutations allows inclusion of advanced clinical data. Hum Mutat 2015; 36(1): 20–25. [DOI] [PubMed] [Google Scholar]
- Cohn AC, Toomes C, Potter C, Towns KV, Hewitt AW, Inglehearn CF et al. Autosomal dominant optic atrophy: penetrance and expressivity in patients with OPA1 mutations. Am J Ophthalmol 2007; 143(4): 656–662.e651. [DOI] [PubMed] [Google Scholar]
- Ferré M, Bonneau D, Milea D, Chevrollier A, Verny C, Dollfus H et al. Molecular screening of 980 cases of suspected hereditary optic neuropathy with a report on 77 novel OPA1 mutations. Hum Mutat 2009; 30(7): E692–E705. [DOI] [PubMed] [Google Scholar]
- Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissière A et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy 'plus' phenotypes. Brain 2008; 131(2): 338–351. [DOI] [PubMed] [Google Scholar]
- Bonneau D, Colin E, Oca F, Ferré M, Chevrollier A, Guéguen N et al. Early-onset Behr syndrome due to compound heterozygous mutations in OPA1. Brain 2014; 137(10): e301–e301. [DOI] [PubMed] [Google Scholar]
- Spiegel R, Saada A, Flannery PJ, Burté F, Soiferman D, Khayat M et al. Fatal infantile mitochondrial encephalomyopathy, hypertrophic cardiomyopathy and optic atrophy associated with a homozygous OPA1 mutation. J Med Genet 2016; 53(2): 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer P. Infantile optic atrophy with dominant mode of inheritance: a clinical and genetic study of 19 Danish families. Acta Ophthalmol Suppl 1959; 164(Supp 54): 1–147. [PubMed] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Burke A, Sellar PW, Clarke MP, Gnanaraj L et al. The prevalence and natural history of dominant optic atrophy due to <em>OPA1</em> mutations. Ophthalmology 2010; 117(8): 1538–1546.e1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Ruiz A, Neuhaus C, Bergmann C, Bolz H. First cases of dominant optic atrophy in Saudi Arabia: report of two novel OPA1 mutations. J Neuroophthalmol 2013; 33(4): 349–353. [DOI] [PubMed] [Google Scholar]
- Dadgar S, Hagens O, Dadgar SR, Haghighi EN, Schimpf S, Wissinger B et al. Structural model of the OPA1 GTPase domain may explain the molecular consequences of a novel mutation in a family with autosomal dominant optic atrophy. Exp Eye Res 2006; 83(3): 702–706. [DOI] [PubMed] [Google Scholar]
- Liskova P, Ulmanova O, Tesina P, Melsova H, Diblik P, Hansikova H et al. Novel OPA1 missense mutation in a family with optic atrophy and severe widespread neurological disorder. Acta Ophthalmol 2013; 91(3): e225–e231. [DOI] [PubMed] [Google Scholar]
- Hamahata T, Fujimaki T, Fujiki K, Miyazaki A, Mizota A, Murakami A. OPA1 mutations in Japanese patients suspected to have autosomal dominant optic atrophy. Jpn J Ophthalmol 2012; 56(1): 91–97. [DOI] [PubMed] [Google Scholar]
- Yen M-Y, Wang A-G, Lin Y-C, Fann M-J, Hsiao K-J. Novel mutations of the OPA1 gene in Chinese dominant optic atrophy. Ophthalmology 2010; 117(2): 392–396.e1. [DOI] [PubMed] [Google Scholar]
- Chen Y, Jia X, Wang P, Xiao X, Li S, Guo X et al. Mutation survey of the optic atrophy 1 gene in 193 Chinese families with suspected hereditary optic neuropathy. Mol Vis 2013; 19: 292–302. [PMC free article] [PubMed] [Google Scholar]
- Rukmini AV, Milea D, Baskaran M, How AC, Perera SA, Aung T et al. Pupillary responses to high-irradiance blue light correlate with glaucoma severity. Ophthalmology 2015; 122(9): 1777–1785. [DOI] [PubMed] [Google Scholar]
- Toomes C, Marchbank NJ, Mackey DA, Craig JE, Newbury-Ecob RA, Bennett CP et al. Spectrum, frequency and penetrance of OPA1 mutations in dominant optic atrophy. Hum Mol Genet 2001; 10(13): 1369–1378. [DOI] [PubMed] [Google Scholar]
- Leo-Kottler B, Luberichs J, Besch D, Christ-Adler M, Fauser S. Leber's hereditary optic neuropathy: clinical and molecular genetic results in a patient with a point mutation at np T11253C (isoleucine to threonine) in the ND4 gene and spontaneous recovery. Graefes Arch Clin Exp Ophthalmol 2002; 240(9): 758–764. [DOI] [PubMed] [Google Scholar]
- Schimpf S, Schaich S, Wissinger B. Activation of cryptic splice sites is a frequent splicing defect mechanism caused by mutations in exon and intron sequences of the OPA1 gene. Hum Genet 2006; 118(6): 767–771. [DOI] [PubMed] [Google Scholar]
- Zhang A-M, Bi R, Hu Q-X, Fan Y, Zhang Q, Yao Y-G. The OPA1 gene mutations are frequent in Han Chinese patients with suspected optic neuropathy. Mol Neurobiol 2016; 1–9. [DOI] [PubMed]
- Nissen C, Ronnback C, Sander B, Herbst K, Milea D, Larsen M et al. Dissociation of pupillary post-illumination responses from visual function in confirmed OPA1 c.983A >G and c.2708_2711delTTAG autosomal dominant optic atrophy. Front Neurol 2015; 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki A, Herbst K, Sander B, Milea D. Selective wavelength pupillometry in Leber hereditary optic neuropathy. Clin Exp Ophthalmol 2010; 38(3): 322–324. [DOI] [PubMed] [Google Scholar]
- Leruez S, Milea D, Defoort-Dhellemmes S, Colin E, Crochet M , Procaccio V et al. Sensorineural hearing loss in OPA1-linked disorders. Brain 2013; 136(7): e236–e236. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 2010; 20(1): 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. Fast, scalable generation of high‚ quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11(4): 361–362. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res 2001; 11(5): 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7(4): 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29(1): 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exome Aggregation Consortium (ExAC), Cambridge, MA, USA. Available at: http://exac.broadinstitute.org (accessed on February 2016).
- Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, USA. Available at: http://evs.gs.washington.edu/EVS/ (accessed on February 2016).
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al. An integrated map of genetic variation from 1092 human genomes. Nature 2012; 491(7422): 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]