Abstract
Haloarchaeon Natrinema sp. J7, the first reported archaeon harboring both plasmid and chromosome-based temperate viruses, is a useful model for investigating archaeal virus-host and virus-virus interactions. However, the lack of genetic tools has limited such studies. On the basis of the automatically replicating sequences of the J7 chromosome and the pyrF marker, we constructed seven vectors, six of which were confirmed to possess replication ability in a pyrF-deletion derivative of J7 (J7-F). Among these vectors, pFJ1, pFJ4, and pFJ6 could be transformed into the host strain with relatively high efficiency (approximately 103 colony-forming units/μg DNA) and were present at about one copy per chromosome. These three vectors could be stably maintained in J7-F without selection and were used for heterologous protein expression. Only pFJ6 was found to be present in the transformed cells in an exclusively episomal, nonintegrated state (one copy per chromosome). In contrast, some pFJ1 and pFJ4 DNA was probably integrated into the J7-F chromosome. In addition, pFJ6 was found to be compatible with pYCJ in J7 cells, suggesting that these two vectors could be used for further studies of virus-virus and virus-host interactions.
1. Introduction
Extremely halophilic archaea constitute a group of microorganisms that thrive in hypersaline environments, including solar salterns and natural salt lakes. This group consists of approximately 48 genera and 177 species [1], forming an important part of archaea [2]. Compared to other groups of archaea, these haloarchaea are particularly suitable for research purposes, as they are easily cultured and manipulated in the laboratory. They also serve as useful models for studying archaeal viruses, which exhibit striking morphological diversity and unique gene contents. However, the study of these haloarchaea and their viruses is limited by the availability of appropriate genetic tools. Among the 48 genera of haloarchaea, genetic tools have been developed for only three genera, namely, Haloferax [3, 4], Halobacterium [5], and Haloarcula [6]. Considering that no single archaeal species is representative of the domain as a whole, or even its own specific group, it will be important to develop genetic tools for the many haloarchaea that still lack such resources.
Most haloarchaeal shuttle vectors are derived from endogenous plasmids, such as pGRB1 [7], pHH1 [8], and pNRC100 [9] from Halobacterium salinarum or pHK2 [4, 10], pHV2 [11, 12], and pHV1/4 [13] from Haloferax volcanii. The replicative function of some shuttle vectors originates from phage elements or chromosomal replication origins (oriCs), such as pUBP1 and pUBP2. These are constructed based on the ΦH replicon [5] and pBBori7, which was created using oriC/orc7 from Halobacterium sp. NRC-1 [14]. Most haloarchaea with established genetic tools can be transformed using polyethylene glycol (PEG). However, only a few selectable markers are available for haloarchaea. Novobiocin, which inhibits DNA gyrase (gyrB) [15], and mevinolin, which inhibits HMG-CoA reductase [16] are two widely used antibiotics in archaea. Auxotrophic selectable markers involved in amino acid or nucleotide biosynthesis have also been used. These include the ura3 and pyrE2 genes involved in uracil biosynthesis [17, 18], the trpA gene involved in tryptophan biosynthesis, the leuB gene participating in leucine biosynthesis, and the hdrB gene used in thymidine biosynthesis [12]. Among these markers, ura3 and pyrE2 are particularly useful for gene knockout or replacement studies because they can be counterselected using 5-fluoroorotic acid [18].
The halobacterial genus Natrinema gen. nov., was proposed by McGenity et al. based on the phylogenetic analysis of the 16S rRNA gene sequences and taxonomic properties of its member species. Strains in this genus require that the salt concentration be at least 10% to allow for basic cell growth and 19.8% to 25.1% for optimal growth [19]. Seven species have been discovered in this genus so far. Among them is Natrinema sp. J7, which was isolated in a Yingcheng salt mine in Hubei province, China. It is the only strain for which genetic manipulation tools had been established [20, 21] and whose genome had been sequenced [22]. Studies of J7 derivatives have focused on the proteases SptA [23, 24] and SptC [25], heat shock protein 70 [26, 27], and DNA fragments conferring promoter activity in the three domains of life. Notably, one of its derivatives, J7-1, was the first archaeon reported to harbor two kinds of temperate haloarchaeal viruses: sphaerolipovirus SNJ1 [28, 29] and pleolipovirus SNJ2 [30]. Accordingly, J7-1 could be used as an excellent model for studying virus-host and virus-virus interactions in archaea. However, the lack of genetic tools hampered these studies.
Previously, integrative plasmids based on the isolated auxotrophic mutants of J7 derivatives were constructed and used to express exogenous proteins in J7 [20]. Recently, a shuttle vector (pYCJ) based on the SNJ1 replicon (nucleotides 1–4481) was also constructed and validated for the expression and purification of heterologous proteins [21]. A number of ORFs and genetic elements controlling virus genome replication, maintenance, and copy number were identified in the SNJ1 replicon. Moreover, the key elements responsible for superinfection exclusion and lytic/lysogenic conversion of the SNJ1 virus are also located in this region (unpublished data). Because any mutation to the SNJ1 replicon would impair the self-replicating ability of the plasmid, the pYCJ vector is not suitable for investigating these regulators and elements. Thus, additional genetic tools must be developed to enable research on these viruses and their host. In this study, seven shuttle vectors derived from the predicted oriCs in the J7 chromosome were constructed. Each vector contained the pyrF gene and the autonomously replicating sequence (ARS, one or two predicted oriCs). Six of these vectors (pFJ1, pFJ3, pFJ4, pFJ5, pFJ6, and pFJ7) were successfully transformed into J7-F, but only pFJ1, pFJ4, and pFJ6 could be transformed with relatively high efficiency (103 colony-forming units [cfu]/μg DNA). These three shuttle vectors were studied further and found to be stably maintained at approximately one copy per chromosome in J7-F without selection. pFJ1 and pFJ4 may exist either in the plasmid form or integrated into the host chromosome, whereas pFJ6 replicated only in a nonintegrated state and was compatible with pYCJ in J7 cells. All three vectors were validated for stable expression of the amylase protein in J7-F cells, making them instrumental for further studies on the SNJ1 virus, as well as J7 derivatives.
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
Strains used in this study are listed in Table 1. Different derivatives of Natrinema sp. J7 were grown in Halo-2 medium at 45°C, as previously described [28, 30]. A J7-F strain lacking the chromosomal pyrF gene was a gift from Professor Yuping Huang (College of Life Science, Wuhan University). Minimal medium (MM; 18%) and modified growth medium (MGM; 18%) were prepared as described in the Halohandbook [31]. Escherichia coli strains DH5α and JM110 were used for plasmid DNA construction and were routinely cultured in Luria–Bertani medium at 37°C. Agar (1.5%) was added when solid analogs of the above-mentioned media were needed.
Table 1.
Strains and plasmids used in this study.
| Strain | Description | Source |
|---|---|---|
| Natrinema sp. J7-1 | With SNJ1 proviral genome pHH205, cannot be infected by SNJ1 | [28, 34] |
| Natrinema sp. J7-F | ΔpyrF, can be infected by SNJ1 | Yuping Huang, Wuhan University, Wuhan, China |
| Escherichia coli DH5α | SupE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | CCTCC |
| Escherichia coli JM110 | dam dcm supE44 hsdR17 thi leu rpsL lacY galK galT ara tonA thr tsx Δ(lac-proAB) F′(traD36 proAB+lacIq lacZ ΔM15) | CCTCC |
| Plasmid | ||
| pNBK-F | AmpR, MevR, used to clone Pfdx-pyrF resistance fragment | Yuping Huang, Wuhan University, Wuhan, China |
| pUC-M | pUC19 with SnaBI, MfeI, AgeI inserted between BamHI and Xbal | Our lab |
| pUC-M-pyrF | pUC-M with the insertion of 0.95-kb BamHI-SnaBI Pfdx-pyrF resistance fragment | This study |
| pFJ1 | pUC-M-pyrF with the insertion of the 2.8-kb SnaBI-MfeI predicted chromosomal ARS-1 | This study |
| pFJ2 | pUC-M-pyrF with the insertion of the 3.3-kb SnaBI-MfeI predicted chromosomal ARS-2 | This study |
| pFJ3 | pUC-M-pyrF with the insertion of the 3.2-kb SnaBI-MfeI predicted chromosomal r ARS-3 | This study |
| pFJ4 | pUC-M-pyrF with the insertion of the 2.2-kb SnaBI-MfeI predicted chromosomal ARS-4 | This study |
| pFJ5 | pUC-M-pyrF with the insertion of the 2.3-kb SnaBI-MfeI predicted chromosomal ARS-5 | This study |
| pFJ6 | pUC-M-pyrF with the insertion of the 2.0-kb SnaBI-MfeI predicted chromosomal ARS-6 | This study |
| pFJ7 | pUC-M-pyrF with the insertion of the 1.1-kb SnaBI-MfeI predicted chromosomal ARS-7 | This study |
| pFJ1-Apro-amyH | pFJ1 with the insertion of 1.5-kb MfeI-SphI Apro-amyH fragment containing AflII, SpeI, NotI, NheI, and NsiI | This study |
| pFJ1-M | pFJ1-Apro-amyH digested with NotI and then ligated by itself | This study |
| pFJ4-Apro-amyH | pFJ4 with the insertion of 1.5-kb MfeI-SphI Apro-amyH fragment containing AflII, SpeI, NotI, NheI, and NsiI | This study |
| pFJ4-M | pFJ4-Apro-amyH digested with NotI and then ligated by itself | This study |
| pFJ6-Apro-amyH | pFJ6 with the insertion of 1.5-kb MfeI-SphI Apro-amyH fragment containing AflII, SpeI, NotI, NheI, and NsiI | This study |
| pFJ6-M | pFJ6-Apro-amyH digested with NotI and then ligated by itself | This study |
| pFJ1-M-pro3916 | pFJ1-M with the insertion of 200-bp NsiI-SphI pro3916 | This study |
| pFJ4-M-pro3916 | pFJ4-M with the insertion of 200-bp NsiI-SphI pro3916 | This study |
| pFJ6-M-pro3916 | pFJ6-M with the insertion of 200-bp NsiI-SphI pro3916 | This study |
2.2. Predicting oriCs in the J7 Chromosome
The web-based tool Ori-Finder 2 was employed to predict oriCs in archaeal genomes [32]. OriCs were predicted based on an integrated analysis of base-composition asymmetry using the Z-curve method, the distribution of origin recognition boxes (ORBs) identified using Find Individual Motif Occurrence [33], and the occurrence of genes that are frequently close to oriCs.
2.3. Plasmid Construction
Plasmids and primers used in this study are listed in Tables 1 and 2, respectively. A 0.95 kb BamHI-SnaBI fragment consisting of the pyrF gene and the 200 bp promoter region of the fdx gene (Pfdx) from Haloferax volcanii DS2 was cloned from the pNBK-F plasmid into the BamHI and SnaBI sites of the pUC-M vector to generate pUC-M-pyrF.
Table 2.
Primers used in this study.
| Primers | 5′-3′ sequence | Restriction sites |
|---|---|---|
| Pfdx-pyrF-F | AATGGATCCATCTCGGCTTATTCTTTTGATT | BamHI |
| Pfdx-pyrF-R | TAATACGTATTATTCTCGATACTGATTGAGTCGCTTC | SnaBI |
| PARS-1-fwd | AATTACGTACGCCCCCGGTGCCTCCTCTCGGA | SnaBI |
| PARS-1-rev | ATTCAATTGACTCGCCGCCGACTACCTCCCCGTCG | MfeI |
| PARS-2-fwd | AATTACGTATAGCCCGGGAAATACTATCTTTGAGTTCT | SnaBI |
| PARS-2-rev | ATTCAATTGGATCGACGCTGGGATATGAAAAGC | MfeI |
| PARS-3-fwd | AATTACGTAAACGGCTTTCGGATCGAAAGCAGC | SnaBI |
| PARS-3-rwd | ATTCAATTGTTCGGTCTGCGGTCCCCATTTCC | MfeI |
| PARS-4-fwd | AATTACGTAGACACACACCACTGTTGCAAGTGAAG | SnaBI |
| PARS-4-rev | ATTCAATTGGTGGCCGCACAAGATCGA | MfeI |
| PARS-5-fwd | AATTACGTACGATCGTGCCGACGTTACCCGGT | SnaBI |
| PARS-5-rev | ATTCAATTGCGATCCCGAAGACGACCGCGT | MfeI |
| PARS-6-fwd | AATTACGTAGGAGACGGTCAGAGTTACTGGTCAGT | SnaBI |
| PARS-6-rev | TAACAATTGCAAGGGTTCGTCTGAAACCGTGT | MfeI |
| PARS-7-fwd | ATATACGTAAGGCTGACTGTATGCGAGT | SnaBI |
| PARS-7-rev | ATACAATTGGCACGACAGTAACAGT | MfeI |
| 3916pro(R)-F | AATATGCATGAGTAAAGTTCGTGTTTCCTTGATTA | NsiI |
| 3916pro(R)-R | ATAGCATGCCGGACAAGACGCCCATTTG | SnaBI |
| Apro-AmyH-M-F | AATCAATTGCTTAAGACTAGTGCGGCCGCGGGAGCCGGAAACGCGGTAGAGATA | MfeI, AgeI, SpeI, NotI |
| Apro-AmyH-M-R | AAGCATGCTATGCATAGCTAGCGCGGCCGCAAGGTAGTGGAAAGCGAGCCAGCGC | NotI, NsiI, NdeI, SphI |
| pyrF test-F | CGATCACCGTCAACCCCTACATGG | / |
| pyrF test-R | TACTGATTGAGTCGCTTCTTCAGTCGTTT | / |
| MevR test-F | TCGCCTCCCTCGAAGTCGGCACCGT | / |
| MevR test-R | GAACAACGGCGAAGAAAAGGCAGTCCA | / |
Seven SnaBI-MfeI-digested ARSs containing one or two predicted oriCs amplified from the J7 genome were ligated into the digested pUC-M-pyrF plasmid to create pFJ1, pFJ2, pFJ3, pFJ4, pFJ5, pFJ6, and pFJ7. The specific locations of the 12 predicted oriCs and seven ARSs are shown in Table 3. A 1.7 kb fragment encompassing amyH and its 200 bp promoter (Apro-amyH) was amplified from Haloarcula hispanica DSM4426. Restriction enzyme sequences for MfeI, AflII, SpeI, and NotI were added to the 5′ end of the fragment and NotI, NheI, NsiI, and SphI sites were added to the 3′ end. The MfeI-Apro-amyH-SphI fragment was ligated into pFJ1, pFJ4, and pFJ6 to generate pFJ1-A, pFJ4-A, and pFJ6-A, respectively. After digestion with NotI followed by self-ligation, the Apro-amyH fragment was removed from each vector to construct pFJ1-M, pFJ4-M, and pFJ6-M. The physical maps of pFJ1-M, pFJ4-M, and pFJ6-M are shown in Figure 1. A 200 bp fragment encompassing the upstream region of the 3916 gene (3916 pro) located in the J7 chromosome was amplified and ligated into pFJ1-M, pFJ4-M, and pFJ6-M to generate the plasmids pFJ1-M-3916, pFJ4-M-3916, and pFJ6-M-3916, respectively, which were used for plasmid copy number determination.
Table 3.
Characteristics of predicted oriCs and replication regions used for shuttle vector construction.
(a) Characteristic of predicted oriCs
| OriCs | Locations | Number of ORB elements | GC content (%) | Adjacent to cdc6 gene |
|---|---|---|---|---|
| 1 | 120445–121114 | 2 | 51.27% | Y |
| 2 | 122480–123045 | 1 | 60.30% | Y |
| 3 | 399506–400667 | 3 | 56.44% | N |
| 4 | 434294–435598 | 8 | 54.87% | Y |
| 5 | 1277983–1279633 | 5 | 59.37% | Y |
| 6 | 1280840–1281049 | 1 | 66.67% | Y |
| 7 | 2211660–2211903 | 2 | 60.51% | Y |
| 8 | 2213131–2213664 | 2 | 62.89% | Y |
| 9 | 2573530–2574443 | 1 | 55.78% | Y |
| 10 | 2860326–2861584 | 7 | 52.74% | N |
| 11 | 3151118–3151637 | 3 | 60.17% | Y |
| 12 | 3153862–3154084 | 1 | 68.61% | Y |
(b) Characteristic and replication ability of the shuttle vectors
| Shuttle vectors | ARSs | OriCs contained | Chromosomal locations of ARSs (positions) | Containing cdc6 gene |
Transformation efficiencya |
|---|---|---|---|---|---|
| pFJ1 | 1 | 1, 2 | 120,345–123,145 | Y | (1.7 ± 0.6) × 103 |
| pFJ2 | 2 | 4 | 434,194–437,483 | Y | 0 |
| pFJ3 | 3 | 5, 6 | 1,277,883–1,281,049 | Y | (3.9 ± 0.4) × 101 |
| pFJ4 | 4 | 7, 8 | 2,211,560–2,213,764 | Y | (2.1 ± 0.1) × 103 |
| pFJ5 | 5 | 9 | 2,572,200–2,574,543 | Y | <10 |
| pFJ6 | 6 | 10 | 2,860,000–2,862,000 | N | (2.2 ± 0.5) × 103 |
| pFJ7 | 7 | 3 | 399,506–400,667 | N | <10 |
aColony-forming units/μg DNA.
Figure 1.
Physical maps of the pFJ1-M (a), pFJ4-M (b), and pFJ6-M (c) plasmids.
2.4. Transformation Method
Genetic transformation of J7-F cells was performed at room temperature using PEG, as previously described [21]. MM (18%) was prepared and used to select and propagate the Natrinema sp. J7-F transformants.
2.5. Stability and Maintenance of the Shuttle Vectors
To assess the structural stability of the vectors, we isolated pFJ1, pFJ4, and pFJ6 from J7-F transformants, back-transformed them into E. coli for amplification, extracted the plasmids, and subjected them to restriction enzyme digestion. The plasmid DNA was extracted from E. coli (without being transformed into J7-F cells), digested, and used as a positive control.
Maintenance of the pFJ1, pFJ4, and pFJ6 shuttle vectors was evaluated by calculating the rate at which each plasmid was lost per generation during nonselective growth, as previously described [21]. Briefly, selected clones were cultured to exponential phase for approximately five days at 45°C in 5 mL 18% MM. The cultures were then diluted 1 : 100 in 5 mL of fresh Halo-2 medium and incubated for 24 h. This dilution process was repeated 12 times (the doubling time of J7 cells was ~2.9 h). Every two dilutions, the cultures were spread onto 18% MGM plates (nonselective condition for J7-F cells). Fifty random clones were selected and reseeded onto 18% MGM or 18% MM plates. The numbers of clones that grew on both kinds of plates were counted. Survival on 18% MM plates indicated maintenance of the vector.
2.6. Southern Blot Analysis
Southern blot analysis was performed to determine whether the shuttle vectors (pFJ1, pFJ4, and pFJ6) had integrated into the J7-F chromosome and to measure their copy numbers. J7-F cells transformed with pFJ1, pFJ4, or pFJ6 were cultivated in 200 mL 18% MM and harvested in exponential phase. Their genomic DNA was isolated as previously described [34], digested at the only HindIII site in the shuttle vectors, and separated by 1% (w/v) agarose gel electrophoresis. After denaturation in alkaline solution (1 M NaCl, 0.5 M NaOH), DNA samples were transferred to a positively charged nylon membrane. Subsequently, the DNA was fixed by incubation at 80°C for 2 h and probed using a pyrF fragment labeled with a digoxigenin- (DIG-) dUTP random primer, based on the instructions of the DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche). To determine plasmid copy number, a 200 bp upstream fragment of the J7 3916 gene, present at one copy per chromosome, was amplified and ligated into pFJ1, pFJ4, and pFJ6. Then, total DNA isolated from J7-F/pFJ1-3916pro, J7-F/pFJ4-3916pro, and J7-F/pFJ6-3916pro was digested with Sau3AI and subjected to Southern blot analysis as described above, using the DIG-labeled 3916 pro sequence as a probe.
2.7. Amylase Activity Assay
Specific amylase activity of the supernatants from CJ7/pYCJ-Apro-amyH, CJ7-F/pFJ1-Apro-amyH, CJ7-F/pFJ4-Apro-amyH, and CJ7-F/pFJ6-Apro-amyH was measured as previously described [21]. One unit of amylase activity was defined as the quantity of amylase required to hydrolyze 1 mg of starch in 1 h.
3. Results
3.1. Analyzing and Cloning of Replication Regions of the J7 Chromosome
To construct shuttle vectors for J7 derivatives, oriCs in the J7 chromosome were predicted using the web-based Ori-Finder 2 tool. As shown in Table 3(a), 12 oriCs were predicted in the J7 chromosome; most of them had a relatively low GC content compared to that of the J7 chromosome (64%) and all of them contained ORBs. Except for oriC3 and oriC10, the other oriCs were all adjacent to cdc6 genes, encoding replication-initiating proteins. However, the locations of some oriC pairs, such as oriC1 and oriC2, oriC5 and oriC6, and oriC7 and oriC8, as well as oriC11 and oriC12, were very close and adjacent to the same cdc6 gene (Table 3(a)). Five presumed ARSs containing one or two oriCs and their adjacent cdc6 gene were amplified (Table 3(b)). These were then ligated into pUC-M-pyrF to construct the pFJ1, pFJ2, pFJ3, pFJ4, and pFJ5 shuttle vectors. In addition, oriC12 and oriC3, which were not adjacent to the cdc6 gene, were also amplified and used to construct pFJ6 and pFJ7, respectively. As shown in Table 3(b), all vectors except pFJ2 were successfully transformed into J7-F cells. Transformation efficiency of pFJ1, pFJ4, and pFJ6 was approximately 103 cfu/μg DNA, a little lower than that of the SNJ1 replicon-based vector, pYCJ [21]. Only a few transformants were obtained after transforming JF-7 cells with pFJ3, pFJ5, and pFJ7. These results indicate that at least six regions in the J7 chromosome possess replication ability, which is consistent with the presence of multiple replication origins in the chromosome of most haloarchaeal strains [35].
3.2. Maintenance and Structural Stability of the oriC-Based Vectors in J7-F Cells
To assess the structural stability of all shuttle vectors (pFJ1, pFJ3, pFJ4, pFJ5, pFJ6, and pFJ7) and verify their ability to replicate independently in J7-F cells, plasmid DNA was extracted from each vector transformant and back-transformed into E. coli (posttransformed plasmids), reisolated, and subjected to enzymatic digestion. Shuttle vectors extracted from E. coli (pretransformed into J7-F cells) were used as positive controls. As shown in Figure 2, pretransformed (lanes 2) and posttransformed (lanes 4) plasmids showed similar digestion profiles, indicating that structural stability of all constructs was maintained in J7-F cells and that these six vectors replicated independently of the chromosome. As mentioned above, pFJ3, pFJ5, and pFJ7 could be successfully transformed into J7-F cells, but their transformation efficiencies were too low to be suitable for genetic manipulation of J7 cells. Hence, these three vectors were excluded from further studies.
Figure 2.
Structural stability of the shuttle vectors in J7-F cells. Pretransformed plasmids (extracted from E. coli before being transformed into J7-F, lanes 1 and 2) and back-transformed plasmid (extracted from E. coli following transformation into J7-F, lanes 3 and 4) were digested with restriction enzymes to compare their structural stabilities. M, 2 kb plus II marker; lanes 1 and 3, undigested plasmids; lanes 2 and 4, plasmids digested with SnaBI and MfeI.
Maintenance of pFJ1, pFJ4, and pFJ6 was determined by calculating the survival frequencies on 18% MM during nonselective growth. As shown in Figure 3, all vectors could be stably maintained in J7-F cells after culturing for 12 days (~100 generations), indicating that all of them could be stably segregated into daughter cells during cell growth.
Figure 3.
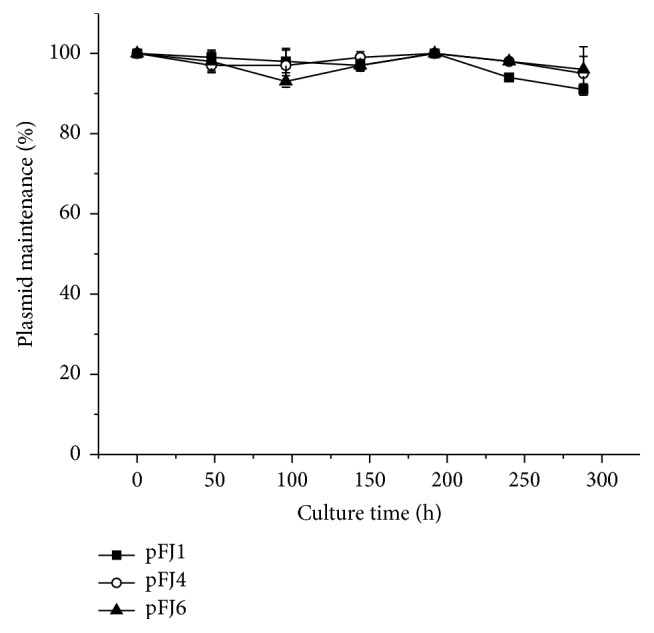
Maintenance of the pFJ1, pFJ4, and pFJ6 shuttle vectors in J7-F cells. J7-F cells containing different shuttle vectors (50 μL) were diluted in 5 mL Halo-2 medium and grown for 24 h. For every two dilutions (12 dilutions in total), aliquots were spread onto Halo-2 plates, and 50 random colonies were selected and spotted onto nonselective 18% MGM plates. Maintenance was measured by calculating the percentage of colonies growing on selective 18% MM plates.
3.3. Presence of pFJ6 in Transformed Cells in an Episomal, Nonintegrated State
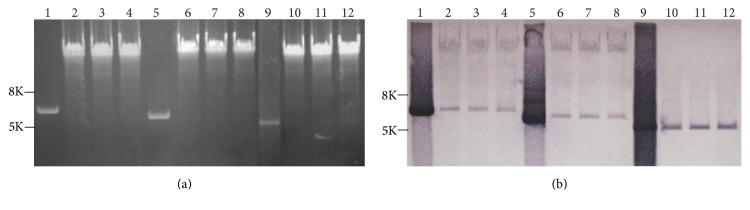
Southern blot analysis was performed to determine whether pFJ1, pFJ4, and pFJ6 integrated into the J7-F chromosome. Total DNA extracted from three independent J7-F transformants of each plasmid was digested with HindIII, separated by 1% (w/v) agarose gel electrophoresis (Figure 4(a)), and then subjected to Southern blot analysis. Plasmids extracted from E. coli were used as controls and the DIG-labeled pyrF gene was used as probe. Only one HindIII restriction site was found in pFJ1, pFJ4, and pFJ6 plasmids. Thus, digested nonintegrated plasmids from total DNA samples were predicted to be linearized to 5.86 kb, 5.66 kb, and 5.35 kb, respectively, distinguishing them from integrated plasmids. As shown in Figure 4(b), hybridization signals could be observed at every position corresponding to the linearized plasmid DNA, either in the HindIII-digested DNA samples extracted from E. coli (lanes 1, 5, and 9) or in the HindIII-digested total DNA preparations extracted from cultures of J7-F clones carrying pFJ1 (lanes 2–4), pFJ4 (lanes 6–8), and pFJ6 (lanes 10–12). However, compared with the single hybridization signal obtained using total DNA extracted from pFJ6-transformed J7-F cells (lanes 10–12), the probe also hybridized with a relatively large fragment in the total DNA samples extracted from pFJ1- and pFJ4-transformed J7-F cells (lanes 2–4 and 6–8, resp.). These data indicate that only pFJ6 was present in an episomal, nonintegrated state in J7 cells, whereas pFJ1 and pFJ4 may have integrated into the CJ7-F chromosome. Additional experiments are required to confirm these results.
Figure 4.
Determination of whether pFJ1, pFJ4, and pFJ6 could integrate into the J7 chromosome by Southern blot analysis. HindIII-digested total DNA extracted from cultures of J7-F clones carrying the pFJ1 (lanes 2–4), pFJ4 (lanes 6–8), or pFJ6 (lanes 10–12) plasmid were (a) separated on a 1% agarose gel at 20 V for >12 h and then (b) subjected to Southern blot analysis. The pFJ1, pFJ4, and pFJ6 plasmids were propagated in E. coli, digested with HindIII, and loaded on lanes 1, 5, and 9 as controls. The DIG-labeled pyrF sequence was used as a hybridization probe.
3.4. Relative Copy Numbers of pFJ1, pFJ4, and pFJ6 in J7-F Cells
Shuttle vectors containing chromosome oriCs are usually present at one copy per chromosome [36]. We tested whether this was true for pFJ1, pFJ4, and pFJ6. The 200 bp single-copy 3916 pro segment was amplified and ligated into the shuttle vectors used in this study. Total DNA from three randomly selected independent J7-F transformants containing pFJ1-M-3916, pFJ4-M-3916, and pFJ6-M-3916, respectively, was digested with Sau3AI, separated by 1% agarose gel electrophoresis, and subjected to Southern blot analysis using a DIG-labeled 3916 pro probe. A 686 bp fragment containing 3916 pro was liberated from the J7 chromosome after digestion with Sau3AI, whereas 1119 bp, 978 bp, and 1266 bp fragments containing 3916 pro were generated from pFJ1-M-3916, pFJ4-M-3916, and pFJ6-M-3916 (Figure 5(a)). As shown in Figure 5(b), the DIG-labeled 3916 pro sequence hybridized to the 686 bp fragment from the J7 chromosome and the corresponding fragment of each plasmid. The intensities of the hybridization signals generated from pFJ6-M-3916 and the chromosomal DNA fragment were almost identical, indicating that pFJ6 was present at approximately one copy per chromosome in J7-F cells. In contrast, the intensity of the hybridization signals generated by pFJ1-M-3916 and pFJ4-M-3916 was slightly stronger than that of the corresponding chromosomal DNA fragment, implying that pFJ1-M-3916 and pFJ4-M-3916 were present at more than one copy per chromosome. These results may be attributed to the fact that a portion of pFJ1 and pFJ4 could integrate into the J7 chromosome; however, additional experiments are required to confirm this thesis.
Figure 5.
Relative copy numbers of the pFJ1, pFJ4, and pFJ6 plasmids in J7-F cells. (a) Schematic diagrams of the J7 chromosome and indicated vectors. The specific locations of Sau3AI sites in the J7 chromosome and plasmids are shown. The 200 bp 3916 pro segment, which was used as a DIG-labeled probe, is indicated with arrows and asterisks. (b) The copy numbers of pFJ1-pro3916, pFJ4-pro3916, and pFJ6-pro3916 were determined by Southern blot analysis. Sau3AI-digested total DNA from J7-F cells transformed with pFJ1 (lanes 2–4), pFJ4 (lanes 6–8), or pFJ6 (lanes 10–12) was hybridized with the DIG-labeled 3916 pro sequence. Total DNA samples from J7-F cells (lane 0), pFJ1-pro3916 (lane 1), pFJ4-pro3916 (lane 5), and pFJ6-pro3916 (lane 9) plasmids propagated in E. coli were used as controls.
3.5. Utility of Shuttle Vectors pFJ1, pFJ4, and pFJ6
To determine the utility of these shuttle vectors, the Haloarcula hispanica DSM 4426 amyH gene and its promoter were inserted into pFJ1, pFJ4, and pFJ6. After transforming CJ7 cells with the respective plasmids (pFJ1-A, pFJ4-A, and pFJ6-A), three randomly selected colonies per plasmid were transferred onto 18% MGM plates supplemented with 2% (w/v) soluble starch. As shown in Figure 6(a), transparent halos were detected around the colonies after flooding the plates with iodine solution, suggesting that amylase was well expressed. In contrast, colonies of J7-F cells harboring only pFJ1, pFJ4, or pFJ6 showed no amylase activity, confirming the role of amylase in starch consumption. As shown in Figure 6(b), amylase activity in the supernatants of CJ7-F/pFJ1-Apro-amyH, CJ7-F/pFJ4-Apro-amyH, and CJ7-F/pFJ6-Apro-amyH cultures at the same OD600 was generally lower than that of CJ7/pYCJ-Apro-amyH. This result can be explained by the copy number of pYCJ (one to three copies per chromosome) [21] being higher than that of pFJ1, pFJ4, and pFJ6. The highest amylase activity occurred in CJ7-F/pFJ6-Apro-amyH, suggesting that pFJ6 drove greater protein expression than pFJ1 and pFJ4.
Figure 6.
Amylase expression in J7-F cells using pFJ1, pFJ4, and pFJ6. (a) The pFJ1-A, pFJ4-A, and pFJ6-A plasmids were transformed into J7-F cells. Three random transformants were transferred to 5 mL of Halo-2 medium, grown to exponential phase, and 2 μL of each culture was spotted onto 18% MGM plates supplemented with 2% (w/v) soluble starch. After five days, iodine solution was added to the plates, and halos formed immediately around the selected transformants indicating that amylase was successfully expressed. 1-1: J7-F transformed with pFJ1 (negative control); 1-2, 1-3, and 1-4: J7-F transformants harboring pFJ1-A; 4-1: J7-F transformed with pFJ4 (negative control); 4-2, 4-3, and 4-4: J7-F transformants harboring pFJ4-A; 6-1: CJ7 cells transformed with pFJ6 (negative control); 6-2, 6-3, and 6-4: J7-F transformants harboring pFJ6-A (negative control). (b) Amylase-specific activities of supernatants collected from CJ7/pFJ1-Apro-amyH, CJ7/pFJ4-Apro-amyH, CJ7/pFJ6-Apro-amyH, and CJ7/pYCJ-Apro-amyH cultures. One unit of amylase activity was defined as the quantity of amylase required to hydrolyze 1 mg of starch in 1 h.
3.6. Compatibility of pFJ6 and SNJ1 Replicon-Based pYCJ
Compatible shuttle vectors are excellent tools for investigating protein-protein and protein-DNA interactions in prokaryotes. Previously, we reported the first Natrinema sp. J7 E. coli shuttle vector, pYCJ, which was constructed based on the SNJ1 replicon and validated for stable expression of heterologous proteins. Because pFJ6 could also be maintained in J7 cells and did not integrate into the J7 chromosome, we tested whether pFJ6 and pYCJ were compatible with each other. pFJ6 and pYCJ were cotransformed into J7-F cells and transformants were selected on 18% MM containing 5 μg/mL mevinolin. The presence of pFJ6 and pYCJ in the transformants was detected by PCR using primers targeting pyrF and mevR, respectively. The result showed that pyrF and mevR were detected in all randomly selected transformants (data not shown), indicating that these two replicons were compatible with each other.
4. Discussion
Haloarchaeon Natrinema sp. J7 is the first archaeon known to harbor both plasmid- and chromosome-based temperate viruses, SNJ1 and SNJ2. These two viruses display many interesting features. First, the efficient production of SNJ2 could only be achieved in J7 strains coinfected with SNJ1, indicating that SNJ1 promoted the replication of SNJ2 [30]. However, little is known about the mechanism behind this virus-virus interaction. Second, SNJ1 could infect CJ7, which does not harbor pHH205, but could not infect J7-1 (which does harbor pHH205), indicating that the lysogenic SNJ1 virus could establish superinfection exclusion or immunity, a phenomenon poorly understood in archaea. Third, several ORFs and genetic elements controlling virus genome replication, maintenance, and copy number were recently identified in the SNJ1 virus [21]. However, the mechanisms of these genetic elements and genes are completely unknown. All of these observations suggest that J7 and its viruses are excellent models for studying virus-host and virus-virus interactions in archaea. To date, SNJ1 replicon-based pYCJ has been the only shuttle vector enabling genetic manipulation in J7 cells. Because the pYCJ shuttle vector contains the SNJ1 replicon [21], it could not be used for most studies on SNJ1, especially for functional studies on the regulatory proteins encoded by the SNJ1 replicon. In addition, given that only one shuttle vector was available for J7 cells, studies on protein-protein or protein-DNA interactions have also been limited. In this study, seven vectors based on the predicted oriCs in the J7 chromosome and the pyrF marker were constructed; six of them could replicate in the uracil auxotrophic J7 strain (J7-F). Three of these plasmids (pFJ1, pFJ4, and pFJ6) could be transformed into J7-F cells with high efficiency (103 cfu/μg DNA). These three vectors were stably maintained in transformed J7-F cells without selection and could be used to express heterologous proteins. Notably, one of these vectors, pFJ6, existed as a plasmid in J7 cells and was compatible with pYCJ. These plasmids should serve as valuable tools for further studies on virus-virus and virus-host interactions in haloarchaea.
The vectors developed in this study displayed many advantages compared to pYCJ. First, they could stably replicate and segregate into daughter cells even under nonselective growth conditions. In contrast, pYCJ has been reported to disappear from CJ7 cells after three days without antibiotic selection [21]. This property is particularly important given that only a few markers are available for archaea. Second, the molecular weights of the vectors constructed here were all around 6 kb, which is much smaller than that of most of the archaeal shuttle vectors previously reported [37], including pYCJ (about 9.9 kb). Consequently, these shuttle vectors should be capable of accommodating larger exogenous fragments and will be much more convenient for genetic manipulation in general. Finally, given their compatibility with SNJ1 (pHH205) and pYCJ, the shuttle vectors constructed in this study and especially pFJ6 could be used for studying the regulators located in the SNJ1 replicon. This will be made possible by expressing only fragments or mutated regions of the SNJ1 replicon. Such kind of molecular manipulation could not be achieved using pYCJ, because any mutation in the 1–4481 region of SNJ1 would impair the stability of pYCJ [21]. In addition, the vectors constructed in this study will provide a useful tool for studying several biological processes in J7 cells. These include superinfection exclusion and lytic/lysogenic conversion of the SNJ1 virus, whose key determinants are located in the 1–4481 region of SNJ1 (unpublished data).
In addition to viral studies, the vectors developed here may be useful for investigating J7 chromosome replication and segregation. Archaeal oriCs normally consist of a long intergenic sequence encompassing an A/T-rich, duplex-unwinding element. They are typically located upstream of a cdc6/orc1 gene, which encodes a putative initiator protein that is homologous to Orc1 of the eukaryotic ORC complex or the helicase loader Cdc6 [38, 39]. Twelve oriCs were predicted in the J7 chromosome using Ori-Finder 2 software; most of them were adjacent to a putative cdc6 gene. Six of the shuttle vectors containing one or two of the oriCs could replicate independently in J7-F cells, suggesting that at least six of the 12 predicted oriCs had the ability to initiate DNA replication. This is not surprising given that most archaea contain multiple replication origins [38]. However, it is remarkable that the homology between the nucleotide sequences in these six regions and the amino acid sequences of replication initiation proteins is low. It will be of interest to test which oriC in these shuttle vectors is required for replication and whether the adjacent cdc6 gene is also essential. pFJ6 carrying the predicted oriC10 does not contain a putative cdc6 gene, but it can replicate, suggesting that some of the cdc genes may be responsible for replication initiation at multiple origins. Furthermore, three of the shuttle vectors are maintained at a single copy per chromosome without selection, indicating that their replication is coordinated with that of the chromosome and that they are faithfully segregated into daughter cells. Investigating the molecular mechanisms underlying such coordination will be particularly interesting because the control and coordination of replication initiation at multiple origins in archaea are poorly understood. These vectors are excellent tools for investigating this important question in archaea.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (no. 31570174), the National Fund for Fostering Talents of Basic Sciences (J1103513), and Research (Innovative) Fund of Laboratory of Wuhan University. The authors thank Dr. Shishen Du (University of Kansas Medical Center, USA) for many helpful discussions and for revising the manuscript.
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Gupta R. S., Naushad S., Baker S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. International Journal of Systematic and Evolutionary Microbiology. 2015;65(3):1050–1069. doi: 10.1099/ijs.0.070136-0. [DOI] [PubMed] [Google Scholar]
- 2.Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam W. L., Doolittle W. F. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(14):5478–5482. doi: 10.1073/pnas.86.14.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes M., Pfeifer F., Dyall-Smith M. Improved shuttle vectors for Haloferax volcanii including a dual-resistance plasmid. Gene. 1994;146(1):117–121. doi: 10.1016/0378-1119(94)90844-3. [DOI] [PubMed] [Google Scholar]
- 5.Blaseio U., Pfeifer F. Transformation of Halobacterium halobium: development of vectors and investigation of gas vesicle synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(17):6772–6776. doi: 10.1073/pnas.87.17.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cline S. W., Doolittle W. F. Transformation of members of the genus Haloarcula with shuttle vectors based on Halobacterium halobium and Haloferax volcanii plasmid replicons. Journal of Bacteriology. 1992;174(3):1076–1080. doi: 10.1128/jb.174.3.1076-1080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krebs M. P., Hauss T., Heyn M. P., RajBhandary U. L., Khorana H. G. Expression of the bacterioopsin gene in Halobacterium halobium using a multicopy plasmid. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(3):859–863. doi: 10.1073/pnas.88.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeifer F., Ghahraman P. Plasmid pHH1 of Halobacterium salinarium: characterization of the replicon region, the gas vesicle gene cluster and insertion elements. MGG Molecular & General Genetics. 1993;238(1-2):193–200. doi: 10.1007/BF00279547. [DOI] [PubMed] [Google Scholar]
- 9.Ng W.-L., DasSarma S. Minimal replication origin of the 200-kilobase Halobacterium plasmid pNRC100. Journal of Bacteriology. 1993;175(15):4584–4596. doi: 10.1128/jb.175.15.4584-4596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes M. L., Dyall-Smith M. L. A plasmid vector with a selectable marker for halophilic archaebacteria. Journal of Bacteriology. 1990;172(2):756–761. doi: 10.1128/jb.172.2.756-761.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlebois R. L., Lam W. L., Cline S. W., Doolittle W. F. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(23):8530–8534. doi: 10.1073/pnas.84.23.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allers T., Ngo H.-P., Mevarech M., Lloyd R. G. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Applied and Environmental Microbiology. 2004;70(2):943–953. doi: 10.1128/aem.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norais C., Hawkins M., Hartman A. L., Eisen J. A., Myllykallio H., Allers T. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLOS genetics. 2007;3(5) doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berquis B. R., DasSarma S. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. Journal of Bacteriology. 2003;185(20):5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes M. L., Dyall-Smith M. L. Mutations in DNA gyrase result in novobiocin resistance in halophilic archaebacteria. Journal of Bacteriology. 1991;173(2):642–648. doi: 10.1128/jb.173.2.642-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam W. L., Doolittle W. F. Mevinolin-resistant mutations identify a promoter and the gene for a eukaryote-like 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the archaebacterium Haloferax volcanii. Journal of Biological Chemistry. 1992;267(9):5829–5834. [PubMed] [Google Scholar]
- 17.Peck R. F., DasSarma S., Krebs M. P. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Molecular Microbiology. 2000;35(3):667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]
- 18.Bitan-Banin G., Ortenberg R., Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. Journal of Bacteriology. 2003;185(3):772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGenity T. J., Gemmell R. T., Grant W. D. Proposal of a new halobacterial genus Natrinema gen. nov., with two species Natrinema pellirubrum nom. nov. and Natrinema pallidum nom. nov. International Journal of Systematic Bacteriology. 1998;48(4):1187–1196. doi: 10.1099/00207713-48-4-1187. [DOI] [PubMed] [Google Scholar]
- 20.Lv J., Wang S., Wang Y., Huang Y., Chen X. Isolation and molecular identification of auxotrophic mutants to develop a genetic manipulation system for the haloarchaeon natrinema sp. J7-2. Archaea. 2015;2015:16. doi: 10.1155/2015/483194.483194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Sima L., Lv J., et al. Identification, characterization, and application of the replicon region of the halophilic temperate sphaerolipovirus SNJ1. Journal of Bacteriology. 2016;198(14):1952–1964. doi: 10.1128/jb.00131-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng J., Liu B., Zhang Z., et al. The complete genome sequence of natrinema sp. J7-2, a haloarchaeon capable of growth on synthetic media without amino acid supplements. PLoS ONE. 2012;7(7) doi: 10.1371/journal.pone.0041621.e41621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Z., Du X., Li T., Gan F., Tang B., Tang X.-F. Functional insight into the C-terminal extension of halolysin SptA from haloarchaeon Natrinema sp. J7. PLoS ONE. 2011;6(8) doi: 10.1371/journal.pone.0023562.e23562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi W., Tang X.-F., Huang Y., Gan F., Tang B., Shen P. An extracellular halophilic protease SptA from a halophilic archaeon Natrinemasp. J7: gene cloning, expression and characterization. Extremophiles. 2006;10(6):599–606. doi: 10.1007/s00792-006-0003-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Wang M., Du X., et al. Chitin accelerates activation of a novel haloarchaeal serine protease that deproteinizes chitin-containing biomass. Applied and Environmental Microbiology. 2014;80(18):5698–5708. doi: 10.1128/AEM.01196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Cui P., Lin L., Shen P., Tang B., Huang Y.-P. Transcriptional analysis of the hsp70 gene in a haloarchaeon Natrinema sp. J7 under heat and cold stress. Extremophiles. 2009;13(4):669–678. doi: 10.1007/s00792-009-0251-5. [DOI] [PubMed] [Google Scholar]
- 27.Chen W., Yang G., He Y., et al. Nucleotides flanking the start codon in hsp70 mRNAs with very short 5’-UTRs greatly affect gene expression in haloarchaea. PLOS ONE. 2015;10(9) doi: 10.1371/journal.pone.0138473.e0138473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Liu Y., Wang S., et al. Temperate membrane-containing halophilic archaeal virus SNJ1 has a circular dsDNA genome identical to that of plasmid pHH205. Virology. 2012;434(2):233–241. doi: 10.1016/j.virol.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 29.Mei Y., He C., Huang Y., et al. Salinity regulation of the interaction of halovirus SNJ1 with its host and alteration of the halovirus replication strategy to adapt to the variable ecosystem. PLoS ONE. 2015;10(4) doi: 10.1371/journal.pone.0123874.e0123874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Wang J., Liu Y., et al. Identification and characterization of SNJ2, the first temperate pleolipovirus integrating into the genome of the SNJ1-lysogenic archaeal strain. Molecular Microbiology. 2015;98(6):1002–1020. doi: 10.1111/mmi.13204. [DOI] [PubMed] [Google Scholar]
- 31.Dyall-Smith M. The Halohandbook: Protocols for Haloarchaeal Genetics. Melbourne, Austrulia: Haloarchaeal Genetics Laboratory; 2008. [Google Scholar]
- 32.Luo H., Zhang C.-T., Gao F. Ori-Finder 2, an integrated tool to predict replication origins in the archaeal genomes. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant C. E., Bailey T. L., Noble W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen P., Chen Y. Plasmid from Halobacterium halobium and its restriction map. Yi Chuan Xue Bao. 1993;21(5):409–416. [PubMed] [Google Scholar]
- 35.Wu Z., Liu H., Liu J., Liu X., Xiang H. Diversity and evolution of multiple orc/cdc6-adjacent replication origins in haloarchaea. BMC Genomics. 2012;13(1, article no. 478) doi: 10.1186/1471-2164-13-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atomi H., Imanaka T., Fukui T. Overview of the genetic tools in the Archaea. Frontiers in Microbiology. 2012;3 doi: 10.3389/fmicb.2012.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farkas J., Picking J., Santangelo T. Genetic techniques for the archaea. Annual Review of Genetics. 2013;47, article 539 doi: 10.1146/annurev-genet-111212-133225. [DOI] [PubMed] [Google Scholar]
- 38.Wu Z., Liu J., Yang H., Xiang H. DNA replication origins in archaea. Frontiers in Microbiology. 2014;5, article no. 179 doi: 10.3389/fmicb.2014.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Peng N., Shah S. A., Huang L., She Q. Archaeal extrachromosomal genetic elements. Microbiology and Molecular Biology Reviews. 2015;79(1):117–152. doi: 10.1128/MMBR.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]