In fibroblasts and myoblasts polarizing for migration, retrograde actin flow moves the nucleus rearward, orienting the centrosome toward the leading edge. The nucleus engages moving dorsal actin cables through linear arrays of nesprin-2G and SUN2 called TAN lines. In this study, Saunders et al. report that the nuclear envelope–localized AAA+ ATPase torsinA and its activator, LAP1, are required for TAN line assembly and retrograde dorsal actin cable flow.
Abstract
The nucleus is positioned toward the rear of most migratory cells. In fibroblasts and myoblasts polarizing for migration, retrograde actin flow moves the nucleus rearward, resulting in the orientation of the centrosome in the direction of migration. In this study, we report that the nuclear envelope–localized AAA+ (ATPase associated with various cellular activities) torsinA (TA) and its activator, the inner nuclear membrane protein lamina-associated polypeptide 1 (LAP1), are required for rearward nuclear movement during centrosome orientation in migrating fibroblasts. Both TA and LAP1 contributed to the assembly of transmembrane actin-associated nuclear (TAN) lines, which couple the nucleus to dorsal perinuclear actin cables undergoing retrograde flow. In addition, TA localized to TAN lines and was necessary for the proper mobility of EGFP-mini–nesprin-2G, a functional TAN line reporter construct, within the nuclear envelope. Furthermore, TA and LAP1 were indispensable for the retrograde flow of dorsal perinuclear actin cables, supporting the recently proposed function for the nucleus in spatially organizing actin flow and cytoplasmic polarity. Collectively, these results identify TA as a key regulator of actin-dependent rearward nuclear movement during centrosome orientation.
Introduction
A hallmark of cell polarity in many migratory cell types is the orientation of the centrosome to a position between the nucleus and leading edge (Luxton and Gundersen, 2011; Gundersen and Worman, 2013). The inability of cells to establish and maintain anterior centrosome orientation results in inefficient migration. Direct imaging of the centrosome in fibroblasts demonstrates that productive protrusion and migration occur only after anterior centrosome orientation is achieved (Gomes et al., 2005). In wounded monolayers of fibroblasts and myoblasts polarizing for migration, anterior centrosome orientation is established by the coordinated positioning of the nucleus and centrosome. During this process, retrograde actin flow moves the nucleus toward the cell rear, whereas the centrosome is maintained at the cell center (Gomes et al., 2005; Chang et al., 2015a). In fibroblasts, the maintenance of the centrosome at the cell center depends on the polarity proteins Par3, Par6, and PKCζ, the minus end–directed microtubule (MT) motor protein dynein, and dynamic MTs (Palazzo et al., 2001; Gomes et al., 2005; Schmoranzer et al., 2009).
In fibroblasts and myoblasts, forces generated by the retrograde flow of dorsal perinuclear actin cables are harnessed by the nucleus through transmembrane actin-associated nuclear (TAN) lines (Luxton et al., 2010; Chang et al., 2015a). TAN lines are composed of the outer nuclear membrane (ONM) and inner nuclear membrane (INM) proteins nesprin-2G (N2G) and SUN2, respectively. N2G is an ∼800-kD spectrin repeat–containing protein, the majority of which extends into the cytosol (Luxton and Starr, 2014). N2G interacts with dorsal perinuclear actin cables through its N-terminal actin-binding calponin homology domains, resulting in the organization of N2G into TAN lines (Zhen et al., 2002; Luxton et al., 2010). The C terminus of N2G contains the conserved KASH (Klarsicht, ANC-1, and Syne homology) domain, which consists of a transmembrane domain (TMD) followed by an ∼20–30-aa KASH peptide that directly interacts with the conserved Sad1/UNC-84 (SUN) domain found at the C terminus of SUN2 (Östlund et al., 2009; Sosa et al., 2013). The N terminus of SUN2 projects into the nucleoplasm and interacts with the nuclear lamina as well as the chromatin (Crisp et al., 2006; Sosa et al., 2013; Tapley and Starr, 2013). The anchorage of TAN lines by A-type lamins is essential for rearward nuclear movement (Folker et al., 2011; Chang et al., 2015a). The fundamental significance of rearward nuclear movement during cell migration is illustrated by the fact that both N2G and SUN2 are required for efficient fibroblast migration (Luxton et al., 2010). Furthermore, N2G depletion inhibits myoblast migration and decreases the efficiency of their fusion into myotubes (Chang et al., 2015a). Despite their importance, the mechanisms of TAN line assembly remain poorly defined.
The KASH–SUN interaction forms an evolutionarily conserved molecular bridge across the perinuclear space, termed the linker of nucleoskeleton and cytoskeleton (LINC) complex (Tapley and Starr, 2013; Chang et al., 2015b). The crystal structure of SUN2 reveals a mushroom-like trimer with a cap composed of SUN domains and a stalk of three coiled-coils (Sosa et al., 2012, 2013; Zhou et al., 2012). Each SUN2 trimer binds three KASH peptides in grooves that are formed between adjacent SUN domains. LINC complexes are typically depicted as being evenly distributed throughout the nuclear envelope (Starr and Fridolfsson, 2010). However, the identification of TAN lines argues that LINC complexes are dynamic within nuclear membranes. This idea is further supported by the observed reorganization of LINC complexes into focal accumulations during meiosis in Caenorhabditis elegans and during nuclear centering in the fission yeast Schizosaccharomyces pombe (King et al., 2008; Sato et al., 2009). Therefore, a molecular mechanism for the regulation of LINC complex dynamics within the nuclear envelope must exist, although currently it is unknown.
Several lines of evidence implicate the conserved ATP-binding protein torsinA (TA) as a key regulator of LINC complex dynamics (Gerace, 2004; Atai et al., 2012; Saunders and Luxton, 2016). TA was originally identified as the protein product encoded by the TOR1A gene, which causes the childhood movement disorder DYT1 dystonia when mutated (Ozelius et al., 1997). The mutation is an in-frame deletion that removes a single glutamic acid (ΔE302/303, or ΔE) that impairs TA function in part by impairing the interaction of TA with its major interacting proteins, LAP1 and luminal domain (LD)–like LAP1 (LULL1; Goodchild and Dauer, 2005; Naismith et al., 2009; Zhao et al., 2013). Although LAP1 is an INM protein (Senior and Gerace, 1988), LULL1 is distributed throughout the ER and, consequently, the ONM (Watson, 1955; Goodchild et al., 2005, 2015).
TA is a member of the AAA+ (ATPase associated with various cellular activities) superfamily, members of which typically function as ring-shaped hexameric molecular chaperones that use energy derived from ATP hydrolysis to remodel protein complexes (Ozelius et al., 1997; Vale, 2000; Hanson and Whiteheart, 2005). Studies of TA exhibiting chaperone-like behaviors both in vitro and in vivo are consistent with the hypothesis that, like AAA+ proteins, TA may act as a molecular chaperone (Burdette et al., 2010; Nery et al., 2011).
Torsin proteins themselves have negligible ATPase activity in vitro (Zhao et al., 2013). However, their activity is stimulated by a direct interaction with the ∼60% identical LD of LAP1 (LAP1LD) or LULL1 (LULL1LD; Goodchild and Dauer, 2005; Zhao et al., 2013; Brown et al., 2014; Sosa et al., 2014). The LAP1LD possesses an atypical AAA+ fold in which R442 is positioned similarly to the arginine finger of canonical AAA+ proteins (Brown et al., 2014; Sosa et al., 2014). Arginine fingers are structural features of AAA+ proteins thought to coordinate nucleotide hydrolysis (Hanson and Whiteheart, 2005). Because TA lacks identifiable arginine fingers, the TA–LAP1 interaction was postulated to result in the formation of an alternating (LAP1-TA)3 heterohexamer, through which the R442 of LAP1 could coordinate ATP hydrolysis with an adjacent TA (Brown et al., 2014; Sosa et al., 2014). This proposed mechanism is supported by the reduced ability of LAP1LD constructs lacking the proposed arginine finger to stimulate the ATPase activity of TA in vitro (Brown et al., 2014; Sosa et al., 2014).
Moreover, TA and the four additional mammalian torsin proteins (torsinB [TB], torsin2A [T2], torsin3A [T3], and torsin4A [T4]) are the only AAA+ proteins known to reside within the ER lumen and contiguous perinuclear space (Goodchild and Dauer, 2004; Naismith et al., 2004; Jungwirth et al., 2010; Kim et al., 2010). Although the functional relevance is unclear, TA is known to interact with TB (Hewett et al., 2004). Among the torsins, T4 is unique because of the presence of a predicted TMD (Rose et al., 2015). Despite these important advances in our understanding of the biochemical mechanism of torsin activity, the protein substrates remodeled by the torsins within the shared ER/perinuclear space remain unknown.
Experimental data suggest that TA has affinity for the KASH domains from nesprin-1, -2, and -3 (Nery et al., 2008), hinting that it might remodel LINC complexes within the nuclear envelope. Studies of the effects of TA inhibition on the architecture of the nuclear envelope provide indirect evidence of TA-mediated LINC complex regulation (Naismith et al., 2004; Goodchild et al., 2005), as both TA and LINC complexes have been implicated in the maintenance of the typical 30–50-nm distance between the INM and ONM (Naismith et al., 2004; Crisp et al., 2006; Cain and Starr, 2015). Furthermore, cells lacking TA redistribute nesprin-3α from the nuclear envelope to the ER, whereas cells with elevated levels of TA exhibit disrupted nuclear envelope localization of N2G, nesprin-3, and SUN2 (Nery et al., 2008; Vander Heyden et al., 2009). Finally, the C. elegans TA homologue OOC-5 is required for the proper localization of the nesprin homologue ZYG-12 in the nuclear envelope of germ cell nuclei (VanGompel et al., 2015). Collectively, these data suggest that TA regulates the LINC complex from within the nuclear envelope.
Although TA has been implicated in centrosome orientation in migrating fibroblasts (Nery et al., 2011, 2014), the mechanism responsible for its role in this process is unclear, as neither centrosomes nor nuclear positioning were analyzed in these studies. Furthermore, the participation of the other torsin proteins, as well as LAP1 and LULL1, in centrosome orientation is unexplored. Therefore, we sought to elucidate the mechanism of TA-mediated centrosome orientation in migrating fibroblasts, hypothesizing that TA is either required for rearward nuclear movement or for centrosome centration during centrosome orientation.
Results
TA is required for rearward nuclear movement during centrosome orientation
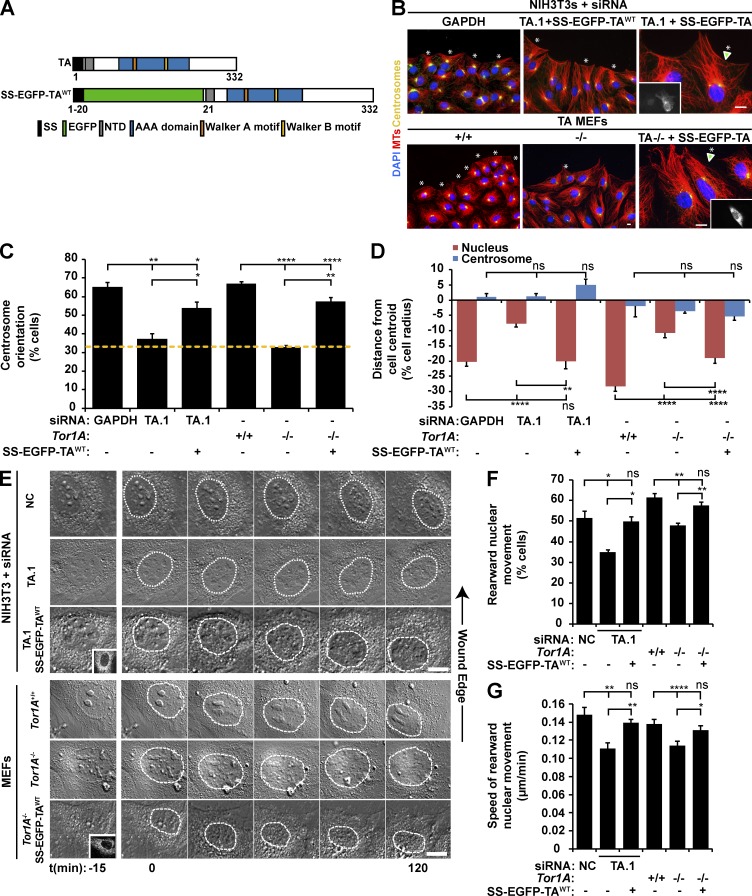
To begin to determine the mechanism of TA-dependent centrosome orientation, we first asked whether acute siRNA-mediated depletion of TA inhibited lysophosphatidic acid (LPA)–stimulated centrosome orientation in serum-starved wound-edge NIH3T3 fibroblasts (Palazzo et al., 2001; Luxton et al., 2010). We tested the ability of four independent siRNAs to efficiently knock down TA mRNA and protein levels relative to control cells treated with previously characterized GAPDH and noncoding (NC) siRNAs (Luxton et al., 2010; Kutscheidt et al., 2014). All four TA-targeting siRNAs significantly depleted TA protein levels (Fig. S1 A). Each of the TA-targeting siRNAs significantly inhibited centrosome orientation and rearward nuclear positioning in NIH3T3 fibroblasts relative to controls without rearward displacement of the centrosome (Fig. 1, B–D; and Fig. S1, B and C). Importantly, the expression of a previously described construct encoding wild-type (WT) TA with EGFP inserted after its signal sequence (SS; SS-EGFP-TAWT; Goodchild and Dauer, 2004) rescued the centrosome orientation and rearward nuclear positioning defects observed in cells treated with siRNA TA.1, indicating that these defects were specifically caused by TA depletion (Fig. 1, A–D). For the remainder of this work, we used siRNA TA.1 to deplete TA. Similar results were obtained after LPA stimulation of serum-starved mouse embryonic fibroblasts (MEFs) from Tor1A−/− mice (Fig. 1, B–D; and Fig. S1 D; Goodchild et al., 2005). Again, both centrosome orientation and rearward nuclear positioning in Tor1A−/− MEFs were rescued by SS-EGFP-TAWT expression (Fig. 1, B–D).
Figure 1.
TA depletion inhibits rearward nuclear movement during centrosome orientation. (A) Diagrams of the human TA protein and SS-EGFP-TAWT. Protein domains were identified using the SMART platform (Schultz et al., 1998). NTD, N-terminal domain (Vander Heyden et al., 2011). (B) Representative epifluorescence images of centrosome orientation in NIH3T3 fibroblasts treated with GAPDH or TA.1 siRNA as well as Tor1A+/+ and Tor1A−/− MEFs. TA-depleted NIH3T3 fibroblasts and Tor1A−/− MEFs expressing SS-EGFP-TAWT are also shown (arrowheads and insets). Asterisks indicate oriented centrosomes. (C) Centrosome orientation in the cells described in B. The dashed yellow line denotes random orientation, which is ∼33% (Palazzo et al., 2001). (D) Mean centrosome and nucleus positions from the cells described in B. The cell center is defined as 0. Positive values are toward the leading edge, and negative values are away. n ≥ 251. (E) Representative montages of DIC images of nuclear movement during centrosome orientation in wound-edge NIH3T3 fibroblasts treated with NC or TA.1 siRNA as well as Tor1A+/+ and Tor1A−/− MEFs. A DIC montage of wound-edge cells expressing SS-EGFP-TAWT (insets) is also provided. Nuclei are outlined by dashed white lines. Time is relative to the addition of LPA (0 min). (F) Percentages of the cells described in E with rearward moving nuclei. (G) Mean velocities of nuclear movement from cells described in E. n ≥ 14. Bars, 10 µm. Error bars show SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant.
To determine whether the nuclear positioning defect was caused by a lack of rearward nuclear movement, we performed live-cell differential interference contrast (DIC) imaging in LPA-stimulated wound-edge TA-depleted and control NIH3T3 fibroblasts. We found that the percentage of cells with rearward-moving nuclei and the speed of rearward nuclear movement were both significantly reduced in cells depleted of TA relative to controls (Fig. 1, E–G). Importantly, the expression of SS-EGFP-TAWT in the TA-depleted cells restored both of these defects to control levels. Also consistent with TA-depleted NIH3T3 fibroblasts, live-cell DIC imaging of Tor1A−/− MEFs confirmed that TA was required for proper rearward nuclear movement because both the percentage of cells with rearward-moving nuclei and the speed of nuclear movement were impaired in Tor1A−/− MEFs relative to Tor1A+/+ MEFs (Fig. 1, E–G). Moreover, both of these defects could be rescued by SS-EGFP-TAWT expression. Collectively, these results demonstrate that TA functions during centrosome orientation to promote rearward nuclear movement.
The redox-regulated ATPase activity of TA is required for rearward nuclear movement during centrosome orientation
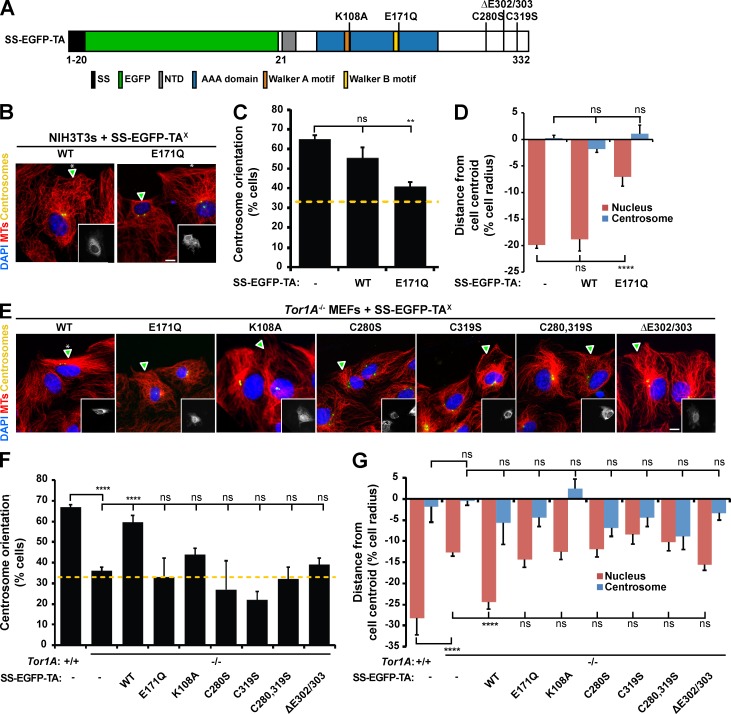
To address the molecular mechanism responsible for TA-dependent rearward nuclear movement and centrosome orientation, we expressed a TA construct encoding a mutation in the conserved Walker B motif (SS-EGFP-TAE171Q; Fig. 2 A; Goodchild and Dauer, 2005) in wound-edge NIH3T3 fibroblasts, which were then stimulated with LPA. TAE171Q does not hydrolyze ATP in the presence of the LD of LAP1 or LULL1 (Zhao et al., 2013), and similar ATP hydrolysis–impaired Walker B mutations are often used as dominant-negative constructs to disrupt AAA+ protein function (Hanson and Whiteheart, 2005). Unlike cells expressing SS-EGFP-TAWT, those expressing SS-EGFP-TAE171Q were significantly reduced in their ability to orient their centrosomes and in rearward nuclear positioning (Fig. 2, B–D). Knowing that SS-EGFP-TAE171Q acts in a dominant-negative fashion to inhibit centrosome orientation, we next asked whether a similar approach could be used to test whether TB, T2, and T3 also functioned during this process. However, expression of SS-EGFP–tagged WT or Walker B mutant constructs of TB, T2, and T3 had no effect on centrosome orientation or rearward nuclear positioning in LPA-stimulated wound-edge cells (Fig. S2). These results further support the requirement for TA-mediated rearward nuclear positioning during centrosome orientation in migrating fibroblasts, and they suggest that the three other luminal torsins are not critically involved in this process.
Figure 2.
The redox-regulated ATPase activity of TA is required for rearward nuclear positioning and centrosome orientation. (A) Diagram indicating the location of the designated mutations in the TA constructs used in this figure. (B) Representative epifluorescence images of centrosome orientation in wound-edge NIH3T3 fibroblasts expressing the indicated construct (arrowheads and insets). (C) Centrosome orientation in the cells described in B. (D) Mean centrosome and nucleus positions in the cells described in B. n ≥ 69. (E) Representative epifluorescence images of centrosome orientation in Tor1A−/− MEFs expressing the indicated SS-EGFP-TAX constructs (arrowheads and inserts). Asterisks indicate oriented centrosomes. (F) Centrosome orientation in the cells described in E. (C and F) Dashed yellow lines denote random centrosome orientation. (G) Mean centrosome and nucleus positions in the cells described in E. n ≥ 115. Bars, 10 µm. Error bars show SEM. **, P < 0.01; ****, P < 0.0001; ns, not significant.
To further test the requirement for the ATPase activity of TA during centrosome orientation, we asked whether expressing SS-EGFP-TAE171Q or SS-EGFP-TAK108A, a construct encoding a mutation within the AAA+ Walker A domain predicted to prevent ATP binding (Goodchild and Dauer, 2004; Hanson and Whiteheart, 2005), could rescue centrosome orientation and rearward nuclear positioning in Tor1A−/− MEFs (Fig. 2, A and E–G). Neither of these constructs nor SS-EGFP-TAΔE, which encodes the ATPase-defective DYT1 dystonia-causing ΔE mutation (Zhao et al., 2013), were able to rescue the centrosome orientation and nuclear positioning defects observed in the Tor1A−/− MEFs (Fig. 2, A and E–G). Next, we tested the requirement for the C-terminal redox-sensitive sensor II motif (RSS), which is composed of two conserved disulfide bond–forming cysteine residues, C280 and C319, and is required for the nucleotide- and partner-binding functions of TA (Zhu et al., 2008, 2010). We mutated each cysteine in the RSS to serine in SS-EGFP-TA either independently (SS-EGFP-TAC280S and SS-EGFP-TAC319S) or together (SS-EGFP-TAC280,319S) and expressed the resulting constructs in Tor1A−/− MEFs (Fig. 3, A and E–G). None of the cysteine mutants rescued centrosome orientation or rearward nuclear positioning (Fig. 3, A and E–G). These results demonstrate that the redox-regulated ATPase activity of TA is required for rearward nuclear positioning during centrosome orientation in wound-edge fibroblasts.
Figure 3.
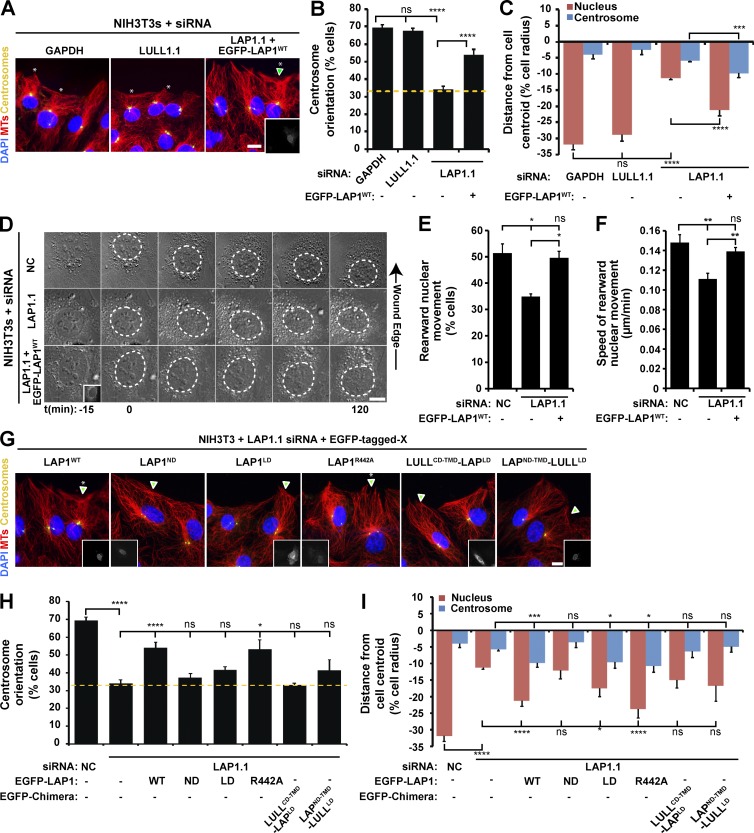
LAP1 is required for rearward nuclear movement during centrosome orientation. (A) Representative epifluorescence images of centrosome orientation in NIH3T3 fibroblasts treated with siRNA. A LAP1-depleted cell expressing EGFP-LAP1WT is shown (arrowhead and inset). (B) Centrosome orientation in the cells described in A. (C) Mean centrosome and nucleus positions in the cells described in B. n ≥ 139. (D) Representative montages of DIC images of nuclear movement during centrosome orientation in NIH3T3 fibroblasts treated with NC or LAP1.1 siRNA. A DIC montage of LAP1-depleted cells expressing EGFP-LAP1WT (inset) is provided. Nuclei are outlined by dashed white lines. (E) Percentages of the cells described in D with rearward-moving nuclei. (F) Mean velocities of nuclear movement from cells described in D. (G) Representative epifluorescence images of centrosome orientation in LAP1-depleted NIH3T3 fibroblasts expressing the indicated EGFP-LAP1X construct (arrowheads and insets). Asterisks indicate oriented centrosomes. (H) Centrosome orientation in the cells described in G. (B and H) Dashed yellow lines denote random centrosome orientation. (I) Mean centrosome and nucleus positions in the cells described in G. n ≥ 11. Bars, 10 µm. Error bars show SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
LAP1 is required for rearward nuclear movement during centrosome orientation
To determine whether LAP1 or LULL1 contributed to rearward nuclear movement during centrosome orientation, we depleted each protein from NIH3T3 fibroblasts with siRNA (Fig. S3, A–C). LAP1 depletion negatively impacted both centrosome orientation and rearward nuclear positioning in cells after LPA stimulation, whereas no significant effect was observed with LULL1 depletion (Fig. 3, A–C). We obtained similar results with independent siRNAs directed against LAP1 and LULL1 (Fig. S3, A–E). Expression of EGFP-tagged LAP1 (EGFP-LAP1WT) in LAP1-depleted cells rescued centrosome orientation and rearward nuclear positioning (Fig. 3, A–C). Live-cell DIC imaging revealed that the percentage of LAP1-depleted cells that displayed rearward nuclear movement and the speed of rearward nuclear movement were both significantly reduced relative to controls (Fig. 3, D–F). Both of these parameters were rescued by EGFP-LAP1WT expression in the LAP1-depleted cells (Fig. 3, D–F). Thus, these results indicate that LAP1, like TA, is required for rearward nuclear movement during centrosome orientation and suggest that LULL1 is largely dispensable for this process.
To begin to explicate the mechanism of LAP1 function during centrosome orientation, we tested the ability of various EGFP-tagged LAP1 constructs to rescue rearward nuclear positioning during centrosome orientation in LAP1-depleted NIH3T3 fibroblasts (Fig. S3 F). Neither the nucleoplasmic domain (ND; EGFP-LAP1ND) nor the LD plus TM domain (EGFP-LAP1LD) of LAP1 rescued centrosome orientation (Fig. 3, G–I). However, EGFP-LAP1LD expression partially rescued rearward nuclear positioning in LAP1-depeleted cells (Fig. 3, G–I). To further explore this result and to test the role of LAP1-mediated stimulation of the ATPase activity of TA during this process, we attempted to rescue LAP1-depleted cells by expressing a LAP1 construct lacking the conserved arginine critical for stimulating the ATPase activity of TA (EGFP-LAP1R442A; Fig 3, G–I; and Fig S3 F; Brown et al., 2014; Sosa et al., 2014). Because EGFP-LAP1R442A expression rescued both centrosome orientation and rearward nuclear positioning in LAP1-depleted cells (Fig. 3, G–I), these results suggest that LAP1 promotes rearward nuclear positioning during centrosome orientation in a manner that is independent of its ability to stimulate the ATPase activity of TA.
As a final test of the differential requirement for LAP1 and LULL1 during centrosome orientation and rearward nuclear positioning, we fused the LULL1LD to the LAP1ND-TMD (EGFP-LAP1ND-TMD-LULL1LD) and attached the LAP1LD to the cytoplasmic domain (CD) of LULL1 (EGFP-LULL1CD-TMD-LAPLD; Fig. S3 F). Similar constructs were previously used to show that the LAP1 ND and LULL1 CD mediate the subcellular localization of LAP1 to the INM and LULL1 to the ONM/ER, respectively (Naismith et al., 2009). Neither chimera rescued centrosome orientation or rearward nuclear positioning in the LAP1-depleted cells (Fig. 3, G–I). Thus, these results show that despite being ∼60% identical (Goodchild and Dauer, 2005), the LAP1LD and LULL1LD are not functionally equivalent. Moreover, these results suggest that the LAP1LD needs to be localized at the INM to function during centrosome orientation.
TA and LAP1 are required for TAN line assembly and persistence
Rearward nuclear movement requires N2G and SUN2 assembly into TAN lines, so we first tested whether TA or LAP1 depletion affected the nuclear envelope localization and/or total protein levels of N2G and SUN2. Quantitative immunofluorescence microscopy revealed that the steady-state levels of the TAN line proteins N2G or other nuclear envelope proteins (lamin A/C, nesprin-3, SUN1) were unchanged in the nuclear envelope of TA- or LAP1-depleted NIH3T3 fibroblasts stimulated with LPA (Fig. S4, A–D). However, a significant increase in the level of SUN2 in the nuclear envelope was observed in LAP1-depleted cells compared with controls (Fig. S4, C and D). In addition, we measured a significant reduction in the nuclear envelope levels of SUN1 and N2G in Tor1A−/− relative to Tor1A+/+ MEFs (Fig. S4, E and F). In contrast, no significant differences were observed in the total protein levels of N2G, nesprin-3, SUN1, and SUN2 in NC-, GAPDH-, TA-, or LAP1-depleted cells (Fig. S4, G–J). Similar results were obtained in Tor1A+/+ versus Tor1A−/− MEFs, though there was a significant decrease in N2G and SUN1 as well as a slight increase in SUN2 levels in the absence of TA (Fig. S4, K and L). Collectively, these results indicate that the role of TA and LAP1 during rearward nuclear movement might be separate from its role in regulating the levels or localization of LINC complex components.
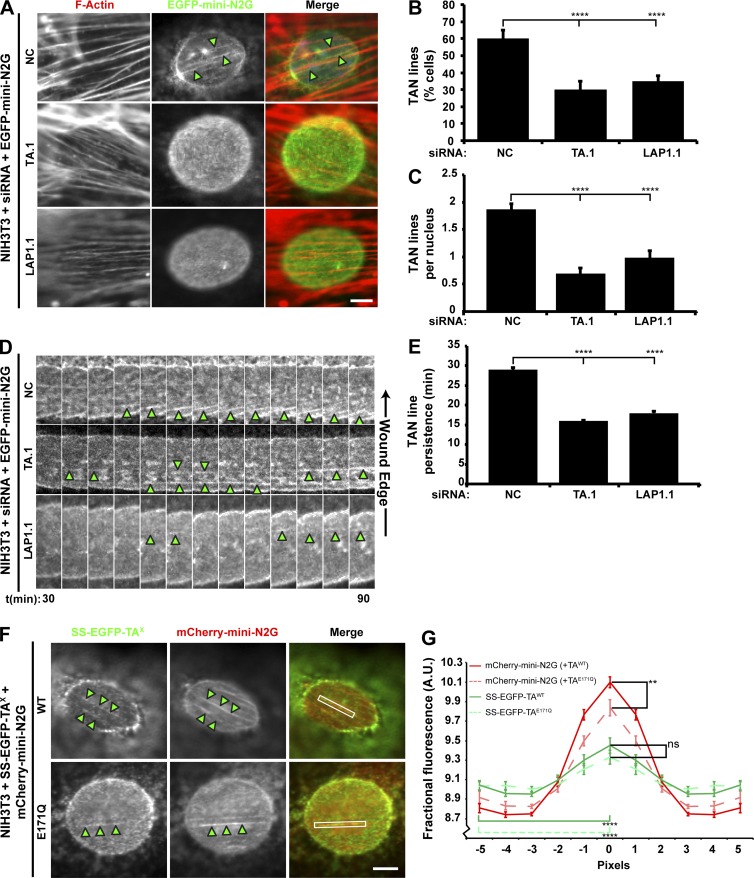
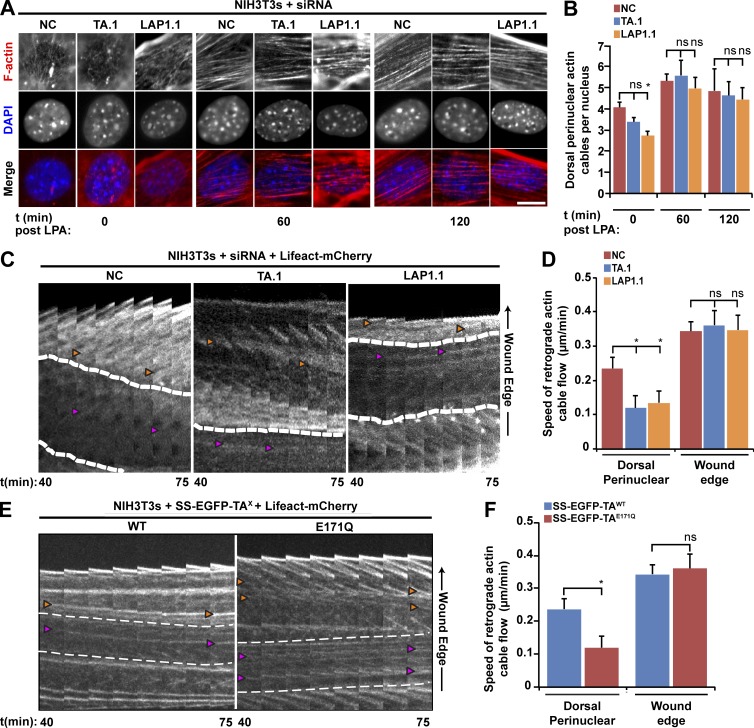
We next examined the effect of TA and LAP1 depletion on the ability of wound-edge NIH3T3 fibroblasts to assemble TAN lines. We expressed the TAN line marker, EGFP–mini-N2G in TA-depleted or control cells stimulated with LPA. Linear arrays of EGFP–mini-N2G were considered TAN lines if they colocalized with dorsal perinuclear actin cables (Fig. 4 A; Luxton et al., 2010). Significantly fewer TA- and LAP1-depleted cells exhibited TAN lines as compared with control cells (Fig. 4, A and B). The number of TAN lines observed per cell was also reduced when either TA or LAP1 was depleted (Fig. 4 C). In addition, live-cell imaging revealed that the persistence of the TAN lines was significantly reduced in TA- and LAP1-depleted cells relative to controls (Fig. 4, D and E). This impaired persistence was not caused by the inability of SUN2 to properly localize to linear arrays of mini-N2G in the absence of TA function because the percentage of TAN lines containing endogenous SUN2 was unaffected by TA depletion or the expression of SS-EGFP-TAE171Q (Fig. S5). Thus, these results demonstrate that both TA and LAP1 are required for the assembly of stable TAN lines.
Figure 4.
The assembly of stable TAN lines requires both TA and LAP1. (A) Representative epifluorescence images of nuclei in wound-edge NIH3T3 fibroblasts treated with siRNA and expressing EGFP–mini-N2G taken 1 h after LPA stimulation. Cells were stained for F-actin (red) and EGFP (green). (B) The mean number of cells with TAN lines quantified from the cells described in A. n ≥ 150. (C) The mean number of TAN lines per nucleus quantified from the cells described in A. (D) Representative kymographs of EGFP–mini-N2G fluorescence in wound-edge NIH3T3 fibroblasts treated with siRNA. Each panel is 0.2 µm. Arrowheads indicate TAN lines. Time after LPA addition is in minutes. (E) The mean TAN line persistence quantified from the experiments described in D. n ≥ 13. (F) Representative epifluorescence images of mCherry–mini-N2G TAN lines in NIH3T3 fibroblasts also expressing the indicated SS-EGFP-TAX construct. White boxes indicate the area used for quantifying TAN line localization in G. (A and F) Arrowheads indicate TAN lines. Bars, 5 µm. (G) Quantification of SS-EGFP-TAX recruitment to TAN lines. Error bars show SEM. **, P < 0.01; ****, P < 0.0001; ns, not significant. A.U., arbitrary units.
To determine whether TAN lines themselves may be sites of TA function, we asked whether SS-EGFP-TAE171Q preferentially localized to TAN lines in comparison with SS-EGFP-TAWT because similar ATP hydrolysis–defective Walker B motif mutations are often used to trap the interaction between AAA+ proteins and their substrates (Hanson and Whiteheart, 2005). Quantification of the localization of either construct to mCherry–mini-N2G TAN lines in serum-starved wound-edge cells stimulated with LPA demonstrated a significant enrichment of TAWT and TAE171Q on TAN lines relative to an adjacent TAN line–free area on the nuclear envelope (Fig. 4, F and G). In addition, we found that the presence of SS-EGFP-TAE171Q resulted in a significant reduction in the amount of mCherry–mini-N2G signal measured in TAN lines as compared with TAN lines with SS-EGFP-TAWT (Fig. 4 G). These results imply that TAN lines may contain substrates of TA, the remodeling of which is necessary for TAN line assembly and persistence.
TA is required for the dynamics of N2G and SUN2 within the nuclear envelope
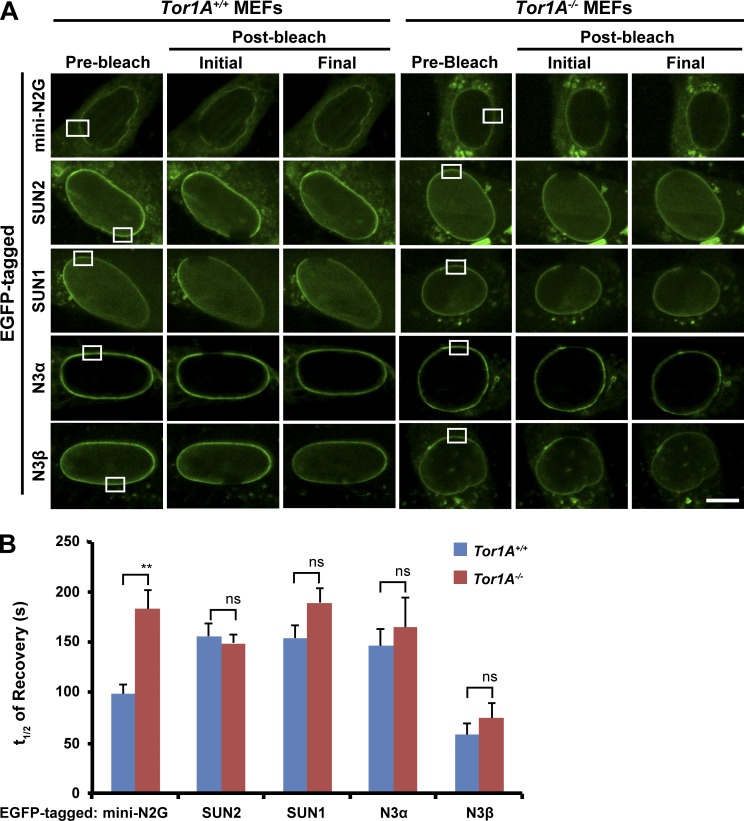
To assess how TA might contribute to TAN line assembly and persistence, we performed FRAP to measure the mobility and dynamics of previously described constructs encoding the EGFP-tagged LINC complex components mini-N2G, nesprin-3α, nesprin-3β, SUN1, and SUN2 within the nuclear envelope of Tor1A+/+ and Tor1A−/− MEFs (Fig. 5 A; Wilhelmsen et al., 2005; Östlund et al., 2009; Luxton et al., 2010). Of the five constructs tested, only EGFP–mini-N2G exhibited a difference in the absence of TA. Specifically, the t1/2 of EGFP–mini-N2G recovery after photobleaching was significantly increased in Tor1A−/− MEFs relative to controls (Fig. 5 B). Consequently, these results suggest that TA is required for the proper mobility of EGFP-N2G within the nuclear envelope.
Figure 5.
TA affects the dynamics of EGFP–mini-N2G in the nuclear envelope. (A) Representative confocal fluorescence images of Tor1A+/+ and Tor1A−/− MEFs expressing the indicated construct and subjected to FRAP. White boxes indicate bleached regions of interest. Bar, 10 µm. (B) t1/2 of recovery quantification from the experiments described in A. n ≥ 3. Error bars show SEM. **, P < 0.01; ns, not significant. N3α/β, nesprin-3α/β.
TA and LAP1 are required for the retrograde flow of dorsal perinuclear actin cables
Because the reorganization of N2G and SUN2 into TAN lines requires the formation of dorsal perinuclear actin cables, which do not require either N2G or SUN2 (Luxton et al., 2010; Folker et al., 2011), we tested the effect of TA depletion on the assembly of these cables in LPA-stimulated NIH3T3 fibroblasts. We did not detect a significant difference in the number of dorsal perinuclear actin cables in TA-depleted or control cells at 0, 60, or 120 min after LPA stimulation (Fig. 6, A and B). Although similar results were obtained in LAP1-depleted cells, they exhibited a significant reduction in cable numbers at the 0-min time point relative to control cells. Thus, neither TA nor LAP1 is required for the assembly of the actin cables responsible for rearward nuclear movement.
Figure 6.
The retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement requires TA and LAP1. (A) Representative epifluorescence images of nuclei in NIH3T3 fibroblasts treated with the indicated siRNAs taken at 0, 60, and 120 min after LPA treatment. Bar, 10 µm. (B) Quantification of the number of dorsal perinuclear actin cables from the cells described in A. n ≥ 3. (C) Kymographs of retrograde actin cable flow in wound-edge NIH3T3 fibroblasts treated with the indicated siRNA and expressing Lifeact-mCherry. (D) Speed of actin cable retrograde flow quantified from the cells described in C. n ≥ 15. (E) Kymographs of retrograde actin flow in NIH3T3 fibroblasts expressing the indicated construct. Time after LPA addition is in mins. (C and E) Each panel is 0.2 µm. Time after LPA addition is in minutes. Dashed lines represent edges of the nucleus. Orange arrowheads indicate actin cables at the leading edge, and pink arrowheads indicate dorsal perinuclear actin cables. (F) Speed of actin cable retrograde flow determined from the cells described in E. n ≥ 10. Error bars are SEM. *, P < 0.05; ns, not significant.
We then tested the ability of dorsal perinuclear actin cables in TA- or LAP1-depleted NIH3T3 fibroblasts to undergo retrograde flow by monitoring actin cables with the live-cell actin probe Lifeact-mCherry (Riedl et al., 2008). The speed of retrograde flow of the dorsal perinuclear actin cables was reduced by >50% in TA- and LAP1-depleted cells relative to control cells, whereas retrograde flow of wound-edge actin cables was unaffected (Fig. 6, C and D). Because overexpression of SS-EGFP-TAE171Q inhibited rearward nuclear positioning in NIH3T3 fibroblasts, we tested the effect of its expression on retrograde actin flow in these cells. Similar to what we observed in TA- or LAP1-depleted cells, the retrograde flow of dorsal perinuclear actin cables was inhibited by the overexpression of SS-EGFP-TAE171Q, whereas expression of SS-EGFP-TAWT had no effect (Fig. 6, E and F). Therefore, TA and LAP1 are also required for the retrograde flow of dorsal perinuclear actin cables during centrosome orientation.
Discussion
We show that rearward nuclear movement during centrosome orientation requires TA and its activator, LAP1. Collectively, TA and LAP1 promote TAN line assembly and the retrograde flow of dorsal perinuclear actin cables, both of which are critical for rearward nuclear movement (Fig. 7). We also demonstrate that TA is necessary for the proper accumulation of EGFP–mini-N2G in TAN lines, perhaps because without TA, the mobility of EGFP–mini-N2G within the nuclear envelope is reduced. Thus, our results establish TA as a key regulator of LINC complex–dependent nuclear–cytoskeletal coupling and actin retrograde flow.
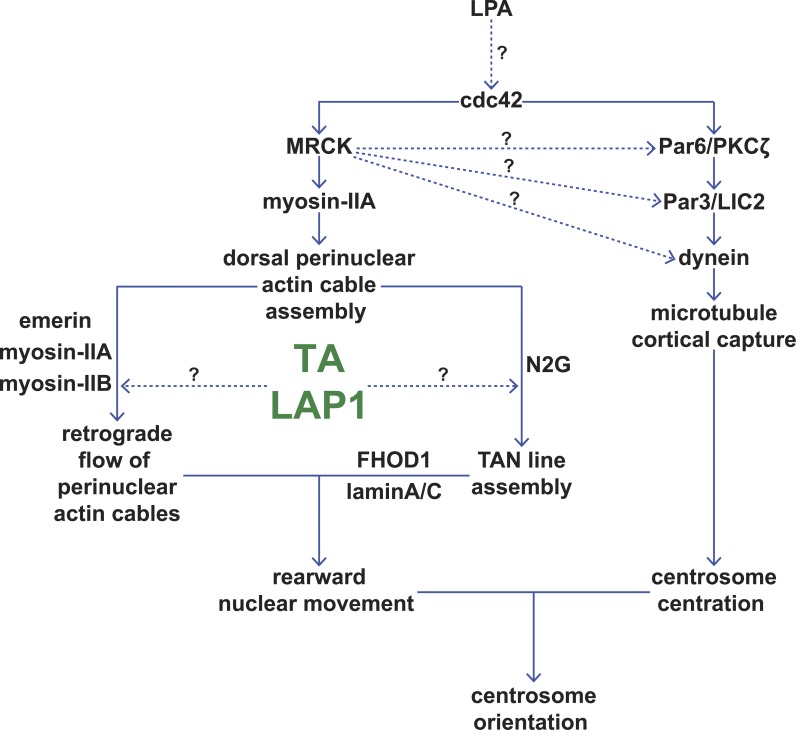
Figure 7.
TA and LAP1 are critical regulators of rearward nuclear movement. Model displaying the potential positions of TA and LAP1 within the two pathways known to contribute to proper centrosome orientation (Gomes et al., 2005; Schmoranzer et al., 2009; Luxton et al., 2010). MRCK is sufficient to stimulate centrosome orientation and so may also regulate the centrosome centration pathway (dotted arrows; Gomes et al., 2005).
Functional specification of torsin proteins
Although we cannot formally conclude that the other luminal torsins we tested are not required for centrosome orientation, our data suggest that TA plays a more significant role than TB, T2, or T3 during this process. This functional specificity for TA is unexpected, as TA and TB are known to function redundantly to maintain normal nuclear membrane morphology in nonneuronal cells (Kim et al., 2010). In addition, TB, T2, and T3 all interact with LAP1 (Hewett et al., 2004; Jungwirth et al., 2010; Kim et al., 2010). However, it is highly likely that different torsins perform specialized cellular functions as indicated by the fact that ATP hydrolysis–defective, substrate-trap TB, T2, and T3 mutants accumulate to varying degrees within the perinuclear space (Hewett et al., 2004; Jungwirth et al., 2010; Kim et al., 2010). Moreover, the ATPase activity of these torsins is differentially stimulated by LAP1LD or LULL1LD in vitro (Zhao et al., 2013). The significant, rearward displacement of the centrosome that we observed in NIH3T3 fibroblasts after the overexpression of SS-EGFP–tagged TBWT, TBE178Q, T2WT, and T3WT may also hint at functional specificity between torsin proteins. We hypothesize that TB, T2, or T3 may act to control MT dynamics, which are critical for proper centrosome positioning in migrating fibroblasts (Gomes et al., 2005; Schmoranzer et al., 2009). Specifically, these torsins may regulate a growing list of known MT- or MT motor–associated membrane proteins that traverse the ER/ONM (Gurel et al., 2014).
TA-LAP1 holoenzyme-mediated rearward nuclear movement
We show that the redox-regulated ATPase activity of TA is required for rearward nuclear movement during centrosome orientation. We also demonstrate that mutations in TA (ΔE and RSS) that impair LAP1 or LULL1 binding (Naismith et al., 2009; Zhu et al., 2010; Zhao et al., 2013) are unable to rescue rearward nuclear movement in Tor1A−/− MEFs. The small amount of centrosome orientation we observed after expression of SS-EGFP-TAE171Q in NIH3T3 fibroblasts versus the inability of this construct to rescue centrosome orientation in Tor1A−/− MEFs above background levels may be explained by the fact that AAA+ proteins typically act as ring-shaped hexamers (Hanson and Whiteheart, 2005). Therefore, the presence of TAWT in NIH3T3 fibroblasts could compensate for the ATP hydrolysis–defective E171Q mutation depending on the ratio of TAWT to TAE171Q in a single ring. However, expression of SS-EGFP-TAE171Q in the Tor1A−/− would result in all TA rings being solely composed of ATP hydrolysis–defective TA and hence nonfunctional.
In addition, we provide evidence for the lack of functional redundancy between LAP1 and LULL1 during rearward nuclear movement despite the ability of both of their LDs to stimulate the ATPase activity of TA in vitro (Zhao et al., 2013). The requirement for LAP1 during rearward nuclear movement necessitated that its LD be tethered at the INM. Unexpectedly, we found that the critical arginine finger in the LAP1LD (R442) was dispensable for LAP1 function during rearward nuclear movement. However, it is important to note that the effect of the arginine finger on the ability of the LAP1LD to stimulate TA’s ATPase activity in vitro varied greatly between two independent studies (Brown et al., 2014; Sosa et al., 2014). Because these measurements were performed in the absence of a TA substrate, they may not accurately reflect TA activity in cells.
Our in vivo results concerning the critical nature of this conserved LAP1 arginine imply that LAP1 may perform other functions in addition to its ability to stimulate TA ATPase activity. It is possible that the stimulation of TA by the LAP1LD is not important for rearward nuclear movement. Unidentified substrates and/or an additional activator within the perinuclear space may stimulate TA within this subcellular compartment, which would be consistent with the inability of either SS-EGFP-TAE171Q or SS-EGFP-TAK108A to rescue rearward nuclear positioning or centrosome orientation in Tor1A−/− MEFs. Another possible explanation for our results would be that EGFP-LAP1R442A retains enough activity to stimulate ATP hydrolysis by TA in LAP1-depleted NIH3T3 fibroblasts. This would be in line with the modest effect of mutating the conserved arginine to alanine, as reported by Sosa et al. (2014) in their in vitro experiments. Recently, a high-resolution crystal structure of TA in a complex with the LULL1LD was solved, which will provide a critical structural framework for future experiments designed to further test the structure–function relationship between TA and LAP1 (Demircioglu et al., 2016).
TA and TAN line assembly
TA has been proposed to be a regulator of LINC complex assembly and function (Gerace, 2004; Atai et al., 2012; Saunders and Luxton, 2016). However, the mechanism underlying this proposed function remains unclear, aside from studies of an interaction between TA and the KASH domains from nesprins-1, -2, and -3 (Nery et al., 2008) as well as the sensitivity of N2G, nesprin-3, and SUN2 nuclear envelope localization to the levels of TA (Nery et al., 2008; Vander Heyden et al., 2009). Although we detected a significant reduction in the nuclear envelope localization of SUN1 and N2G, we did not detect altered levels of nesprin-3 or SUN2 in the complete absence of TA. Furthermore, we were unable to detect any changes in either nuclear envelope localization or the levels of these proteins after TA or LAP1 depletion, with the exception of elevated nuclear envelope SUN2 levels after LAP1 depletion. Our inability to detect a significant reduction in the levels of N2G and SUN1 in TA-depleted NIH3T3 fibroblasts may result from an incomplete knockdown, consistent with TA being a very stable protein (Giles et al., 2008). Because centrosome orientation and rearward nuclear movement were both inhibited in the Tor1A−/− MEFs as well as in TA- or LAP1-depleted NIH3T3 fibroblasts, we do not believe that the changes in N2G, SUN1, or SUN2 levels reflect a general function for TA in controlling the stability or localization of these proteins. Instead, we propose that TA regulates the mobility of N2G by acting as a molecular chaperone to structurally remodel N2G-containing protein complexes within the nuclear envelope.
This hypothesis is supported by our results demonstrating that TA and LAP1 are required for the assembly of stable TAN lines, which are linear arrays of LINC complexes composed of N2G and SUN2 (Luxton et al., 2010, 2011). In addition, the localization of TA to TAN lines further strengthens the connection between TA and N2G-containing protein complexes and raises the possibility that TA may act directly in the assembly of TAN lines. Although we did measure a significant enrichment of TA levels in TAN lines as compared with adjacent TAN line–free regions of the nuclear envelope, we did not see a preferential accumulation of TAE171Q relative to TAWT in TAN lines. Instead, we found a slight but significant decrease in the amount of TAE171Q present in TAN lines as compared with TAWT. This result indicates that the ability of TA to localize to TAN lines may be somewhat sensitive to its nucleotide state and that TAN lines themselves may not be substrates for TA. Nevertheless, the presence of TAE171Q in TAN lines did result in a significant reduction of the amount of mini-N2G in these structures relative to the mini-N2G levels measured in the presence of TAWT.
We offer the following potential explanation for the effect of TAE171Q on mini-N2G levels in TAN lines. TAE171Q may preferentially interact with and structurally remodel a previously unidentified TA substrate important for controlling the mobility of N2G within the nuclear envelope and hence the assembly of stable TAN lines. We predict that this substrate is either N2G itself, which would be consistent with the previous study of an interaction between TA and the KASH domains from nesprin-1, -2, and -3 (Nery et al., 2008), or another N2G-interacting protein. Supporting evidence for this hypothesis is provided by our FRAP experiments, which revealed a significantly increased t1/2 of recovery for EGFP–mini-N2G in the absence of TA, suggesting that TA is needed for the proper assembly dynamics of N2G-containing LINC complexes required for TAN line assembly.
Previously, we showed that the KASH peptide is necessary and sufficient to immobilize EGFP–mini-N2G in the nuclear envelope of MEFs (Östlund et al., 2009). In addition, we demonstrated that SUN2 depletion significantly decreases the t1/2 of EGFP–mini-N2G recovery, whereas SUN1 depletion had no effect (Östlund et al., 2009). Although the total levels of N2G, SUN1, and SUN2 were significantly altered in the Tor1A−/− MEFs, we do not believe that these alterations are responsible for the increased t1/2 of EGFP–mini-N2G that we measured in these cells. For example, if reduced N2G levels were to create free binding sites within the perinuclear space for EGFP–mini-N2G, then this would apply to all nesprin proteins, given the promiscuous ability of KASH peptides to interact with SUN1 and SUN2 (Stewart-Hutchinson et al., 2008). A similar argument could apply to the elevated SUN2 levels we observed in the Tor1A−/− MEFs, as this would also increase the number of potential nesprin-binding sites. Our inability to detect significant differences in EGFP–nesprin-3α/β mobility in the Tor1A−/− MEFs, which also interact with SUN1 and SUN2 (Ketema et al., 2007), strongly suggests that TA selectively regulates the mobility of N2G within the perinuclear space. Furthermore, the lack of change in EGFP-SUN1 or EGFP-SUN2 mobility demonstrates that the absence of TA does not influence the number of EGFP–mini-N2G–binding sites, despite elevated SUN2 and decreased SUN1 levels.
Based on these results, we postulate two potential direct mechanisms for how TA controls TAN line assembly. In the first, TA may promote the turnover of N2G-containing LINC complexes by dissociating the luminal N2G KASH peptide from SUN2. In the second, the previously studied TA–KASH domain interaction (Nery et al., 2008) may preclude SUN protein binding, which would contribute to N2G mobility within the nuclear envelope. It is important to note that in order for either of these hypotheses to be feasible, TA would have to act from a position near the ONM. However, using our EGFP-tagged LAP1/LULL1 chimeric constructs, we show that the LAP1LD needs to be anchored at the INM via the LAP1ND-TMD to rescue rearward nuclear positioning and centrosome orientation in LAP1-depleted NIH3T3 fibroblasts. Because the distance between the ONM and INM is ∼30–50 nm (Naismith et al., 2004; Crisp et al., 2006; Cain and Starr, 2015), this would necessitate TA being able to dissociate from the inner leaflet of the INM and then diffuse to the ONM, where it could access the N2G KASH domain and/or the SUN domain of SUN2. Interestingly, a small population of TA has been shown to be soluble within the ER lumen/perinuclear space in addition to a large membrane-associated population of TA using cellular fractionation (Vander Heyden et al., 2009, 2011). Furthermore, TA was recently shown to undergo a proteolytic processing event during B cell activation, resulting in the cleavage of its hydrophobic N terminus, which mediates the monotopic association of TA with the inner leaflet of the ER/nuclear envelope membrane (Zhao et al., 2016). We hypothesize that in addition to stimulating the ATPase activity of TA, the interaction between TA and the LAP1LD or the LULL1LD may serve to regulate the membrane association of TA, thereby allowing it access to the ONM.
Alternatively, TA may indirectly influence TAN line assembly by its recently described role in regulating lipid metabolism (Grillet et al., 2016). For example, changes in the lipid composition of the nuclear envelope could potentially explain the reduced mobility of EGFP–mini-N2G that we observed in the Tor1A−/− MEFs. However, such a global alteration in the biophysical nature of the nuclear envelope would most likely cause pleiotropic effects on the mobility of multiple nuclear envelope membrane proteins. Although we did not find alterations in the mobility of the five other nuclear envelope membrane proteins we measured here, it remains a formal possibility that TA regulates the metabolism of lipids that are specifically involved in N2G mobility. Clearly, future in vitro biochemical experiments are necessary to differentiate between these direct and indirect potential mechanisms of TA-mediated regulation of N2G.
TA is a novel regulator of dorsal perinuclear retrograde actin flow
Previously, TA had been implicated in the regulation of the actin cytoskeleton (Hewett et al., 2006; Muraro and Moffat, 2006). Our finding that TA is required for the retrograde flow of dorsal perinuclear actin cables and not their assembly extends these results and offers two new insights: (1) TA controls the actin cytoskeleton from within the perinuclear space, and/or (2) retrograde actin flow at different subcellular locations is subject to differential regulation. To control retrograde actin flow from its luminal residence, TA must interact with and regulate a transmembrane ER or ONM protein involved in the regulation of the actin cytoskeleton. LAP1 is a strong candidate because our results show that it is also required for this process. Recently, LAP1ND was reported to directly interact with the ND of emerin, which is primarily concentrated in the INM but also found at the ONM (Salpingidou et al., 2007; Chang et al., 2013; Shin et al., 2013). Emerin interacts with actin and nonmuscle myosin IIB within the cytoplasm and is also required for rearward nuclear movement during centrosome orientation in migrating fibroblasts (Chang et al., 2013). Future efforts toward mapping the emerin-binding site or sites in LAP1ND will enable the role of the LAP1–emerin interaction during centrosome orientation to be addressed. Alternatively, TA may control the actin cytoskeleton through a different LAP1-binding partner or a non-LAP1 nuclear membrane protein. Analysis of the recently published LAP1 interactome (Serrano et al., 2016), together with proteomics-based approaches to identify novel LAP1- and TA-interacting proteins, should help lead to the identification of potential TA substrates required for rearward nuclear movement during centrosome orientation.
TA-mediated cell polarity and DYT1 dystonia
In summary, our data support the hypothesis that TA plays a critical evolutionarily conserved role in the establishment and/or maintenance of cell polarity (Basham and Rose, 1999; Nery et al., 2008). When considered together with the recent study of fibroblasts derived from DYT1 dystonia patients being defective in centrosome orientation and directional migration (Nery et al., 2014), our results highlight the possible connection between defective cell polarity and DYT1 dystonia. Although DYT1 dystonia is caused by neuronal dysfunction (Liang et al., 2014), fibroblasts may provide an experimental model for investigating the molecular and cellular mechanisms underlying TA-dependent cell polarity. Ultimately, a better understanding of these mechanisms will guide the rational design of potential therapeutics that may be used to combat DYT1 dystonia pathogenesis.
Materials and methods
Antibodies
β-Catenin mouse mAb and rabbit polyclonal antibody were purchased from Invitrogen. GAPDH mouse mAb was from Sigma-Aldrich. TA rabbit polyclonal antibody was from Abcam. EGFP mouse mAb and dsRed rabbit polyclonal antibody were purchased from Sigma-Aldrich and Takara Bio Inc., respectively. Lamin A/C (MANLAC1) mouse mAb was a gift from G.E. Morris (The Wolfson Centre for Inherited Neuromuscular Disease, Shropshire, England, UK). LAP1 and LULL1 rabbit polyclonal antibodies were previously described (Goodchild and Dauer, 2005). Pericentrin mouse mAb and rabbit polyclonal antibody were purchased from Covance or BD. The N2G rabbit polyclonal antibody was previously described (Luxton et al., 2010), as was the nesprin-3 rabbit polyclonal antibody (Morgan et al., 2011), which was a gift from D. Starr (University of California, Davis, Davis, CA). Tyrosinated α-tubulin rat mAb (YL1/2) was collected in house from a supernatant generated by hybridomas purchased from the European Collection of Animal Cell Cultures. SUN1 and SUN2 rabbit polyclonal antibodies were purchased from Abcam. Vinculin mouse mAb was from Sigma-Aldrich. Secondary antibodies were from three different sources. From Jackson ImmunoResearch Laboratories, Inc., we purchased goat anti–mouse secondary antibodies conjugated to Alexa Fluor 488 or 650 as well as rhodamine. We purchased goat anti–rat and anti–rabbit secondary antibodies conjugated to Dylight 488, 561, or 649 from Thermo Fischer Scientific. IRDye 650 nm–conjugated goat anti–mouse secondary antibodies were from LI-COR Biosciences.
Cell culture and monolayer wounding
NIH3T3 fibroblasts were cultured in DMEM with 10% bovine calf serum purchased from Thermo Fischer Scientific. Fibroblasts were serum starved for 2 d, wounded, and stimulated with 10 µM LPA as previously described (Palazzo et al., 2001; Gomes et al., 2005). MEFs were grown in DMEM with 15% bovine calf serum, serum starved for 3 d, wounded, and stimulated with 10 µM LPA.
DNA constructs
The Lifeact-mCherry plasmid was a gift from R. Wedlich-Soldner (Max Planck Institute of Biochemistry, Martinsried, Germany; Riedl et al., 2008). The previously described EGFP-N3α and -N3β constructs (Wilhelmsen et al., 2005) were gifts from A. Sonnenberg (Netherlands Cancer Institute, Amsterdam, Netherlands). We have previously described the SS-EGFP-TAWT, -TAK108A, -TAE171Q, -TAΔE302/303, -TBWT, -TBE178Q, -T2WT, -T2E162Q, -T3WT, -T3E236Q, and –mini-N2G constructs (Goodchild and Dauer, 2004; Kim et al., 2010; Luxton et al., 2010).
The following constructs were generated during the course of this investigation using the primers described in Table S1. All constructs were confirmed by sequencing performed at the University of Minnesota Genomics Center. The following mutations were introduced into SS-EGFP-TAWT: C280S, C319S, and C280,319S using QuikChange (Agilent Technologies) following the manufacturer’s instructions. SS-EGFP-TAC280S was created using the TAC280S-forward and TAC280S-reverse primer pair, whereas the primer pair TAC319S-forward and TAC319S-reverse was used to create SS-EGFP-TAC319S. SS-EGFP-TAC280,319S was generated using the TAC319S-forward and TAC319S-reverse primer pair to mutagenize SS-EGFP-TAC280S. To make the EGFP-LAP1WT construct, we used PCR to amplify EGFP from pEGFP-N1 using the primers EGFP-LAP1WT-forward and EGFP-LAP1WT-reverse, both of which contain KpnI cut sites. The resulting PCR product was then purified and digested alongside the previously described myc-LAP1 construct (Goodchild and Dauer, 2005) with RE1 and RE2. The digested PCR product and plasmid were then purified and ligated to make EGFP-LAP1WT. EGFP-LAP1R442A was created via kinase-ligase-DpnI treatment, where 2 µl of PCR product, amplified using the primer pair EGFP-LAP1R442A-forward and EGFP-LAP1R442A-reverse, was treated with T4 ligase, T4 polynucleotide kinase, and DpnI in T4 ligase buffer in a 20-µl reaction for 20 min at room temperature.
The chimeric EGFP-LAP1ND-TMD-LULL1LD and EGFP-LULL1CD-TMD-LAP1LD constructs were generated as follows. The first step toward creating EGFP-LAP1ND-TMD-LULL1LD was to amplify the ND through the TMD of LAP1 by PCR, using the primer pair LAP1ND-TMD-forward and LAP1ND-TMD-reverse. After purification, the PCR product was used in combination with the LAP1LD-reverse primer to amplify the LULL1LD by PCR from a previously described myc-LULL1 construct (Goodchild and Dauer, 2005). The resulting PCR product was then purified, digested with HindIII and BamHI, and cloned into pEGFP-C1, which was then digested with the same restriction enzymes. A similar protocol was followed to generate EGFP-LULL1CD-TMD-LAP1LD. However, the first PCR product was produced using the primer pair LULL1CD-TMD-forward and LULL1CD-TMD-reverse and myc-LULL1 as a template. The second PCR product was then generated using the first PCR product and LAP1LD-reverse as primers and myc-LAP1 as a template. After purification, the second PCR product was digested with HindIII and BamHI and cloned into pEGFP-C1, which was digested with the same restriction enzymes.
DNA microinjections
Plasmid DNA was purified using the Plasmid Midi kit (QIAGEN) and was microinjected into nuclei at concentrations between 5 and 30 µg/ml as previously described (Gomes et al., 2005). Injections were performed on a TS100 microscope (Nikon) equipped with a chrome-free infinity Apochromat long working distance apodized dark low 20×/NA 0.4 objective (3-mm working distance) and a Narishige NT-88 manipulator set (Nikon).
Fixed-cell fluorescence imaging
All fixed-cell imaging was performed on an Eclipse Ni-E microscope driven by NIS-Elements software using an oil immersion 40×/1.30 NA Plan Fluor Eco-Glass oil immersion objective lens (0.20-mm working distance; Nikon), a SOLA solid state white-light excitation subsystem (Lumencor), and a CoolSNAP ES2 12-bit 20-MHz digital monochrome charge-coupled device camera (Photometrics). A custom DAPI filter for the SOLA light source was used, which consisted of an ET 395/25× excitation filter and an ET 460/50-m emission filter purchased from Chroma Technology Corp. EGFP (C-FL EGFP hard coat high signal to noise Zero Shift), Texas Red (C-FL Texas Red hard coat high signal to noise Zero Shift), and Cy5 (C-FL CY5 hard coat high signal to noise Zero Shift; Nikon) filter sets were also used.
FRAP
FRAP was performed with an LSM 510 META scanning confocal microscope equipped with a 30-mW argon 488-nm laser and a 40×/1.3 NA Plan-Apochromat objective (ZEISS) as previously described (Östlund et al., 2009). In brief, a region of interest was selected and photobleached for 25 iterations at 100% laser power, after which recovery of fluorescence was monitored with 2-s intervals at 5% laser power. ImageJ (National Institutes of Health) was then used to quantify the mean intensity of the fluorescence within the region of interest, which was normalized relative to the change in total fluorescence using the formula, Irel = T0It/Ttl0, where T0 is the total cellular intensity during the prebleach, Tt is the total cellular intensity at time point t, I0 is the mean intensity in the bleached area during prebleach, and It is the mean intensity in the region of interest at time point t. The normalized fluorescence was then plotted against the time after bleaching.
Because the immobile fraction (the difference between the fluorescence intensity in the bleached area prebleach and the intensity at infinity after bleach) differed between the different EGFP-tagged constructs, we used a modified t1/2 value, where t1/2 is the time after bleach required for the fluorescence levels to reach the median between levels immediately after bleach and prebleach, rather than using the median between prebleach levels and steady-state levels. To determine t1/2, a modification of the method described by Harrington et al. (2002) was used. We plotted ln(1 − it) versus time after bleach, where it is the mean normalized fluorescence intensity in the bleach region at time t and 1 is the mean normalized fluorescence intensity in the bleach region prebleach. The curves were fitted using KaleidaGraph (Synergy Software), and t1/2 was calculated as t1/2 = ln2 × (−1/slope). Data from the first 31 s after bleach were used in all experiments. The t1/2 of recovery was calculated from at least three independent experiments.
Immunofluorescence
Cells grown on no. 1.5 coverslips were fixed in either −20°C methanol or room temperature 4% paraformaldehyde as previously described (Palazzo et al., 2001; Gomes et al., 2005). Coverslips were mounted on slides using Fluoromount purchased from Thermo Fisher Scientific.
Live-cell DIC and fluorescence imaging
NIH3T3 fibroblasts or MEFs were grown on 35-mm dishes with glass coverslip bottoms (0.16–0.19 mm; no. 1.5) purchased from In Vitro Scientific. Confluent monolayers were serum starved and wounded as described in the Cell culture and monolayer wounding section. Cells were then washed twice with live imaging media (GIBCO HBSS [purchased from Invitrogen] containing essential and nonessential MEM amino acids purchased from Thermo Fisher Scientific), 2.5 g/liter glucose, 2 mM glutamine, 1 mM sodium pyruvate, and 20 mM Hepes, pH 7.4) and transferred to the 37°C full-enclosure incubator (Okolab USA Inc.) with temperature control attached to a Marianas 200 Microscopy Workstation (Intelligent Imaging Innovations) built on an AxioObserver Z.1 stand (ZEISS) and driven by SlideBook 6.0 software (Intelligent Imaging Innovations). After selecting the regions of the coverslip to be imaged, the cells were stimulated with 20 µM LPA. DIC images were captured with a CoolSNAP HQ2 charge-coupled device camera. Fluorescence confocal imaging was performed with a 5,000-rpm CSU-X1 M1 spinning-disk confocal head (Yokogawa Electric Corporation) and an Evolve electron-multiplying charge-coupled device camera (Photometrics). A LaserStack laser launch containing two 50-mW solid-state lasers (488 nm and 561 nm; Intelligent Imaging Innovations) was used as a light source for the live-cell fluorescence imaging. A CSUX filter wheel (Yokogawa Electric Corporation) containing Semrock filters for 405-, 488-, 561-, or 640-nm excitation and with a quad emitter and individual emitters was used as the filter set. All live-cell imaging was performed using a Plan-Apochromat 63×/1.4 NA oil objective (ZEISS) containing DIC III prisms.
Quantification of dorsal perinuclear actin cables
LPA- and serum-stimulated wound-edge NIH3T3 fibroblasts that had been stained for F-actin and nuclei were used to assess the effect of siRNA-mediated TA and LAP1 depletion on dorsal perinuclear actin cables. Dorsal actin cables above the nucleus were manually counted from single-plane images taken of F-actin and nuclei, counting only those actin cables that passed over the nucleus.
Quantification of nuclear envelope protein levels
11 z-section epifluorescence images of LPA-stimulated wound-edge fibroblasts stained for DNA, MTs, and a nuclear envelope protein were obtained with a 300-nm step size. After acquiring the images, the “select best planes” function from NIS-Elements software was used to select a single z section from each imaged area of cells, using the MT channel as the guide for selecting the in-focus section. The selected images were then exported as 16-bit greyscale TIFF files and imported into custom-made MATLAB software (MathWorks) for analysis. This software applies a mask to the DAPI-stained DNA channel using the “canny edge detection” algorithm. The user then identifies the nuclei on the wound edge to be analyzed, and the program integrates the intensity of the nuclear envelope protein channel within the DAPI mask and then divides that intensity by the area of the mask.
Quantification of rearward nuclear movement
Multipage TIFF files were obtained for each cell imaged using DIC microscopy, with a single z section (or maximum intensity projection) and time along the third axis of each image. TIFF files were then imported into custom MATLAB software for quantifying nuclear movement. Using this software, the user defined the wound edge and then manually traced an outline of the nucleus in the cell of interest at each time step. Next, the “region props” function was used, which calculates the centroid, area, eccentricity, and absolute angle of the major axis of the traced object. Using Excel (Microsoft), the coordinates of our wound clicks were then rotated so that each cell analyzed was parallel to the x axis, providing a similar point of reference for each nucleus. Once rotated, the changes in nuclear centroid position over time were used to determine overall speed (displacement) during the 2-h time course. Nuclei that moved ≥10% of the cell radius toward the cell rear were defined as rearward-moving nuclei. Experiments were repeated at least twice.
Quantification of nucleus and centrosome position
Fluorescence images of cells stained for centrosomes (antipericentrin), cell–cell contacts (anti–β-catenin), DNA (DAPI), expression tag, and MTs (anti-Tyr–α-tubulin) were acquired as previously described (Palazzo et al., 2001; Gomes et al., 2005). Centrosome orientation was determined as previously described (Palazzo et al., 2001), and experiments were repeated up to three times. Analysis of the nucleus and centrosome position was performed as previously described (Gomes et al., 2005). In brief, images were pseudocolored, combined, and aligned so that the wound edge was parallel to the x axis using MetaMorph software (Molecular Devices). Cell perimeters were drawn over the cell–cell contacts (β-catenin) and the wound edge (Tyr-tubulin or injection markers). From this information, the cell centroid and equivalent radius were calculated using the “integrated morphometry analysis” program in MetaMorph. The MetaMorph “measure pixel” function was used to identify the position of the centrosome (pericentrin) and the approximate centroid of the nucleus. A vector representing the distances from the nuclear centroid and the centrosome to the cell centroid was drawn and resolved into x and y coordinates (parallel and perpendicular to the leading edge, respectively). Measurements were normalized to cell size to allow for a comparison between cells. Only the y coordinate was used in plots, as the x coordinate (position of the nuclear centroid or centrosome along the x axis) did not change significantly. The difference between the cell centroid and the nucleus centroid or centrosome was then divided by the radius to determine the percentage of the cell radius either organelle traveled.
Quantification of retrograde actin flow
Serum-starved wound-edge NIH3T3 fibroblasts treated with either NC or TA-targeting siRNAs were microinjected with the Lifeact-mCherry construct (20 ng/µl) and incubated for 1.5–2.0 h at 37°C to allow for expression. Cells were then washed with recording media and moved to the heated stage of our Marianas 200 Microscopy Workstation (described in the Live-cell DIC and fluorescence imaging section). Both Lifeact-mCherry expression and nuclear position (DIC) were imaged every 5 min for 2.0 h shortly after the addition of 20 µM LPA to allow tracking of both nuclear movement and F-actin. The resulting images were then rotated so that each wound edge was parallel to the x axis to keep the point of reference similar between cells. Nuclear tracking using the DIC channel was completed as described in the Quantification of rearward nuclear movement section. To track F-actin over the nucleus, we isolated a single plane on the dorsal side of the nucleus and tracked the y position of the center of individual actin cables between 35 and 90 min after LPA stimulation. To track F-actin in the cytoplasm, we created maximum-intensity projections of the cells and identified a region of the cell that was ∼2 µm directly rearward from the center of the wound edge, where we tracked individual cables between 35 and 90 min after LPA stimulation. Experiments were repeated three times.
Quantification of TAN line localization
After acquisition as described in the Fixed-cell fluorescence imaging section, epifluorescence images were opened in ImageJ FIJI. A maximum-intensity projection of the image planes containing the top half of the nuclear envelope for each channel was created. Using mCherry–mini-N2G lines as a guide, the image was rotated without interpolation so that the TAN line was parallel to the x axis. The rotation was then propagated to the TA channel. An 11-pixel-tall box was then drawn to cover the length of the TAN line, with the central pixel centered on the TAN line. The mean intensity for each row of pixels was then calculated for each channel, and then each mean intensity was expressed as a percentage of the sum intensity of the whole measured region. The regions were then mirrored by taking the means of the 1st and the 11th, the 2nd and 10th, etc., pixel. This process was repeated for all TAN lines. Fractional fluorescence of each pixel row was then calculated, and the mean and SEM were plotted as a function of the distance from the center of the TAN lines. n ≥ 35 cells for each construct.
Quantification of TAN line persistence
Serum-starved wound-edge NIH3T3 fibroblasts treated with either NC or TA-targeting siRNAs were microinjected with the EGFP–mini-N2G construct (5.5 ng/µl) and incubated for 45 min at 37°C to allow for expression. Cells were then washed with live-imaging media and moved to the heated stage of our Marianas 200 Microscopy Workstation (described in the Live-cell DIC and fluorescence imaging section). 19 z-section confocal stacks with a 0.4-µm step size of expressing cells were collected every 5 min for 2 h using a 63×/1.4 NA Plan-Apochromat DIC objective after stimulation with 20 µM LPA. After the acquisition time course was completed, videos were inspected in the 3D time-lapse view within SlideBook 6.0 to monitor the EGFP–mini-N2G signal on the dorsal nuclear surface. A maximum-intensity projection of the dorsal half of the nucleus was then generated using SlideBook 6.0 for the time course, in which TAN line persistence was analyzed by tracking individual TAN lines (linear EGFP–mini-N2G signals ≥2 µm in length) over time using ImageJ. Experiments were repeated three times.
Quantification of total protein levels by Western blotting
Whole-cell lysate samples were prepared by lysing cells with nonreducing sample buffer. Protein concentration was determined, and 15 µg protein was loaded onto protein gels (Novex; Thermo Fisher Scientific). Samples were subjected to SDS-PAGE and then transferred to nitrocellulose and probed with the indicated antibodies. Blots were imaged, and bands were quantified using the Odyssey Imaging System and Image Studio software (LI-COR Biosciences). All experiments were repeated three times.
Reagents
F-actin was stained with rhodamine-phalloidin, which was purchased from Cytoskeleton, Inc. DAPI was purchased from Thermo Fisher Scientific. LPA was purchased from Avanti Polar Lipids, Inc. Restriction enzymes were purchased from either New England Biolabs, Inc. or Promega. The QuikChange kit was purchased from Agilent Technologies. Phusion DNA polymerase, T4 DNA ligase, and T4 polynucleotide kinase were also purchased from New England Biolabs, Inc. All other chemicals were from Sigma-Aldrich unless otherwise specified. The Wizard SV Gel and PCR Clean-Up system was from Promega. The GeneJet Plasmid Midiprep kit was from Thermo Fisher Scientific.
RNAi
All RNAi experiments were performed using 50-nM siRNA duplexes purchased from Shanghai GenePharma, which were transfected using Lipofectamine RNAiMAX purchased from Thermo Fisher Scientific according to the manufacturer’s instructions. To identify functional siRNA duplexes, four sequences were selected using the artificial neural network algorithm BIOPREDsi (Huesken et al., 2005). The duplexes used in this study were the most effective at depleting the target protein based on Western blotting or RT-PCR analysis. The sequences of the duplexes used are described in Table S2. The GAPDH and N2G siRNA sequences were previously described (Luxton et al., 2010).
Statistics
Two-tailed t tests were used to calculate p-values (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) throughout the manuscript. The error bars presented in bar graphs throughout the manuscript show SEM.
Online supplemental material
Fig. S1 shows the validation of torsin protein depletion from NIH3T3 fibroblasts by various siRNAs. Fig. S2 shows the testing of the roles of TB, T2, and T3 during rearward nuclear positioning and centrosome orientation in NIH3T3 fibroblasts. Fig. S3 shows the validation of LAP1 and LULL1 depletion from NIH3T3 fibroblasts by various siRNAs. Fig. S4 shows quantification of the nuclear and biochemical levels of TAN line components in TA- and LAP1-depleted NIH3T3 fibroblasts as well as T1A−/− MEFs. Fig. S5 shows quantification of endogenous SUN2 recruitment to TAN lines in NIH3T3 fibroblasts with impaired TA function. Table S1 shows the primers used to generate constructs used in this paper. Table S2 shows the sequences of siRNA duplexes used in this paper.
Supplementary Material
Acknowledgments
We thank members of the Dauer, Gundersen, Luxton, and Worman laboratories for helpful discussions as well as M. Li for excellent technical assistance. Special acknowledgments go to B. Lauring and E. Gomes for initiating this work.
This work was funded in part by the National Institutes of Health (grants AR068636-01 to G.G. Gunderson and H.J. Worman; GM094481 to G.G. Gunderson; R01NS07773 to W.T. Dauer; AR57220, R21NS095109-01, and R42DA037622-03 to G.W.G. Luxton; and AR007612 to N.J. Harris and C.A. Saunders) and the Dystonia Medical Research Foundation (to G.G. Gunderson and G.W.G. Luxton).
The authors declare no competing financial interests.
Author contributions: C.A. Saunders and N.J. Harris designed experiments, acquired data, and analyzed results as well as contributed to the generation of figures and the writing of the manuscript. P.T. Willey, B.M. Woolums, Y. Wang, A.J. McQuown, and A. Schoenhofen acquired data and analyzed results. H.J. Worman, W.T. Dauer, and G.G. Gundersen designed experiments, helped analyze results, and edited the manuscript. G.W.G. Luxton contributed to all aspects of experimental design, data acquisition, data analysis, and figure generation as well as manuscript writing and editing.
Footnotes
Abbreviations used:
- AAA+
- ATPase associated with various cellular activities
- CD
- cytoplasmic domain
- DIC
- differential interference contrast
- INM
- inner nuclear membrane
- KASH
- Klarsicht, ANC-1, and Syne homology
- LD
- luminal domain
- LINC
- linker of nucleoskeleton and cytoskeleton
- LPA
- lysophosphatidic acid
- MEF
- mouse embryonic fibroblast
- MT
- microtubule
- NC
- noncoding
- ND
- nucleoplasmic domain
- ONM
- outer nuclear membrane
- RSS
- redox-sensitive sensor II motif
- SS
- signal sequence
- SUN
- Sad1/UNC-84
- TAN
- transmembrane actin-associated nuclear
- TMD
- transmembrane domain
- WT
- wild type
References
- Atai N.A., Ryan S.D., Kothary R., Breakefield X.O., and Nery F.C.. 2012. Untethering the nuclear envelope and cytoskeleton: biologically distinct dystonias arising from a common cellular dysfunction. Int. J. Cell Biol. 2012:634214 10.1155/2012/634214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basham S.E., and Rose L.S.. 1999. Mutations in ooc-5 and ooc-3 disrupt oocyte formation and the reestablishment of asymmetric PAR protein localization in two-cell Caenorhabditis elegans embryos. Dev. Biol. 215:253–263. 10.1006/dbio.1999.9447 [DOI] [PubMed] [Google Scholar]
- Brown R.S., Zhao C., Chase A.R., Wang J., and Schlieker C.. 2014. The mechanism of Torsin ATPase activation. Proc. Natl. Acad. Sci. USA. 111:E4822–E4831. 10.1073/pnas.1415271111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette A.J., Churchill P.F., Caldwell G.A., and Caldwell K.A.. 2010. The early-onset torsion dystonia-associated protein, torsinA, displays molecular chaperone activity in vitro. Cell Stress Chaperones. 15:605–617. 10.1007/s12192-010-0173-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain N.E., and Starr D.A.. 2015. SUN proteins and nuclear envelope spacing. Nucleus. 6:2–7. 10.4161/19491034.2014.990857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Folker E.S., Worman H.J., and Gundersen G.G.. 2013. Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol. Biol. Cell. 24:3869–3880. 10.1091/mbc.E13-06-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Antoku S., Östlund C., Worman H.J., and Gundersen G.G.. 2015a Linker of nucleoskeleton and cytoskeleton (LINC) complex-mediated actin-dependent nuclear positioning orients centrosomes in migrating myoblasts. Nucleus. 6:77–88. 10.1080/19491034.2015.1004947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Worman H.J., and Gundersen G.G.. 2015b Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 208:11–22. 10.1083/jcb.201409047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., and Hodzic D.. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53. 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demircioglu F.E., Sosa B.A., Ingram J., Ploegh H.L., and Schwartz T.U.. 2016. Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia. eLife. 5:e17983 10.7554/eLife.17983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E.S., Ostlund C., Luxton G.W., Worman H.J., and Gundersen G.G.. 2011. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc. Natl. Acad. Sci. USA. 108:131–136. 10.1073/pnas.1000824108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L. 2004. TorsinA and torsion dystonia: Unraveling the architecture of the nuclear envelope. Proc. Natl. Acad. Sci. USA. 101:8839–8840. 10.1073/pnas.0402441101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles L.M., Chen J., Li L., and Chin L.S.. 2008. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum. Mol. Genet. 17:2712–2722. 10.1093/hmg/ddn173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., and Gundersen G.G.. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463. 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Goodchild R.E., and Dauer W.T.. 2004. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc. Natl. Acad. Sci. USA. 101:847–852. 10.1073/pnas.0304375101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild R.E., and Dauer W.T.. 2005. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell Biol. 168:855–862. 10.1083/jcb.200411026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild R.E., Kim C.E., and Dauer W.T.. 2005. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 48:923–932. 10.1016/j.neuron.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Goodchild R.E., Buchwalter A.L., Naismith T.V., Holbrook K., Billion K., Dauer W.T., Liang C.C., Dear M.L., and Hanson P.I.. 2015. Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J. Cell Sci. 128:2854–2865. 10.1242/jcs.167452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet M., Dominguez Gonzalez B., Sicart A., Pöttler M., Cascalho A., Billion K., Hernandez Diaz S., Swerts J., Naismith T.V., Gounko N.V., et al. . 2016. Torsins are essential regulators of cellular lipid metabolism. Dev. Cell. 38:235–247. 10.1016/j.devcel.2016.06.017 [DOI] [PubMed] [Google Scholar]
- Gundersen G.G., and Worman H.J.. 2013. Nuclear positioning. Cell. 152:1376–1389. 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel P.S., Hatch A.L., and Higgs H.N.. 2014. Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr. Biol. 24:R660–R672. 10.1016/j.cub.2014.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P.I., and Whiteheart S.W.. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529. 10.1038/nrm1684 [DOI] [PubMed] [Google Scholar]
- Harrington K.S., Javed A., Drissi H., McNeil S., Lian J.B., Stein J.L., Van Wijnen A.J., Wang Y.-L., and Stein G.S.. 2002. Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically associate with stationary subnuclear domains. J. Cell Sci. 115:4167–4176. 10.1242/jcs.00095 [DOI] [PubMed] [Google Scholar]
- Hewett J.W., Kamm C., Boston H., Beauchamp R., Naismith T., Ozelius L., Hanson P.I., Breakefield X.O., and Ramesh V.. 2004. TorsinB--perinuclear location and association with torsinA. J. Neurochem. 89:1186–1194. 10.1111/j.1471-4159.2004.02404.x [DOI] [PubMed] [Google Scholar]
- Hewett J.W., Zeng J., Niland B.P., Bragg D.C., and Breakefield X.O.. 2006. Dystonia-causing mutant torsinA inhibits cell adhesion and neurite extension through interference with cytoskeletal dynamics. Neurobiol. Dis. 22:98–111. 10.1016/j.nbd.2005.10.012 [DOI] [PubMed] [Google Scholar]
- Huesken D., Lange J., Mickanin C., Weiler J., Asselbergs F., Warner J., Meloon B., Engel S., Rosenberg A., Cohen D., et al. . 2005. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 23:995–1001. 10.1038/nbt1118 [DOI] [PubMed] [Google Scholar]
- Jungwirth M., Dear M.L., Brown P., Holbrook K., and Goodchild R.. 2010. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum. Mol. Genet. 19:888–900. 10.1093/hmg/ddp557 [DOI] [PubMed] [Google Scholar]
- Ketema M., Wilhelmsen K., Kuikman I., Janssen H., Hodzic D., and Sonnenberg A.. 2007. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J. Cell Sci. 120:3384–3394. 10.1242/jcs.014191 [DOI] [PubMed] [Google Scholar]
- Kim C.E., Perez A., Perkins G., Ellisman M.H., and Dauer W.T.. 2010. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci. USA. 107:9861–9866. 10.1073/pnas.0912877107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Drivas T.G., and Blobel G.. 2008. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 134:427–438. 10.1016/j.cell.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutscheidt S., Zhu R., Antoku S., Luxton G.W., Stagljar I., Fackler O.T., and Gundersen G.G.. 2014. FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat. Cell Biol. 16:708–715. 10.1038/ncb2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C.C., Tanabe L.M., Jou S., Chi F., and Dauer W.T.. 2014. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J. Clin. Invest. 124:3080–3092. 10.1172/JCI72830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., and Gundersen G.G.. 2011. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr. Opin. Cell Biol. 23:579–588. 10.1016/j.ceb.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., and Starr D.A.. 2014. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr. Opin. Cell Biol. 28:69–75. 10.1016/j.ceb.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., Gomes E.R., Folker E.S., Vintinner E., and Gundersen G.G.. 2010. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 329:956–959. 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., Gomes E.R., Folker E.S., Worman H.J., and Gundersen G.G.. 2011. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2:173–181. 10.4161/nucl.2.3.16243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J.T., Pfeiffer E.R., Thirkill T.L., Kumar P., Peng G., Fridolfsson H.N., Douglas G.C., Starr D.A., and Barakat A.I.. 2011. Nesprin-3 regulates endothelial cell morphology, perinuclear cytoskeletal architecture, and flow-induced polarization. Mol. Biol. Cell. 22:4324–4334. 10.1091/mbc.E11-04-0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro N.I., and Moffat K.G.. 2006. Down-regulation of torp4a, encoding the Drosophila homologue of torsinA, results in increased neuronal degeneration. J. Neurobiol. 66:1338–1353. 10.1002/neu.20313 [DOI] [PubMed] [Google Scholar]
- Naismith T.V., Heuser J.E., Breakefield X.O., and Hanson P.I.. 2004. TorsinA in the nuclear envelope. Proc. Natl. Acad. Sci. USA. 101:7612–7617. 10.1073/pnas.0308760101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith T.V., Dalal S., and Hanson P.I.. 2009. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J. Biol. Chem. 284:27866–27874. 10.1074/jbc.M109.020164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery F.C., Zeng J., Niland B.P., Hewett J., Farley J., Irimia D., Li Y., Wiche G., Sonnenberg A., and Breakefield X.O.. 2008. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J. Cell Sci. 121:3476–3486. 10.1242/jcs.029454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery F.C., Armata I.A., Farley J.E., Cho J.A., Yaqub U., Chen P., da Hora C.C., Wang Q., Tagaya M., Klein C., et al. . 2011. TorsinA participates in endoplasmic reticulum-associated degradation. Nat. Commun. 2:393 10.1038/ncomms1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery F.C., da Hora C.C., Atai N.A., Kim E.Y., Hettich J., Mempel T.R., Breakefield X.O., and Irimia D.. 2014. Microfluidic platform to evaluate migration of cells from patients with DYT1 dystonia. J. Neurosci. Methods. 232:181–188. 10.1016/j.jneumeth.2014.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östlund C., Folker E.S., Choi J.C., Gomes E.R., Gundersen G.G., and Worman H.J.. 2009. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 122:4099–4108. 10.1242/jcs.057075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozelius L.J., Hewett J.W., Page C.E., Bressman S.B., Kramer P.L., Shalish C., de Leon D., Brin M.F., Raymond D., Corey D.P., et al. . 1997. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 17:40–48. 10.1038/ng0997-40 [DOI] [PubMed] [Google Scholar]
- Palazzo A.F., Joseph H.L., Chen Y.J., Dujardin D.L., Alberts A.S., Pfister K.K., Vallee R.B., and Gundersen G.G.. 2001. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr. Biol. 11:1536–1541. 10.1016/S0960-9822(01)00475-4 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. . 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A.E., Brown R.S., and Schlieker C.. 2015. Torsins: not your typical AAA+ ATPases. Crit. Rev. Biochem. Mol. Biol. 50:532–549. 10.3109/10409238.2015.1091804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpingidou G., Smertenko A., Hausmanowa-Petrucewicz I., Hussey P.J., and Hutchison C.J.. 2007. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 178:897–904. 10.1083/jcb.200702026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., and Dernburg A.F.. 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 139:907–919. 10.1016/j.cell.2009.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C.A., and Luxton G.W.G.. 2016. LINCing defective nuclear-cytoskeletal coupling and DYT1 dystonia. Cell. Mol. Bioeng. 9:207–216. 10.1007/s12195-016-0432-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J., Fawcett J.P., Segura M., Tan S., Vallee R.B., Pawson T., and Gundersen G.G.. 2009. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr. Biol. 19:1065–1074. 10.1016/j.cub.2009.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., and Ponting C.P.. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 95:5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A., and Gerace L.. 1988. Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J. Cell Biol. 107:2029–2036. 10.1083/jcb.107.6.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano J.B., da Cruz E Silva O.A., and Rebelo S.. 2016. Lamina Associated Polypeptide 1 (LAP1) interactome and its functional features. Membranes (Basel). 6:E8 10.3390/membranes6010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.Y., Méndez-López I., Wang Y., Hays A.P., Tanji K., Lefkowitch J.H., Schulze P.C., Worman H.J., and Dauer W.T.. 2013. Lamina-associated polypeptide-1 interacts with the muscular dystrophy protein emerin and is essential for skeletal muscle maintenance. Dev. Cell. 26:591–603. 10.1016/j.devcel.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B.A., Rothballer A., Kutay U., and Schwartz T.U.. 2012. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 149:1035–1047. 10.1016/j.cell.2012.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B.A., Kutay U., and Schwartz T.U.. 2013. Structural insights into LINC complexes. Curr. Opin. Struct. Biol. 23:285–291. 10.1016/j.sbi.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B.A., Demircioglu F.E., Chen J.Z., Ingram J., Ploegh H.L., and Schwartz T.U.. 2014. How lamina-associated polypeptide 1 (LAP1) activates Torsin. eLife. 3:e03239 10.7554/eLife.03239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., and Fridolfsson H.N.. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26:421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson P.J., Hale C.M., Wirtz D., and Hodzic D.. 2008. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res. 314:1892–1905. 10.1016/j.yexcr.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley E.C., and Starr D.A.. 2013. Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr. Opin. Cell Biol. 25:57–62. 10.1016/j.ceb.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D. 2000. AAA proteins. Lords of the ring. J. Cell Biol. 150:F13–F19. 10.1083/jcb.150.1.F13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heyden A.B., Naismith T.V., Snapp E.L., Hodzic D., and Hanson P.I.. 2009. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell. 20:2661–2672. 10.1091/mbc.E09-01-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heyden A.B., Naismith T.V., Snapp E.L., and Hanson P.I.. 2011. Static retention of the lumenal monotopic membrane protein torsinA in the endoplasmic reticulum. EMBO J. 30:3217–3231. 10.1038/emboj.2011.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGompel M.J., Nguyen K.C., Hall D.H., Dauer W.T., and Rose L.S.. 2015. A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol. Biol. Cell. 26:1752–1763. 10.1091/mbc.E14-07-1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.L. 1955. The nuclear envelope; its structure and relation to cytoplasmic membranes. J. Biophys. Biochem. Cytol. 1:257–270. 10.1083/jcb.1.3.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., and Sonnenberg A.. 2005. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171:799–810. 10.1083/jcb.200506083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Brown R.S., Chase A.R., Eisele M.R., and Schlieker C.. 2013. Regulation of Torsin ATPases by LAP1 and LULL1. Proc. Natl. Acad. Sci. USA. 110:E1545–E1554. 10.1073/pnas.1300676110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Brown R.S., Tang C.H., Hu C.C., and Schlieker C.. 2016. Site-specific proteolysis mobilizes TorsinA from the membrane of the endoplasmic reticulum (ER) in response to ER stress and B cell stimulation. J. Biol. Chem. 291:9469–9481. 10.1074/jbc.M115.709337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y.Y., Libotte T., Munck M., Noegel A.A., and Korenbaum E.. 2002. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115:3207–3222. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Du X., Cai Z., Song X., Zhang H., Mizuno T., Suzuki E., Yee M.R., Berezov A., Murali R., et al. . 2012. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J. Biol. Chem. 287:5317–5326. 10.1074/jbc.M111.304543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Wrabl J.O., Hayashi A.P., Rose L.S., and Thomas P.J.. 2008. The torsin-family AAA+ protein OOC-5 contains a critical disulfide adjacent to Sensor-II that couples redox state to nucleotide binding. Mol. Biol. Cell. 19:3599–3612. 10.1091/mbc.E08-01-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Millen L., Mendoza J.L., and Thomas P.J.. 2010. A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J. Biol. Chem. 285:37271–37280. 10.1074/jbc.M110.123471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.