VDAC2 controls the stable localization of BAK to mitochondria and its ability to mediate mitochondrial outer membrane permeabilization. Hosoi et al. now report that BAK shifts from mitochondria to peroxisomes under VDAC2-deficient conditions, giving rise to the mislocalization of peroxisomal matrix proteins such as catalase, which suggests that BAK can also regulate the permeability of peroxisomal membranes.
Abstract
Peroxisomal biogenesis disorders (PBDs) are fatal genetic diseases consisting of 14 complementation groups (CGs). We previously isolated a peroxisome-deficient Chinese hamster ovary cell mutant, ZP114, which belongs to none of these CGs. Using a functional screening strategy, VDAC2 was identified as rescuing the peroxisomal deficiency of ZP114 where VDAC2 expression was not detected. Interestingly, knockdown of BAK or overexpression of the BAK inhibitors BCL-XL and MCL-1 restored peroxisomal biogenesis in ZP114 cells. Although VDAC2 is not localized to the peroxisome, loss of VDAC2 shifts the localization of BAK from mitochondria to peroxisomes, resulting in peroxisomal deficiency. Introduction of peroxisome-targeted BAK harboring the Pex26p transmembrane region into wild-type cells resulted in the release of peroxisomal matrix proteins to cytosol. Moreover, overexpression of BAK activators PUMA and BIM permeabilized peroxisomes in a BAK-dependent manner. Collectively, these findings suggest that BAK plays a role in peroxisomal permeability, similar to mitochondrial outer membrane permeabilization.
Introduction
Peroxisomes are single membrane–bound organelles that participate in many metabolic pathways, including β oxidation of fatty acids (Wanders and Waterham, 2006). Many metabolic pathways of peroxisomes lead to the production of hydrogen peroxide, which is subsequently decomposed by catalase (Titorenko and Terlecky, 2011). Peroxisomal functions are highlighted by the existence of fatal human genetic peroxisomal biogenesis disorders (PBDs) such as Zellweger syndrome. Genetic heterogeneity comprising 14 complementation groups (CGs) is found in PBDs (Matsumoto et al., 2003; Steinberg et al., 2006; Ebberink et al., 2012). To date, all of the 14 genes responsible for PBDs (called peroxin genes or PEX) have been identified (Matsumoto et al., 2003; Ebberink et al., 2012; Fujiki et al., 2014).
Much progress has been made in the identification and characterization of peroxins by genetic and biochemical analysis with peroxisome-deficient mutants of yeast and mammalian cells (Fujiki et al., 2006; van der Klei and Veenhuis, 2006). According to recent models for mammalian peroxisome biogenesis, peroxisomal matrix proteins harboring peroxisome targeting signal types 1 and 2 (PTS1/2) are recognized and imported by their receptors, Pex5p and Pex7p, respectively (Otera et al., 2002; Lazarow, 2006). Pex1p, Pex2p, Pex6p, Pex10p, Pex12p, Pex13p, Pex14p, and Pex26p are also involved in the matrix protein import pathway (Matsumoto et al., 2003; Rayapuram and Subramani, 2006; Schliebs et al., 2010; Francisco et al., 2013; Fujiki et al., 2014). With respect to membrane assembly, newly synthesized peroxisomal membrane proteins (PMPs) are recognized by Pex19p in the cytosol, and the Pex19p–PMP complex is targeted to peroxisomes via the Pex19p receptor, Pex3p (Fang et al., 2004; Fujiki et al., 2014). Pex3p is also imported by Pex19p via the Pex3p receptor, Pex16p (Matsuzaki and Fujiki, 2008; Fujiki et al., 2014).
Mitochondria are essential organelles functioning in many metabolic pathways as well as in energy production. Under irreparable cellular stress, such as oxidative stress, the cells are committed to apoptosis through compromising the integrity of the mitochondrial outer membrane by inducing mitochondrial outer membrane permeabilization (MOMP; Wang, 2001; Newmeyer and Ferguson-Miller, 2003; Tait and Green, 2010). MOMP leads to the release of intermembrane space proteins, including cytochrome c, followed by the activation of caspases responsible for the progression of apoptosis (Wang, 2001; Newmeyer and Ferguson-Miller, 2003; Tait and Green, 2010). The BCL-2 family proteins regulate apoptosis by controlling MOMP (Czabotar et al., 2014). Proapoptotic BCL-2 effector proteins, BAK and BAX, undergo homooligomerization to induce MOMP (Wei et al., 2001). Antiapoptotic BCL-2 proteins, including BCL-2, BCL-XL, and MCL-1, promote survival by sequestering the proapoptotic BH3-only molecules, BID, BIM, and PUMA, thus preventing the initiation of BAX and BAK activation (Cheng et al., 2001). The antiapoptotic BCL-2 members can also sequester BH3-exposed BAX and BAK monomers to prevent the homooligomerization of BAX and BAK (Chen et al., 2015). Alternatively, it has been reported that BH3-only proteins activate BAK and BAX by neutralizing antiapoptotic BCL-XL and MCL-1 (O’Neill et al., 2016). BH3-only proteins are activated by various cellular stress signals and promote MOMP (Kim et al., 2006; Czabotar et al., 2014). The mitochondrial outer membrane channel voltage-dependent anion channel 2 (VDAC2) is reported to interact with BAK and keeps BAK as an inactive monomer at mitochondria (Cheng et al., 2003). VDAC2 is also reported to stabilize the mitochondrial targeting of BAK (Setoguchi et al., 2006).
We earlier isolated a peroxisome-deficient cell mutant, ZP114, which belongs to a novel CG (Tateishi et al., 1997). ZP114 cells show a defect in matrix protein import with normal PMP assembly. In the present study, we found that VDAC2 is a complementing gene of ZP114 cells. In ZP114 cells, BAK distribution shifted from mitochondria to peroxisomes and cytosol. BAK inactivation by RNA interference or overexpression of BAK inhibitors BCL-XL and MCL-1 restored peroxisome biogenesis in ZP114 cells, suggesting that BAK is the element responsible for peroxisome deficiency in ZP114 cells. Furthermore, knockdown of BAK in the wild-type cells increased catalase latency. Conversely, activation of BAK by overexpression of either of the proapoptotic BH3-only proteins, PUMA or BIM, released catalase from peroxisomes. Collectively, our results strongly suggest that BAK potentially localizes to peroxisomes and is involved in peroxisomal membrane permeability.
Results
VDAC2 deficiency abrogates peroxisome biogenesis
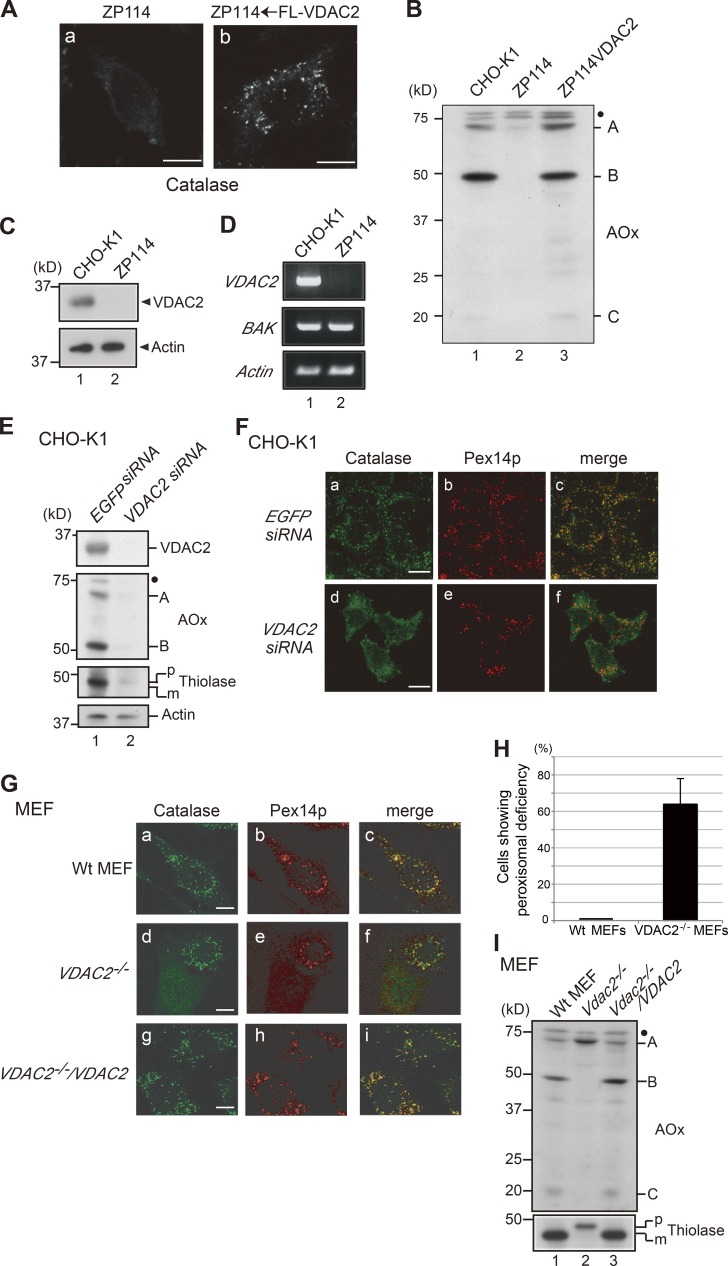
We previously isolated a peroxisome-deficient CHO cell mutant, ZP114, which belonged to a novel CG. ZP114 cells show the impaired import of matrix proteins but not of PMPs (Tateishi et al., 1997). In ZP114 cells, endogenous catalase failed to localize to peroxisomes and showed a diffused staining pattern (Fig. 1 A, a). In contrast, Pex14p, one of the PMPs, has normal peroxisomal localization (not depicted). To search for a complementing gene of ZP114 cells, a human kidney cDNA library was transiently expressed in ZP114 cells that stably express EGFP-catalase (Matsumoto et al., 2003). The peroxisome-restoring positive cDNA clone was isolated by monitoring peroxisomal localization of EGFP-catalase in the ZP114 cells. To our surprise, the positive cDNA clone encoded a mitochondrial outer membrane channel, VDAC2, suggesting that VDAC2 is deficient in ZP114 cells and that VDAC2 deficiency likely affects the peroxisomal import of catalase. To confirm this functional screening result, Flag-tagged VDAC2 (FL-VDAC2) was expressed in ZP114 cells, which were then immunostained with anticatalase antibody. Upon transfection with FL-VDAC2, catalase showed a punctate staining pattern, suggesting that the impaired matrix protein import was restored (Fig. 1 A, b). Matrix protein imports in these cells were also assessed by the processing of acyl-CoA oxidase (AOx), the first enzyme of the fatty acid β oxidation pathway (Miyazawa et al., 1989). AOx-B and -C are generated from 75-kD AOx-A by intraperoxisomal conversion in wild-type CHO-K1 cells, whereas AOx processing was not discernible in ZP114 cells (Fig. 1 B, lanes 1 and 2). Upon transfection of FL-VDAC2 into ZP114 cells, AOx processing became discernible (Fig. 1 B, lane 3), indicating that matrix protein import was restored in ZP114.
Figure 1.
VDAC2 deficiency leads to peroxisomal dysfunction. (A) ZP114 cells were mock transfected (a) or transfected with Flag-VDAC2 (b). After 48 h, cells were fixed and immunostained with anticatalase antibody. FL, Flag. (B) Total cell lysates from CHO-K1 cells, ZP114 cells, and ZP114 cells transfected with Flag-VDAC2 were analyzed by SDS-PAGE and Western blotting with an anti-AOx antibody. A full-length 75-kD AOx-A chain and intraperoxisomal processed 52-kD B and 22-kD C chains are indicated. (C) Total cell lysates from CHO-K1 and ZP114 cells were analyzed by Western blotting with antibodies to VDAC2 (top) and actin (bottom). (D) Total RNA was prepared from wild-type CHO-K1 cells and ZP114 cells. RT-PCR was performed with a set of primers for VDAC2, BAK, and Actin. (E) CHO-K1 cells transfected with EGFP siRNA and VDAC2 siRNA were subjected to Western blotting with antibodies to VDAC2, AOx, thiolase, and actin. AOx bands A and B are as in panel B; p and m denote a larger precursor and mature forms of thiolase, respectively. (F) CHO-K1 cells transfected with EGFP siRNA and VDAC2 siRNA were immunostained with antibodies to catalase and Pex14p. Merged views of panels a plus b and panels d plus e are shown in c and f, respectively. (G) Normal control wild-type (Wt) MEFs, Vdac2−/− MEFs, and Vdac2−/− MEFs transfected with VDAC2 (Vdac2−/−/VDAC2) were immunostained with antibodies to catalase and Pex14p. Merged views of panels a plus b, panels d plus e, and panels g plus h are shown in c, f, and i, respectively. Bars: (A and F) 10 µm; (G) 20 µm. (H) Normal control wild-type MEFs and Vdac2−/− MEFs lacking peroxisomal structure as assessed by immunostaining with anti-Pex14p and anticatalase antibodies are counted (200 cells each). The percentages of the cells showing abnormal peroxisome assembly are presented. Data represent means ± SD. n = 3. (I) Total cell lysates from control wild-type MEFs, Vdac2−/− MEFs, and Vdac2−/−/VDAC2 MEFs were analyzed by Western blotting with antibodies to AOx and thiolase. Note that peroxisomal AOx conversion from the A chain to B and C chains and processing of the larger precursor to mature thiolase were restored in Vdac2−/−/VDAC2 MEFs (lane 3). (B, E, and I) Dots indicate nonspecific bands.
To verify whether ZP114 is a VDAC2-deficient mutant, we investigated the expression of VDAC2. Total protein extracts from CHO-K1 and ZP114 cells were analyzed by Western blotting with anti-VDAC2 antibody. VDAC2 was not detected in ZP114 cells (Fig. 1 C). Furthermore, VDAC2 mRNA was not detectable in ZP114 cells as analyzed by RT-PCR (Fig. 1 D), indicating that ZP114 is a VDAC2-null mutant.
Next, we investigated whether VDAC2 deficiency actually caused the defect in peroxisomal biogenesis by a reverse genetic approach. Vdac2 was knocked down with siRNA in wild-type CHO-K1 cells. VDAC2 expression was efficiently knocked down by transfection of VDAC2 siRNA to under detectable levels, as assessed by anti-VDAC2 Western blotting (Fig. 1 E). In EGFP siRNA–transfected cells, normal peroxisomal morphology was observed by immunostaining with antibodies to catalase and Pex14p (Fig. 1 F, a and b). In VDAC2 siRNA–transfected cells, catalase was mislocalized to the cytosol (Fig. 1 F, d), whereas Pex14p showed normal localization (Fig. 1 F, e). In addition, proteolytic processing of newly synthesized matrix proteins, AOx and 3-ketoacyl-CoA thiolase (hereafter referred to as thiolase), was compromised in VDAC2 siRNA–transfected cells, where both proteins were rather destabilized and apparently degraded (Fig. 1 E). Of note, matrix proteins except for catalase are unstable and degraded in the cytosol. Hence, the phenotype of VDAC2 knockdown cells clearly reproduced the phenotype of ZP114 (Fig. 1 A). Similarly, VDAC2 knockdown in rat astrocytoma (RCR-1) cells induced mislocalization of catalase but not of Pex14p (Fig. S1 A). We also analyzed peroxisomal biogenesis in mouse embryonic fibroblasts (MEFs) from the Vdac2 knockout mouse (Cheng et al., 2003). Vdac2−/− MEFs appeared heterogeneous in peroxisomal biogenesis based on the immunostaining with antibodies to PTS1 and Pex14p. Distinct from ZP114 showing complete deficiency in the peroxisomal localization of matrix proteins (Fig. 1 A; Tateishi et al., 1997), ∼60% of the cells showed abnormal localization of both catalase and Pex14p, but morphologically normal peroxisomes were discernible in the rest of cells (Fig. 1, G [d and e] and H). Interestingly, heterogeneous peroxisomal biogenesis was still observed in Vdac2-deficient MEFs after single-cell subcloning, suggesting that the impact of VDAC2 deficiency on peroxisomal biogenesis may be stochastic. Reexpression of VDAC2 in Vdac2−/− MEFs rescued the peroxisomal defect phenotype. Normal peroxisomes were exclusively observed in Vdac2−/− MEFs reconstituted with VDAC2, termed Vdac2−/−/VDAC2 (Fig. 1 G, g and h). Moreover, proteolytic processing of AOx and thiolase was completely abrogated in Vdac2−/− MEFs (Fig. 1 I, lane 2) and was restored in Vdac2−/−/VDAC2 reconstituted with VDAC2 (lane 3), suggesting that the peroxisomal matrix protein processing protease, Tysnd1 (Okumoto et al., 2011a), may not be active in Vdac2−/− MEFs. Disappearance of Pex14p-positive structures and deficiency in the processing of AOx were also observed in wild-type MEFs treated with Vdac2 siRNA (Fig. S1 B). Collectively, both Vdac2−/− MEFs and ZP114 cells showed impaired localization of peroxisomal matrix proteins such as catalase and AOx. However, Vdac2−/− MEFs might have more severe defects in peroxisomal biogenesis than ZP114 and VDAC2 knockdown RCR-1 cells because the PMP Pex14p was also affected.
BAK inactivation restores the impaired peroxisomal localization of matrix proteins in ZP114 cells
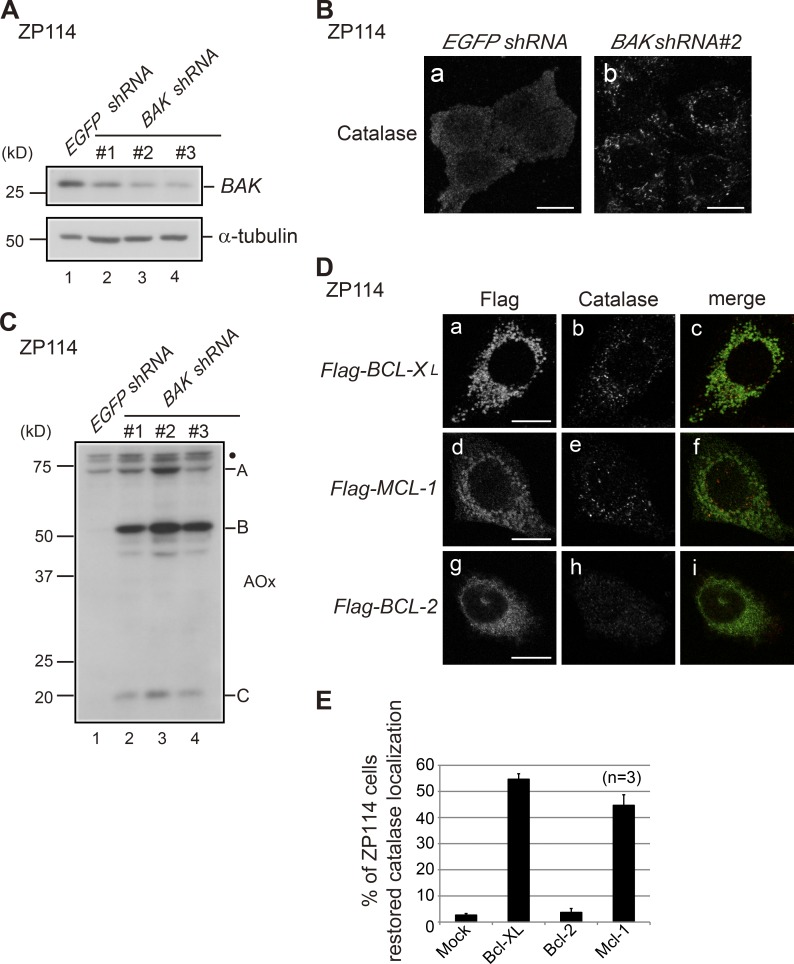
Given that VDAC2 is reported to be exclusively localized to the mitochondrial outer membrane (Yu et al., 1995), the observation that VDAC2 deficiency led to abnormal peroxisomal biogenesis was surprising. We next investigated whether VDAC2 is localized to peroxisomes. However, no data supporting peroxisomal localization of VDAC2 could be obtained (unpublished data). VDAC2 is shown not only to stabilize the mitochondrial targeting of BAK but also to inhibit BAK activity by direct interaction (Cheng et al., 2003; Setoguchi et al., 2006). BAK is a C-tail–anchored protein that is inserted into the membrane with a transmembrane domain located at its C terminus. Notably, C-tail–anchored proteins such as Fis1 and Mff show dual localization to mitochondria and peroxisomes (Kobayashi et al., 2007; Gandre-Babbe and van der Bliek, 2008). Thus, we hypothesized that BAK might be localized to peroxisomes to affect peroxisomal biogenesis in ZP114 cells. To test this hypothesis, shRNA-mediated knockdown of BAK was performed in ZP114 cells. The BAK level was significantly reduced in ZP114 cells with three different BAK shRNAs (Fig. 2 A). Remarkably, immunostaining of catalase showed punctate staining in most shRNA-transfected ZP114 cells, suggesting that localization of matrix proteins to peroxisomes was efficiently restored in ZP114 cells when BAK was knocked down (Fig. 2 B, b). As a marker for normal biogenesis of peroxisomes, intraperoxisomal processing of AOx was also restored in BAK knockdown ZP114 cells (Fig. 2 C). In contrast, BAX knockdown did not restore the peroxisomal formation in ZP114 cells (Fig. S2). Furthermore, overexpression of BAK inhibitors BCL-XL and MCL-1 in ZP114 cells also partly restored peroxisomal biogenesis in ZP114 (Fig. 2, D [b and e] and E), whereas overexpression of BCL-2, another antiapoptotic protein, did not compensate peroxisomal assembly in ZP114 cells (Fig. 2, D [h] and E). Of note, BCL-2 is reported to interact with BAX but not with BAK (Willis et al., 2005). Collectively, these results strongly suggest that BAK but not BAX is directly involved in peroxisomal deficiency in VDAC2-deficient ZP114 cells.
Figure 2.
BAK inactivation restores peroxisome biogenesis in ZP114 cells. (A) The effect of BAK RNA interference was verified by Western blotting. ZP114 cells transfected with EGFP shRNA and each of three different BAK shRNA plasmids were analyzed by Western blotting with antibodies to BAK and α-tubulin. (B) ZP114 cells transfected with EGFP shRNA and BAK shRNA were immunostained with anticatalase antibody. Note that numerous peroxisomes are detectable in b. (C) ZP114 cells transfected with EGFP shRNA and three BAK shRNA plasmids were subjected to Western blotting with anti-AOx antibody as described in Fig. 1 B. A, B, and C denote A, B, and C chains, respectively; the dot indicates nonspecific bands. (D) ZP114 cells were transfected with cDNAs each for Flag–BCL-XL, Flag–MCL-1, and Flag–BCL-2. The cells were immunostained with antibodies to Flag and catalase. Merged views of panels a plus b, panels d plus e, and panels g plus h are shown in c, f, and i, respectively. (B and D) Bars, 10 µm. (E) Percentages of the cells restored in the catalase and the localization of catalase to peroxisomes in total cells expressing Flag-tagged proteins (100 cells each) are presented. Data represent means ± SD.
BAK is localized to peroxisomes and the cytosol in addition to mitochondria
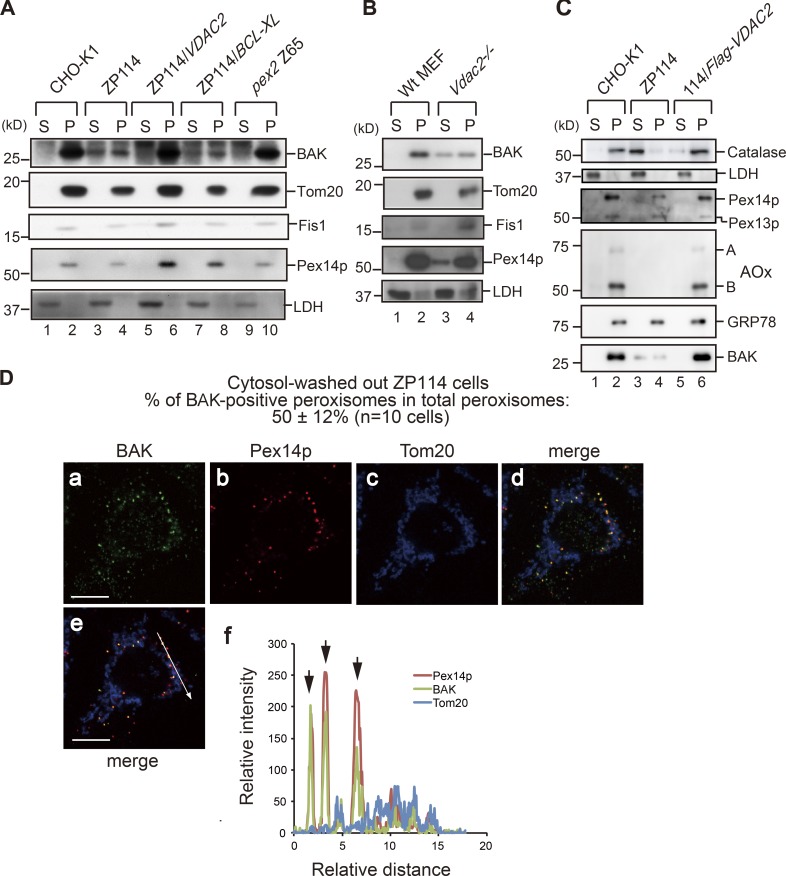
The data shown in Fig. 2 support that the VDAC2 interaction partner BAK, but not VDAC2 itself, may be directly involved in peroxisome biogenesis. Accordingly, we investigated whether BAK is localized to peroxisomal remnants in ZP114 cells. Cells were homogenized and separated into cytosol and organelle fractions by centrifugation. In CHO-K1 and pex2 CHO mutant Z65 cells, BAK was predominantly detected in the organelle fraction (Fig. 3 A, lanes 1, 2, 9, and 10). In contrast, ∼40% of BAK was localized in the cytosolic fraction in ZP114 cells (Fig. 3 A, lanes 3 and 4). However, other mitochondrial proteins, including Tom20 and Fis1, were exclusively detected in the organelle fraction in ZP114 cells (Fig. 3 A, lanes 3 and 4). In ZP114 cells stably expressing VDAC2, named ZP114/VDAC2 cells, BAK became predominantly detected in the organelle fraction (Fig. 3 A, lanes 5 and 6). As shown in Fig. 2, BCL-XL expression restored peroxisomal biogenesis in ZP114 cells. Nevertheless, BCL-XL expression did not restore the organelle localization of BAK in ZP114 cells (Fig. 3 A, lanes 7 and 8), suggesting that BCL-XL did not function redundantly with VDAC2 with respect to the organelle localization of BAK. In Vdac2−/− MEFs, ∼50% of BAK was also localized to the cytosol (Fig. 3 B, lanes 3 and 4). In addition, the PMP Pex14p was partly detected in the cytosolic fraction in Vdac2−/− MEFs, suggesting that Vdac2−/− MEFs might have more severe defects in peroxisomal biogenesis than ZP114 cells. The subcellular localization of BAK was also analyzed by immunostaining. By normal cell fixation methods, a diffuse cytosolic staining pattern of BAK was discernible in ZP114 cells (not depicted), consistent with the subcellular fractionation results shown in Fig. 3 A. To further assess the BAK distribution within the organelles, cells were semipermeabilized with digitonin, and cytosolic proteins were washed out before the fixation. Immunostaining of ZP114 cells washed out with cytosol revealed that BAK was localized to ∼50% of peroxisomes in ZP114 cells with reduced localization with mitochondria (Fig. 3 D). However, BAK was mostly localized to mitochondria in wild-type CHO-K1 cells along with a small number of BAK-positive peroxisomes (Fig. S3). Collectively, these data are consistent with a model in which a shift of subcellular localization of BAK from mitochondria to peroxisomes as a consequence of VDAC2 loss likely affects peroxisomal biogenesis in ZP114 cells.
Figure 3.
BAK is localized to peroxisomes and the cytosol in VDAC2-deficient cells. (A) Various CHO cells indicated at the top were homogenized and fractionated into cytosol (S) and organelle (P) fractions. Equal aliquots of cytosol and organelle fractions were analyzed by SDS-PAGE and Western blotting with antibodies against BAK, Tom20, Fis1, Pex14p, and lactate dehydrogenase (LDH). (B) Normal control wild-type (Wt) MEFs and Vdac2−/− MEFs were analyzed as in A. (C) CHO-K1, ZP114, and Flag-VDAC2–transfected ZP114 were likewise analyzed as in A using antibodies to respective proteins indicated on the right. A and B denote AOx-A and -B chains, respectively. (D) After permeabilization with digitonin and washing out the cytosol, ZP114 cells were fixed and immunostained with antibodies to BAK, Pex14p, and Tom20. Merged views of a–c are shown in d and e. Signal intensities of BAK, Pex14p, and Tom20 along the arrow in e are plotted in f. Downward arrows in f indicate the superimposed points where BAK is colocalized with Pex14p, but not with Tom20. Bars, 10 µm.
BAK is localized to the ER (Scorrano et al., 2003; Zong et al., 2003) in addition to mitochondria. Therefore, VDAC2 deficiency may give rise to BAK accumulation in the ER, thereby affecting the integrity of the ER as well as that of peroxisomes. Therefore, to investigate ER integrity in VDAC2-deficient cells, CHO-K1, ZP114, and ZP114/VDAC2 cells were fractionated to cytosol and organelle fractions. The peroxisomal matrix protein, catalase, was predominantly detected in the cytosol fraction in ZP114 cells (Fig. 3 C, lanes 3 and 4), whereas most of the catalase was discernible in the organelle fraction in CHO-K1 and ZP114/VDAC2 cells (Fig. 3 C, lanes 1, 2, 5, and 6), consistent with the results shown in Fig. 1. In contrast to catalase, GRP78, an ER-luminal soluble protein, was detected in the organelle fraction in ZP114 cells (Fig. 3 C, lane 4), hence suggesting that ER integrity was maintained under the VDAC2-deficient condition.
Peroxisome-targeted BAK abrogates peroxisomal localization of matrix proteins
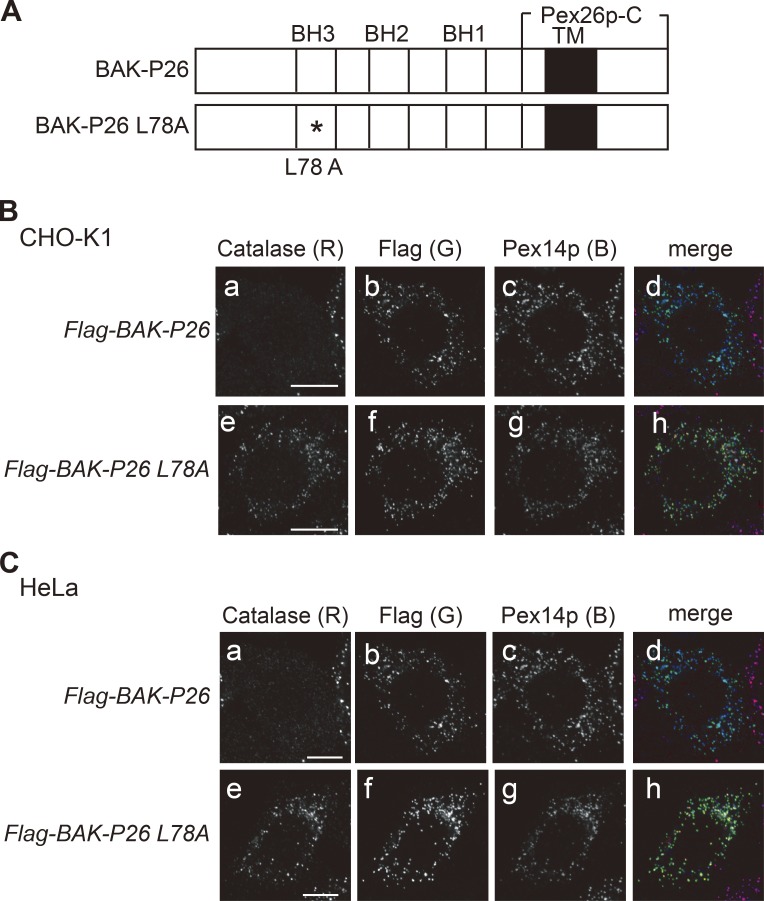
To determine whether BAK translocated to peroxisomes is able to affect the localization of peroxisomal matrix proteins in wild-type cells, we generated a BAK mutant specifically targeted to peroxisomes by replacing the C-terminal transmembrane domain of BAK with that of Pex26p (BAK-P26; Fig. 4 A; Matsumoto et al., 2003). BAK-P26 was indeed localized to peroxisomes when it was expressed in CHO-K1 and HeLa cells (Fig. 4, B and C, b and c). Importantly, cells expressing BAK-P26 displayed mislocalization of catalase to the cytosol (Fig. 4, A and B, a), which parallels the phenotype of ZP114 cells. In contrast, BAK-P26–L78A harboring the L78A mutation in the BH3 domain failed to affect the catalase localization (Fig. 4, B and C, e) despite its peroxisomal localization (Fig. 4, B and C, f and g). These results suggest that BAK localization to peroxisomes autonomously induces mislocalization of peroxisomal matrix proteins, including catalase, in the wild-type CHO-K1 and HeLa cells. This activity requires the BH3 domain, which is a prerequisite for the homooligomerization and proapoptotic activity of BAK (Dewson et al., 2008; Kim et al., 2009). These observations help explain how peroxisome deficiency is induced in the VDAC2-defective cells where the subcellular location of BAK was shifted from mitochondria to peroxisomes (Fig. 3).
Figure 4.
Peroxisome-targeted BAK abrogates peroxisome biogenesis. (A) Schematic presentation of peroxisome-targeting BAK-P26. The asterisk designates an L78A mutation in the BH3 domain of BAK. Solid bars indicate a transmembrane segment (TM). (B and C) Flag–BAK-P26 and Flag–BAK-P26–L78A were expressed in CHO-K1 (B) and HeLa (C) cells. The cells were immunostained with antibodies to catalase, Flag, and Pex14p. Merged views of a–c and e–g are shown in d and h, respectively, where three fluorescent colors, red (R), green (G), and blue (B), were merged. Bars, 10 µm.
Peroxisome-targeted BAK releases catalase from peroxisomes to cytosol
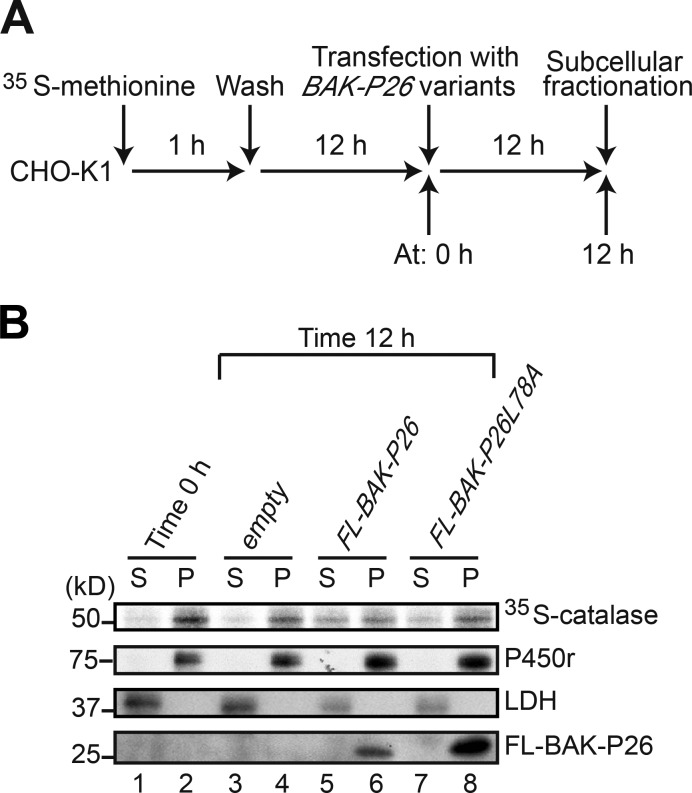
During apoptosis, BAK oligomerizes and permeabilizes the mitochondrial outer membrane, thereby releasing intermembrane-space proteins like cytochrome c to the cytosol (Wei et al., 2001). Accordingly, it is plausible that BAK may permeabilize the peroxisomal membrane and release peroxisomal matrix proteins. Alternatively, BAK may inhibit protein import into the peroxisome matrix. To discern these two possibilities, we performed a pulse-chase experiment during BAK-P26–induced peroxisomal disruption in CHO-K1 cells. CHO-K1 cells were pulse labeled with [35S]methionine for 1 h. After a 12-h incubation (time 0; Fig. 5 A), BAK-P26, BAK-P26–L78A, and empty vector were transfected to the 35S-labeled cells. At 12 h after transfection (time 12 h), the cells were fractionated into organelle and cytosol fractions. Each fraction was solubilized and subjected to immunoprecipitation with anticatalase antibody. At time 0, [35S]catalase was detected predominantly in the organelle fraction (Fig. 5 B, lanes 1 and 2), indicating that almost all [35S]catalase proteins were imported into peroxisomes at this time point. If BAK-P26 permeabilizes peroxisomes, [35S]catalase will be detected in the cytosol fraction. At 12 h, subcellular distribution of [35S]catalase did not change in cells transfected with the empty vector (Fig. 5 B, lanes 3 and 4). In contrast, ∼40% of the imported [35S]catalase was detected in the cytosol fraction in BAK-P26–transfected cells (Fig. 5 B, lanes 5 and 6). However, the imported [35S]catalase remained in the organelle fraction in BAK-P26–L78A–transfected cells (Fig. 5 B, lanes 7 and 8). Collectively, these results strongly suggest that BAK releases catalase to the cytosol rather than inhibits the import of catalase. Notably, the ability of BAK to permeabilize either mitochondria or peroxisomes requires an intact BH3 domain.
Figure 5.
Peroxisome-targeted BAK releases catalase from peroxisomes to cytosol. (A) Time flow of pulse-chase experimentation of catalase translocation. (B) Cells at 0 h (before transfection) and 12 h after transfection with the indicated plasmids (time 12 h) were fractionated into cytosol (S) and organelle (P) fractions. Equal aliquots of cytosol and organelle fractions were solubilized, and catalase was immunoprecipitated with anticatalase antibody. [35S]methionine– and [35S]cysteine–labeled catalase was detected by a Fujix FLA-5000 autoimaging analyzer. As marker proteins for cytosol and the ER, lactate dehydrogenase (LDH) and P450 reductase (P450r) in S and P fractions were detected with the respective antibodies. BAK-P26 variants were detected with anti-Flag antibody.
BAK potentially localizes to peroxisomes and regulates matrix protein localization in wild-type cells
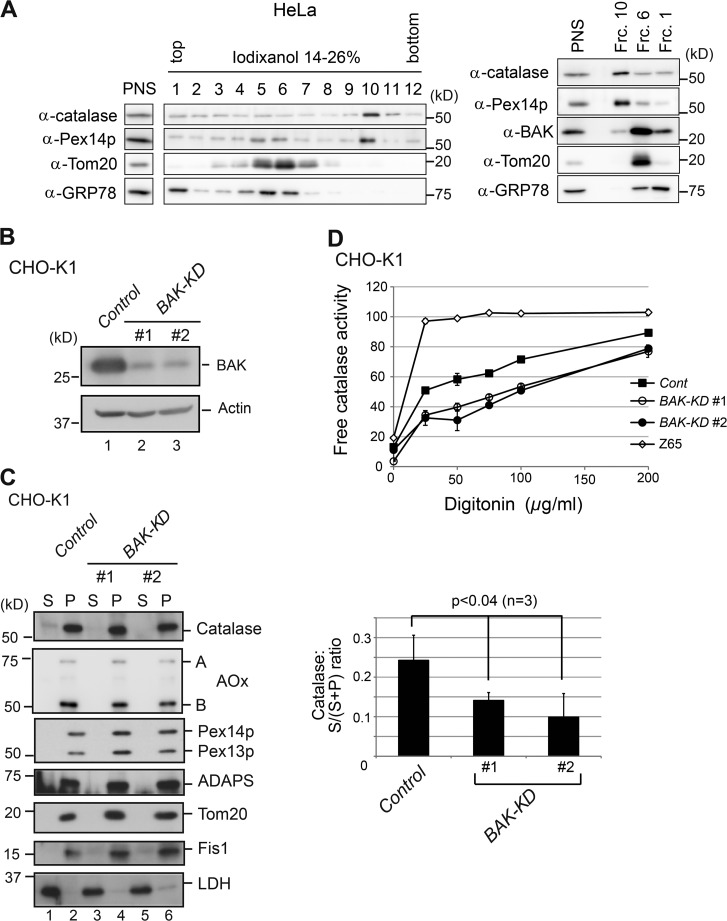
We have presented evidence that BAK localizes to peroxisomes and affects the localization of peroxisomal matrix proteins such as catalase in VDAC2-deficient ZP114 cells. These findings raise an issue as to whether BAK targets to peroxisomes under normal conditions. We attempted to detect BAK on peroxisomes in wild-type cells by immunostaining (Fig. S3). However, because of the close proximity of mitochondria and peroxisomes in cells, it is technically challenging to convincingly demonstrate the localization of BAK to both mitochondria and peroxisomes by microscopic observation. To overcome this difficulty, we performed a subcellular fractionation study. The postnuclear supernatant (PNS) fraction from wild-type HeLa cells was subjected to ultracentrifugation on an iodixanol density gradient to separate peroxisomes from other organelles. A highly purified peroxisome fraction apparently devoid of mitochondria and ER (Fig. 6 A, left, fraction 10) was subjected to Western blotting with anti-BAK antibody. In the peroxisome fraction, BAK was clearly present, whereas Tom20 (a mitochondria marker) and GRP78 (an ER marker) were hardly detectable (Fig. 6 A, right). Moreover, BAK was detected at higher levels in fractions 6 and 1 (Fig. 6 A, right) where Tom20 and GRP78 were distinctly detected, indicative of mitochondria and ER, respectively (Scorrano et al., 2003; Zong et al., 2003). This result suggests that BAK is localized to peroxisomes in addition to VDAC2-mediated mitochondria and other compartments in wild-type cells. Under a VDAC2-deficient condition, the targeting of BAK to mitochondria is reduced and more readily directed to the peroxisomes and the cytosol (Fig. 3).
Figure 6.
BAK localizes to peroxisomes and regulates catalase latency in wild-type cells. (A, left) The PNS fraction from HeLa cells was separated by ultracentrifugation on an iodixanol density gradient into 12 fractions. Same-volume aliquots of gradient fractions and the PNS fraction (one twentieth of the loaded amount) were analyzed by Western blotting with antibodies against catalase, Pex14p, Tom20, and GRP78. (Right) The PNS fraction, along with fractions (Frc.) 1, 6, and 10 representing the ER, mitochondria, and peroxisomes, respectively, were likewise analyzed by Western blotting with antibodies against catalase, Pex14p, BAK, Tom20, and GRP78. (B) CHO-K1 cells stably expressing empty vector (control) and two independent BAK shRNA vectors (BAK-KD#1 and #2) were analyzed by Western blotting with antibodies to BAK and actin. (C, left) Control and BAK-KD cells were treated with digitonin to selectively permeabilize the plasma membrane. Permeabilized cells were centrifuged to separate into cytosol (S) and organelle (P) fractions. Equal aliquots of S and P fractions were analyzed by Western blotting with antibodies to proteins, including peroxisomal matrix proteins, catalase, AOx, and ADAPS, as indicated. (Right) Catalase in cytosol and organelle fractions from control and BAK-KD cells was quantified and shown as ratios of cytosol/total (cytosol plus organelle). (D) Control, BAK-KD, and pex2 Z65 cells were treated with digitonin at the indicated concentrations, and free catalase activity was determined in the isotonic medium. Free catalase activity is shown as a percentage of the total catalase activity detected in the presence of 1% Triton X-100. (C and D) Data represent means ± SD. n = 3.
To investigate whether endogenous BAK affects the localization of peroxisomal matrix protein in wild-type cells, we established two independent lines of BAK-KD cells, named BAK-KD#1 and BAK-KD#2, each expressing distinct shRNA against BAK. BAK was efficiently knocked down in BAK-KD cells in comparison with CHO-K1 cells transfected with empty vector (control cells), as verified by Western blotting with an anti-BAK antibody (Fig. 6 B). BAK-KD and control cells were treated with digitonin to selectively permeabilize the plasma membrane and were subsequently separated into cytosol and organelle fractions. In control cells, catalase was mainly detected in the organelle fraction, whereas a part of catalase was detected in the cytosol fraction (Fig. 6, C [lane 1] and D), suggesting that a part of catalase resided in the cytosol. To our surprise, the amount of cytosolic catalase was significantly reduced in BAK-KD cells (Fig. 6 C, lanes 3 and 5 on left and quantification on right). In contrast, other matrix proteins, AOx and alkyl-dihydroxyacetone phosphate synthase (ADAPS), were exclusively localized to the organelle fraction in both control and BAK-KD cells (Fig. 6 C, lanes 3 and 5 on the left and quantification on the right). Catalase latency in these cells was also evaluated by digitonin titration assay (Tateishi et al., 1997). Catalase was more latent in BAK-KD cells as compared with control cells. In contrast, a peroxisome-deficient CHO cell mutant, pex2 Z65 (Tsukamoto et al., 1991), showed the lowest catalase latency, indicating that the digitonin titration assay adequately reflected subcellular catalase distribution (Fig. 6 D). Furthermore, BAK knockdown also reduced cytosolic catalase in HeLa cells (Fig. S4). These results strongly suggest that endogenous BAK may reduce the retention of catalase in peroxisomes of normal cells, likely through permeabilization of the peroxisomal membrane.
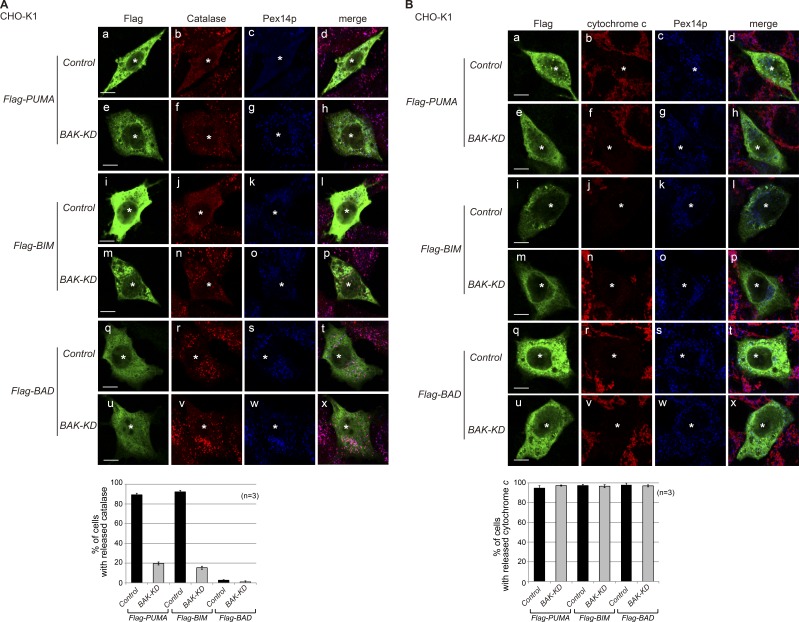
BH3-only molecules, including BID, BIM, and PUMA, are known to induce MOMP by either directly activating BAK and BAX (Kim et al., 2006, 2009; Ren et al., 2010) or neutralizing the BAK/BAX inhibitors MCL-1 and BCL-XL (O’Neill et al., 2016). Accordingly, we subsequently investigated whether activation of endogenous BAK by proapoptotic BH3-only proteins affects peroxisomal permeabilization and the localization of peroxisomal matrix proteins. Upon overexpression of a ΒAK activator, PUMA or BIM, catalase was diffused to the cytosol and was no longer visible as a punctate staining pattern in ∼80% of PUMA- and BIM-overexpressing cells (Fig. 7 A). Importantly, BAK knockdown mitigated the impaired catalase localization induced by the overexpression of PUMA and BIM (Fig. 7 A), suggesting that BAK functions downstream of PUMA and BIM in permeabilizing peroxisomes. Of note, overexpression of another BH3-only molecule, BAD, that could not activate BAK showed no effect on catalase localization (Fig. 7 A). Furthermore, PUMA, BIM, and BAD induced cytochrome c release from mitochondria in BAK-KD cells as well as in control cells, suggesting that the BH3-only protein induced MOMP via BAX in BAK-KD cells (Fig. 7 B). Collectively, these results suggest that peroxisome-resident BAK is activated by BH3-only molecules, such as PUMA and BIM, to permeabilize peroxisomes. A subset of BAK appears to localize to peroxisomes and regulate the latency of peroxisomal matrix proteins, including catalase, by altering peroxisomal membrane permeability.
Figure 7.
Overexpression of BH3-only proteins PUMA and BIM, but not of BAD, disrupts peroxisomes in a BAK-dependent manner. (A and B) Control and BAK-KD#2 cells (see Fig. 6, B–D) were transfected with Flag-PUMA, Flag-BIM, and Flag-BAD in the presence of a caspase inhibitor, Q-VD-Oph. At 48 h after transfection, cells were immunostained with antibodies to Flag, Pex14p, and catalase (A) or cytochrome c (B). Asterisks indicate respective transfectants. Merged views of a–c, e–g, i–k, m–o, q–s, and u–w are shown in d, h, l, p, t, and x, respectively. Bars, 10 µm. Bottom graphs show percentages of the cells showing released catalase (A) or cytochrome c (B) in cells expressing Flag-tagged proteins. Data represent means ± SD.
Discussion
The BCL-2 family proteins, consisting of both antiapoptotic and proapoptotic members, are the major regulators of mammalian apoptosis (Czabotar et al., 2014). BAK and BAX are mutually redundant effectors of cell death, and either protein is sufficient to trigger apoptosis in response to apoptotic signals (Wei et al., 2001). The BCL-2 family proteins are known to localize to mitochondria and the ER (Scorrano et al., 2003; Zong et al., 2003). Most studies have focused on how the BCL-2 proteins function on mitochondria. Upon apoptotic stimuli, BAK and BAX are activated by activator BH3-only molecules to induce MOMP, thereby causing the release of apoptogenic factors such as cytochrome c (Cheng et al., 2001; Wei et al., 2001; Kim et al., 2006, 2009). BH3-only molecules activate BAK and BAX either by their direct binding to BAK and BAX (Cheng et al., 2001; Wei et al., 2001; Kim et al., 2006, 2009) or by neutralizing antiapoptotic MCL-1 and BCL-XL (O’Neill et al., 2016). BAK and BAX are also shown to localize to the ER for the regulation of Ca2+ homeostasis (Scorrano et al., 2003; Zong et al., 2003). The BCL-2 family proteins contain a transmembrane domain at their C-terminal regions, called C-tail–anchored proteins, and predominantly localize to mitochondria and partially localize to the ER. Although the import mechanism of BCL-2 family proteins to mitochondria or ER is not fully elucidated, VDAC2 is reported to stabilize the mitochondrial targeting of BAK (Cheng et al., 2003). In addition, FKBP38 is reported to be essential for the mitochondrial targeting of BCL-2 and BCL-XL (Shirane and Nakayama, 2003).
In this study, we showed that VDAC2 deficiency leads to impaired peroxisome biogenesis. BAK distribution shifts from the mitochondria to the peroxisomes and cytosol in VDAC2-deficient CHO ZP114 cells, giving rise to the release of peroxisomal matrix proteins to the cytosol. Introduction of VDAC1 and VDAC3 does not complement the peroxisomal defect in ZP114 (not depicted), indicative of a VDAC2-specific function in regulating peroxisome biogenesis. Similar results were obtained with Vdac2 knockdown cells as well as with Vdac2−/− MEFs (Fig. 1). However, the phenotype of Vdac2−/− MEFs appears to be distinct from that of ZP114 cells. ZP114 cells have peroxisome membrane structures devoid of matrix proteins, forming so-called “peroxisome ghosts” (Tateishi et al., 1997). In Vdac2−/− MEFs, ∼60% of cells contain no peroxisomes, and the rest of cells have morphologically normal peroxisomes. Notably, this heterogeneity is maintained in Vdac2-deficient MEFs after single-cell subcloning, suggesting that the impact of VDAC2 deficiency on peroxisomal biogenesis may be stochastic. Interestingly, AOx processing is completely impaired in Vdac2−/− MEFs, suggesting that the peroxisomal matrix protein–processing protease, Tysnd1 (Okumoto et al., 2011a), may not be active in Vdac2−/− MEFs. Hence, peroxisomal function may be impaired even in normal-appearing peroxisomes.
The impact of VDAC2 loss in peroxisomal biogenesis appears to be indirect through its interaction partner BAK because VDAC2 itself is not localized to peroxisomes. Knockdown of BAK but not of BAX in ZP114 cells restores peroxisome biogenesis, supporting the direct contribution of BAK to peroxisomal deficiency in ZP114 cells (Fig. 2). Furthermore, BAK targeted to peroxisomes with the transmembrane region of Pex26p releases peroxisomal matrix proteins to the cytosol in wild-type cells, suggesting that overexpressed BAK-P26 is spontaneously activated to permeabilize peroxisomes (Fig. 4). Overexpression of antiapoptotic BCL-2 family members BCL-XL and MCL-1 restores peroxisome formation in ZP114 cells (Fig. 2) even though the localization of BAK is not altered (Fig. 3 A), suggesting that some BCL-XL and MCL-1 may target to peroxisomes to inhibit BAK, at least upon overexpression.
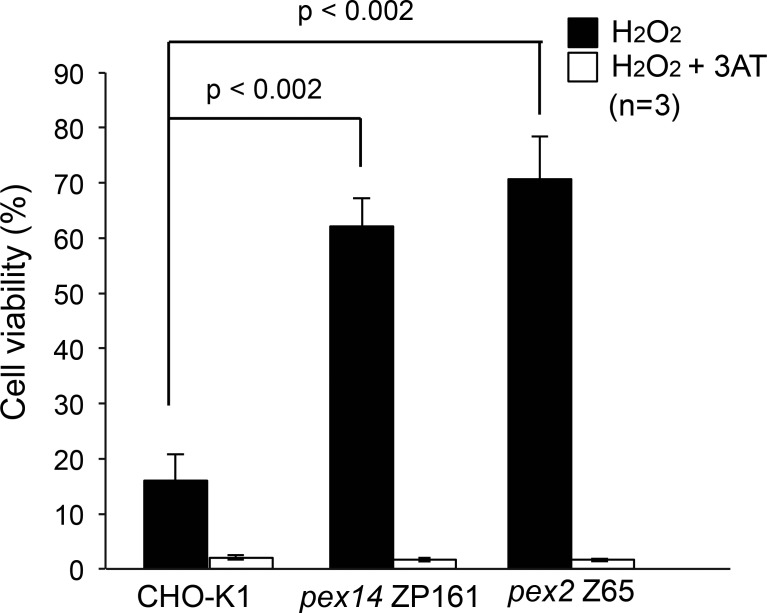
We implied that BAK is involved in the intracellular localization of catalase even in normal cells. In wild-type CHO-K1 cells, a portion of catalase remains in the cytosol at steady state. Interestingly, catalase latency is elevated upon stable knockdown of BAK. Overexpression of the proapoptotic BH3-only molecules PUMA and BIM resulted in the release of peroxisomal matrix proteins in a BAK-dependent manner. These results strongly suggest that BAK may target to peroxisomes to play a role in the permeability of the peroxisomal membrane and thereby the localization of peroxisomal matrix proteins. Catalase is partly localized to the cytosol, whereas the other matrix proteins, including AOx and ADAPS, are exclusively localized to peroxisomes. Partial localization of catalase to the cytosol might result from a lower efficiency of catalase import to the peroxisomes as compared with that of the other matrix proteins. Catalase retains its activity in the cytosol in peroxisome-deficient cells (Wanders et al., 1984). Catalase harbors the noncanonical PTS1, KANL, at its C terminus (Purdue and Lazarow, 1996; Otera and Fujiki, 2012). The peroxisome-targeting signal of catalase possesses a weaker binding affinity to Pex5p than a canonical PTS1 such as SKL (Otera and Fujiki, 2012). The C-terminal KANL sequence of catalase is highly conserved between mammalian species, implying that the weak targeting signal plays a physiological role. Yeast and worms have cytosolic catalase, which is thought to have an important role for the elimination of cellular reactive oxygen species together with glutathione peroxidase (Hartman et al., 2003). Mammals lack cytosolic catalase and have only peroxisomal catalase. Given these findings, we propose that mammalian catalase also has a physiological function in the cytosol. The inefficient import signal and BAK-mediated release from the peroxisomal matrix would increase cytosolic catalase, which may help eliminate the cytosolic reactive oxygen species. Consistent with such a possible functional model, we found that peroxisome-defective mutant pex2 Z65 and pex14 ZP161 cells in which catalase is diffused to the cytosol are more resistant to exogenous H2O2 than the wild-type CHO-K1 cells (Fig. 8). The differential sensitivity to H2O2 between CHO-K1 and pex mutant cells is abolished by the addition of the catalase inhibitor 3-aminotriazole (3-AT). These observations strongly suggest that cytosolic catalase could more efficiently eliminate oxidative stress than peroxisomal catalase.
Figure 8.
Peroxisome-deficient cell mutants show resistance to exogenous H2O2. CHO-K1, pex14 ZP161, and pex2 Z65 cells were treated with 1 mM H2O2 in the presence and absence of 50 mM 3-AT, a catalase inhibitor. Cell viability was determined by MTT assay and represented as percentages relative to those of mock-treated, H2O2-untreated cells. Data represent means ± SD.
Overexpression of antiapoptotic BCL-2 family proteins such as BCL-XL and MCL-1 restores the impaired peroxisome biogenesis in ZP114 cells (Fig. 2). Conversely, overexpression of proapoptotic BH3-only proteins, including PUMA and BIM, disrupts peroxisomes in wild-type CHO-K1 cells (Fig. 7). These results suggest that a similar mechanism of BAK-dependent membrane permeabilization likely operates in both mitochondria and peroxisomes. However, BAK is likely constitutively activated at a low level on the peroxisomal membrane to keep a part of catalase in cytosol, whereas it is strictly inhibited at a steady state in mitochondria because of the lethal effect of cytochrome c efflux. BH3-only proteins are activated by various cellular stress signals, such as genotoxic and ER stress, and promote MOMP by directly inducing the homooligomerization of BAK and BAX or inactivating the antiapoptotic BCL-2 members (Kim et al., 2006; Czabotar et al., 2014). Thus, peroxisome membrane permeability may also be regulated by the BCL-2 family proteins in response to various cellular stress signals. Further studies regarding the physiological roles of peroxisomal BAK and cytosolic catalase could open a way to delineate the mechanisms underlying regulation in the localization of peroxisomal matrix proteins.
Materials and methods
Chemicals
Restriction enzymes and DNA-modifying enzymes were purchased form Nippon Gene and Takara Bio, Inc. FBS, Ham’s F12, and DMEM were purchased from Invitrogen. Caspase inhibitors including Zvad-fmk (Sigma-Aldrich) and Q-VD-Oph (EMD Millipore), digitonin (Wako Pure Chemical Industries), Paclitaxel (Sigma-Aldrich), and molecular mass standards (Precision Plus Protein All Blue Standards; Bio-Rad Laboratories) were also purchased.
Antibodies
We used guinea pig antisera to Pex14p (Itoh and Fujiki, 2006), rabbit antisera to catalase (Tsukamoto et al., 1990), AOx (Tsukamoto et al., 1990), PTS1 (Tsukamoto et al., 1990), ADAPS (Honsho et al., 2008), BAK (EMD Millipore), VDAC2 (Cheng et al., 2003), Flag M2 (Sigma-Aldrich), and human Fis1 (Apotech). Mouse antibodies to Flag M2 (Sigma-Aldrich), Tom20 (F-10; Santa Cruz Biotechnology, Inc.), chicken α-tubulin (Seikagaku Kogyo), actin (EMD Millipore), P450 reductase (Santa Cruz Biotechnology, Inc.), and cytochrome c (BD), as well as goat antibodies to lactate dehydrogenase antibody (Rockland) and GRP78 (Santa Cruz Biotechnology, Inc.), were purchased.
Cell culture and transfection
Various CHO cells were cultured in Ham’s F12 medium supplemented with 10% FCS under 5% CO2/95% air. MEFs and HeLa cells were cultured in DMEM supplemented with 10% FCS. cDNA was introduced using Lipofectamine 2000 (Invitrogen) as described in the manufacturer’s protocols. Cells transfected with Flag–BAK-P26, Flag–BAK-P26–L78A, Flag-PUMA, Flag-BIM, or Flag-BAD were further cultured in the presence of caspase inhibitor, Zvad-fmk, after transfection.
Expression screening of a cDNA library
Expression screening of a complementing gene of ZP114 was performed as described previously (Matsumoto et al., 2003). Human kidney cDNA library was divided into small pools, each containing ∼1,300 clones. Each pool was transfected into ZP114 stably expressing EGFP-catalase. Restoration of peroxisome biogenesis was evaluated by localization of EGFP-catalase. Among the cDNA pools examined, a positive pool that restored peroxisomal biogenesis in ZP114 was further divided into subpools and then was further analyzed. A single positive clone was isolated, and we determined its nucleotide sequence. Consequently, VDAC2 was identified as a complementing gene of ZP114.
Construction of plasmids
Flag-VDAC2, Flag-PUMA, Flag-BIM, Flag-BAD, Flag–BCL-XL, Flag–MCL-1, and Flag–BCL-2 were cloned into BamHI–NotI sites of pcDNA3.1 Zeo-FLAG vector (Okumoto et al., 2011b). BAK/PEX26 chimera genes (BAK-P26) encoding 2–187 aa of BAK and 237–305 aa of Pex26p were generated by PCR and cloned into BamHI–NotI sites of the pcDNA3.1 Zeo-Flag vector. L78A mutation was introduced into Flag–BAK-P26 using site-directed mutagenesis (Hutchison et al., 1978).
For three BAK shRNA (Lim et al., 2006) constructs and one control EGFP construct, the following oligonucleotides were cloned into the pSilencer vector (Okumoto et al., 2011a). The underlining indicates sense and antisense sequences of the target genes: shBAK#1, 5′-GATCCTTTACTGGAGCAGCTGCAGTTCAAGAGACTGCAGCTGCTCCAGTAAATTTTTGGAAA-3′; shBAK#2, 5′-GATCCTGCCTACGAACTCTTCACCTTCAAGAGAGGTGAAGAGTTCGTAGGCATTTTTGGAAA-3′; shBAK#3, 5′-GATCCCTCTTCACCAAGATTGCCTTTCAAGAGAAGGCAATCTTGGTGAAGAGTTTTTGGAAA-3′; and shEGFP, 5′-GATCCGGTTATGTACAGGAACGCATTCAAGAGATGCGTTCCTGTACATAACCTTTTTGGAAA-3′.
RT-PCR
The total RNA was prepared from wild-type CHO-K1 and ZP114 cells using the NucleoSpin RNA II kit (Takara Bio, Inc.), and first-strand cDNA was obtained by reverse transcription, respectively. ClVDAC2 was amplified by PCR with primers ClVDAC2 forward, 5′-ATGGCGGACTGTTGTGTACCGATATGCCCA-3′, and ClVDAC2 reverse, 5′-TTAAGCCTCCAGTTCCAAGGCAAGCCCAAG-3′, using reverse transcription products as a template. BAK was similarly amplified by PCR with primers ClBAK forward, 5′- ATGGCGTCTGGACAAGGACCAGGTCCTCCT-3′, and ClBAK reverse, 5′-TCAAGATCTGAAGAATCTGTGTACCACGTA-3′.
Pulse-chase experiment
CHO-K1 cells growing in 35-mm dishes were washed twice with PBS, incubated in cysteine- and methionine-free DMEM for 30 min, and then pulse labeled for 1 h with 10 mCi/ml [35S]methionine plus [35S]cysteine (American Radiolabeled Chemicals). To chase the 35S-labeled proteins, cells were washed with PBS and further incubated with complete Ham's F12 medium containing 10% FBS for 12 h. Cells were transfected with empty vector, BAK-P26, or BAK-P26–L78A, and were further incubated for 12 h. 35S-labeled cells were harvested and incubated in homogenizing buffer containing 25 µg/ml digitonin for 5 min at room temperature as described previously (Natsuyama et al., 2013; or origin). After centrifugation at 100,000 g for 30 min at 4°C, cytosolic and organellar fractions were subjected to immunoprecipitation with anticatalase antibody as described previously (Tsukamoto et al., 1990). 35S-labeled proteins were separated on SDS-PAGE (9% gel) and detected with an Autoimaging analyzer (FLA-5000; Fujifilm).
siRNA treatment
For siRNA experiments, CHO-K1 cells were transfected with control and VDAC2 siRNA (EMD Millipore) using Lipofectamine 2000.
Catalase latency
Catalase latency was evaluated as described previously (Tsukamoto et al., 1990). In brief, trypsinized cells were washed and suspended at 106 cells/ml in 0.25 M sucrose and 10 mM Hepes-NaOH, pH 7.4. The cells were treated with different concentrations of digitonin or 1% Triton X-100. After detergent treatment, 50 µl of cell suspensions were added to 500 µl H2O2 solution (20 mM imidazole-HCl, pH 7.0, 0.25 M sucrose, 0.1% BSA, and 0.01% H2O2) and further incubated for 10 min on ice. After incubation, the catalase reaction was halted by addition of 500 µl Ti(SO4)2 solution (2.0 M H2SO4 and 1.25% Ti(SO4)2). The concentration of residual H2O2 was determined by absorbance at 410 nm H2TiO4. Catalase activity was calculated from the decrease in H2O2 concentration after the enzymatic reaction.
Subcellular fractionation
For differential centrifugation, to fractionate into the cytosol and organelle proteins, CHO-K1, ZP114, ZP114/VDAC2, ZP114/BCL-XL, pex2 Z65, and CHO-K1 stably expressing EGFP shRNA or BAK shRNA were harvested and incubated in homogenizing buffer containing 25 µg/ml digitonin for 5 min at room temperature as described previously (Natsuyama et al., 2013) followed by ultracentrifugation at 100,000 g for 30 min. Proteins were separated by SDS-PAGE and analyzed by immunoblotting.
For isopycnic ultracentrifugation using iodixanol gradient, ∼107 HeLa cells were homogenized with a Potter-Elvehjem Teflon homogenizer (Wheaton) in 800 µl homogenized buffer (20 mM Hepes-KOH, pH 7.4, 0.25 M sucrose, and 1 mM DTT containing complete protease inhibitor cocktail [Roche]) and centrifuged at 800 g for 10 min at 4°C to obtain the PNS fraction. Homogenized buffer containing 50% iodixanol (OptiPrep; Axis-shield; Sigma-Aldrich) was added to the PNS fraction to yield a final 30% iodixanol solution. 5 ml of 14% iodixanol solution in homogenized buffer was layered on an equal volume of 26% iodixanol solution in a 14 × 89–mm centrifuge tube (Beckman Coulter), and linear gradient was made by Gradient Master (Biocomp) according to the manufacturer’s protocol. PNS fraction in 30% iodixanol solution (1.2 ml) was loaded under the linear gradient and centrifuged in a SW41Ti rotor (Beckman Coulter) at 100,000 g for 90 min at 4°C. The gradient was collected to 12 fractions. The equal aliquots were diluted by adding homogenized buffer and centrifuged at 20,000 g for 20 min at 4°C. Purified organelles in the pellet were dissolved in SDS-PAGE sample buffer for immunoblot analysis.
H2O2 sensitivity assay
Cell viability was measured with an MTT-based toxicology assay kit (Roche). CHO cells were grown in 24-well dishes to 80% confluence. Cells were treated with 1 mM H2O2 alone or together with 50 mM 3-AT for 12 h. After treatment, cells were incubated with MTT solution for an additional 4 h as described in the manufacturer’s protocols.
Morphological analysis
Cells were fixed with ice-cold methanol on ice for 5 min or with 4% PFA at room temperature for 20 min and then permeabilized with PBS containing 1% Triton X-100 and 1% BSA for 5 min at room temperature. To visualize organelle-associated BAK, ZP114 cells were semipermeabilized with 25 µg/ml digitonin as described previously, washed to remove cytosolic fraction, and then subjected to immunostaining as described previously (Matsuzaki and Fujiki, 2008). Especially, the mixture of rabbit anti-BAK, guinea pig anti-Pex14p, and mouse anti-Tom20 antibodies diluted in Can Get Signal (TOYOBO) solution was reacted with ZP114 and semiintact ZP114 cells. Indirect immunostaining using secondary antibodies labeled with Alexa Fluor 488, 568, and 633 (Invitrogen) was performed as described previously (Matsuzaki and Fujiki, 2008).
Immunoblotting
Western blotting analysis was performed using electrophoretically transferred samples on polyvinylidene difluoride membranes (Bio-Rad Laboratories) with primary antibodies and second antibodies of donkey anti-rabbit, mouse, or goat immunoglobulin G antibody conjugated to horseradish peroxidase (GE Healthcare). Antigen–antibody complexes were visualized with an ECL Western blotting detection reagent (GE Healthcare).
Online supplemental material
Fig. S1 shows that Vdac2 knockdown induces the defects of peroxisomal biogenesis in RCR-1 and MEF cells. Fig. S2 shows that BAX knockdown did not restore the impaired peroxisome formation in ZP114 cells. Fig. S3 shows that BAK is mostly localized to mitochondria in wild-type CHO-K1 cells. Fig. S4 shows that BAK knockdown decreases cytosolic catalase in HeLa cells.
Supplementary Material
Acknowledgments
We thank M.Y. El-Shemerly for participation at the initial stage of this work, K. Shimizu for preparing figures, and the other members of our laboratory for discussions.
This work was supported in part by a Core Research for Evolutional Science and Technology grant to Y. Fujiki from the Science and Technology Agency of Japan; the Global Centers of Excellence Program and Grants for Excellent Graduate Schools from The Ministry of Education, Culture, Sports, Science and Technology of Japan; Grants-in-Aid for Scientific Research 20370039, 23570236, 24247038, 25112518, 25116717, 26116007, 15K14511, and 15K21743 (to Y. Fujiki); Kyushu University Interdisciplinary Programs in Education and Projects in Research Development (to Y. Fujiki); and grants from the Takeda Science Foundation (to Y. Fujiki), the Naito Foundation (to Y. Fujiki), and Japan Foundation for Applied Enzymology (to Y. Fujiki); and the National Institutes of Health grant R01CA125562 (to E.H. Cheng).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 3-AT
- 3-aminotriazole
- ADAPS
- alkyl-dihydroxyacetonephophate synthase
- AOx
- acyl-CoA oxidase
- CG
- complementation group
- MEF
- mouse embryonic fibroblast
- MOMP
- mitochondrial outer membrane permeabilization
- PBD
- peroxisomal biogenesis disorder
- PMP
- peroxisomal membrane protein
- PNS
- postnuclear supernatant
References
- Chen H.C., Kanai M., Inoue-Yamauchi A., Tu H.C., Huang Y., Ren D., Kim H., Takeda S., Reyna D.E., Chan P.M., et al. . 2015. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat. Cell Biol. 17:1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E.H.-Y.A., Wei M.C., Weiler S., Flavell R.A., Mak T.W., Lindsten T., and Korsmeyer S.J.. 2001. BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell. 8:705–711. 10.1016/S1097-2765(01)00320-3 [DOI] [PubMed] [Google Scholar]
- Cheng E.H.-Y., Sheiko T.V., Fisher J.K., Craigen W.J., and Korsmeyer S.J.. 2003. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 301:513–517. 10.1126/science.1083995 [DOI] [PubMed] [Google Scholar]
- Czabotar P.E., Lessene G., Strasser A., and Adams J.M.. 2014. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 15:49–63. 10.1038/nrm3722 [DOI] [PubMed] [Google Scholar]
- Dewson G., Kratina T., Sim H.W., Puthalakath H., Adams J.M., Colman P.M., and Kluck R.M.. 2008. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol. Cell. 30:369–380. 10.1016/j.molcel.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Ebberink M.S., Koster J., Visser G., Spronsen F., Stolte-Dijkstra I., Smit G.P.A., Fock J.M., Kemp S., Wanders R.J.A., and Waterham H.R.. 2012. A novel defect of peroxisome division due to a homozygous non-sense mutation in the PEX11β gene. J. Med. Genet. 49:307–313. 10.1136/jmedgenet-2012-100778 [DOI] [PubMed] [Google Scholar]
- Fang Y., Morrell J.C., Jones J.M., and Gould S.J.. 2004. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164:863–875. 10.1083/jcb.200311131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco T., Rodrigues T.A., Freitas M.O., Grou C.P., Carvalho A.F., Sá-Miranda C., Pinto M.P., and Azevedo J.E.. 2013. A cargo-centered perspective on the PEX5 receptor-mediated peroxisomal protein import pathway. J. Biol. Chem. 288:29151–29159. 10.1074/jbc.M113.487140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Okumoto K., Kinoshita N., and Ghaedi K.. 2006. Lessons from peroxisome-deficient Chinese hamster ovary (CHO) cell mutants. Biochim. Biophys. Acta-Mol. Cell Res. 1763:1374–1381. 10.1016/j.bbamcr.2006.09.012 [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Okumoto K., Mukai S., Honsho M., and Tamura S.. 2014. Peroxisome biogenesis in mammalian cells. Front. Physiol. 5:307 10.3389/fphys.2014.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S., and van der Bliek A.M.. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19:2402–2412. 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman P., Belmont P., Zuber S., Ishii N., and Anderson J.. 2003. Relationship between catalase and life span in recombinant inbred strains of Caenorhabditis elegans. J. Nematol. 35:314–319. [PMC free article] [PubMed] [Google Scholar]
- Honsho M., Yagita Y., Kinoshita N., and Fujiki Y.. 2008. Isolation and characterization of mutant animal cell line defective in alkyl-dihydroxyacetonephosphate synthase: localization and transport of plasmalogens to post-Golgi compartments. Biochim. Biophys. Acta. 1783:1857–1865. 10.1016/j.bbamcr.2008.05.018 [DOI] [PubMed] [Google Scholar]
- Hutchison C.A. III, Phillips S., Edgell M.H., Gillam S., Jahnke P., and Smith M.. 1978. Mutagenesis at a specific position in a DNA sequence. J. Biol. Chem. 253:6551–6560. [PubMed] [Google Scholar]
- Itoh R., and Fujiki Y.. 2006. Functional domains and dynamic assembly of the peroxin Pex14p, the entry site of matrix proteins. J. Biol. Chem. 281:10196–10205. 10.1074/jbc.M600158200 [DOI] [PubMed] [Google Scholar]
- Kim H., Rafiuddin-Shah M., Tu H.-C., Jeffers J.R., Zambetti G.P., Hsieh J.J.-D., and Cheng E.H.-Y.. 2006. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 8:1348–1358. 10.1038/ncb1499 [DOI] [PubMed] [Google Scholar]
- Kim H., Tu H.-C., Ren D., Takeuchi O., Jeffers J.R., Zambetti G.P., Hsieh J.J.-D., and Cheng E.H.-Y.. 2009. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 36:487–499. 10.1016/j.molcel.2009.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Tanaka A., and Fujiki Y.. 2007. Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 313:1675–1686. 10.1016/j.yexcr.2007.02.028 [DOI] [PubMed] [Google Scholar]
- Lazarow P.B. 2006. The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta. 1763:1599–1604. 10.1016/j.bbamcr.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Lim S.F., Chuan K.H., Liu S., Loh S.O.H., Chung B.Y.F., Ong C.C., and Song Z.. 2006. RNAi suppression of Bax and Bak enhances viability in fed-batch cultures of CHO cells. Metab. Eng. 8:509–522. 10.1016/j.ymben.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Matsumoto N., Tamura S., and Fujiki Y.. 2003. The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat. Cell Biol. 5:454–460. 10.1038/ncb982 [DOI] [PubMed] [Google Scholar]
- Matsuzaki T., and Fujiki Y.. 2008. The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183:1275–1286. 10.1083/jcb.200806062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa S., Osumi T., Hashimoto T., Ohno K., Miura S., and Fujiki Y.. 1989. Peroxisome targeting signal of rat liver acyl-coenzyme A oxidase resides at the carboxy terminus. Mol. Cell. Biol. 9:83–91. 10.1128/MCB.9.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsuyama R., Okumoto K., and Fujiki Y.. 2013. Pex5p stabilizes Pex14p: a study using a newly isolated pex5 CHO cell mutant, ZPEG101. Biochem. J. 449:195–207. 10.1042/BJ20120911 [DOI] [PubMed] [Google Scholar]
- Newmeyer D.D., and Ferguson-Miller S.. 2003. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 112:481–490. 10.1016/S0092-8674(03)00116-8 [DOI] [PubMed] [Google Scholar]
- O’Neill K.L., Huang K., Zhang J., Chen Y., and Luo X.. 2016. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 30:973–988. 10.1101/gad.276725.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K., Kametani Y., and Fujiki Y.. 2011a Two proteases, trypsin domain-containing 1 (Tysnd1) and peroxisomal lon protease (PsLon), cooperatively regulate fatty acid β-oxidation in peroxisomal matrix. J. Biol. Chem. 286:44367–44379. 10.1074/jbc.M111.285197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K., Misono S., Miyata N., Matsumoto Y., Mukai S., and Fujiki Y.. 2011b Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic. 12:1067–1083. 10.1111/j.1600-0854.2011.01217.x [DOI] [PubMed] [Google Scholar]
- Otera H., and Fujiki Y.. 2012. Pex5p imports folded tetrameric catalase by interaction with Pex13p. Traffic. 13:1364–1377. 10.1111/j.1600-0854.2012.01391.x [DOI] [PubMed] [Google Scholar]
- Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., and Fujiki Y.. 2002. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 22:1639–1655. 10.1128/MCB.22.6.1639-1655.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue P.E., and Lazarow P.B.. 1996. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J. Cell Biol. 134:849–862. 10.1083/jcb.134.4.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram N., and Subramani S.. 2006. The importomer—a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim. Biophys. Acta. 1763:1613–1619. 10.1016/j.bbamcr.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Ren D., Tu H.-C., Kim H., Wang G.X., Bean G.R., Takeuchi O., Jeffers J.R., Zambetti G.P., Hsieh J.J.-D., and Cheng E.H.-Y.. 2010. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 330:1390–1393. 10.1126/science.1190217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliebs W., Girzalsky W., and Erdmann R.. 2010. Peroxisomal protein import and ERAD: variations on a common theme. Nat. Rev. Mol. Cell Biol. 11:885–890. 10.1038/nrm3008 [DOI] [PubMed] [Google Scholar]
- Scorrano L., Oakes S.A., Opferman J.T., Cheng E.H., Sorcinelli M.D., Pozzan T., and Korsmeyer S.J.. 2003. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 300:135–139. 10.1126/science.1081208 [DOI] [PubMed] [Google Scholar]
- Setoguchi K., Otera H., and Mihara K.. 2006. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 25:5635–5647. 10.1038/sj.emboj.7601438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane M., and Nakayama K.I.. 2003. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat. Cell Biol. 5:28–37. 10.1038/ncb894 [DOI] [PubMed] [Google Scholar]
- Steinberg S.J., Dodt G., Raymond G.V., Braverman N.E., Moser A.B., and Moser H.W.. 2006. Peroxisome biogenesis disorders. Biochim. Biophys. Acta-Mol. Cell Res. 1763:1733–1748. [DOI] [PubMed] [Google Scholar]
- Tait S.W.G., and Green D.R.. 2010. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11:621–632. 10.1038/nrm2952 [DOI] [PubMed] [Google Scholar]
- Tateishi K., Okumoto K., Shimozawa N., Tsukamoto T., Osumi T., Suzuki Y., Kondo N., Okano I., and Fujiki Y.. 1997. Newly identified Chinese hamster ovary cell mutants defective in peroxisome biogenesis represent two novel complementation groups in mammals. Eur. J. Cell Biol. 73:352–359. [PubMed] [Google Scholar]
- Titorenko V.I., and Terlecky S.R.. 2011. Peroxisome metabolism and cellular aging. Traffic. 12:252–259. 10.1111/j.1600-0854.2010.01144.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T., Yokota S., and Fujiki Y.. 1990. Isolation and characterization of Chinese hamster ovary cell mutants defective in assembly of peroxisomes. J. Cell Biol. 110:651–660. 10.1083/jcb.110.3.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T., Miura S., and Fujiki Y.. 1991. Restoration by a 35K membrane protein of peroxisome assembly in a peroxisome-deficient mammalian cell mutant. Nature. 350:77–81. 10.1038/350077a0 [DOI] [PubMed] [Google Scholar]
- van der Klei I.J., and Veenhuis M.. 2006. Yeast and filamentous fungi as model organisms in microbody research. Biochim. Biophys. Acta. 1763:1364–1373. 10.1016/j.bbamcr.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Wanders R.J.A., and Waterham H.R.. 2006. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75:295–332. 10.1146/annurev.biochem.74.082803.133329 [DOI] [PubMed] [Google Scholar]
- Wanders R.J.A., Kos M., Roest B., Meijer A.J., Schrakamp G., Heymans H.S.A., Tegelaers W.H.H., van den Bosch H., Schutgens R.B.H., and Tager J.M.. 1984. Activity of peroxisomal enzymes and intracellular distribution of catalase in Zellweger syndrome. Biochem. Biophys. Res. Commun. 123:1054–1061. 10.1016/S0006-291X(84)80240-5 [DOI] [PubMed] [Google Scholar]
- Wang X. 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15:2922–2933. [PubMed] [Google Scholar]
- Wei M.C., Zong W.-X., Cheng E.H.-Y., Lindsten T., Panoutsakopoulou V., Ross A.J., Roth K.A., MacGregor G.R., Thompson C.B., and Korsmeyer S.J.. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 292:727–730. 10.1126/science.1059108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis S.N., Chen L., Dewson G., Wei A., Naik E., Fletcher J.I., Adams J.M., and Huang D.C.S.. 2005. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 19:1294–1305. 10.1101/gad.1304105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.H., Wolfgang W., and Forte M.. 1995. Subcellular localization of human voltage-dependent anion channel isoforms. J. Biol. Chem. 270:13998–14006. 10.1074/jbc.270.23.13998 [DOI] [PubMed] [Google Scholar]
- Zong W.-X., Li C., Hatzivassiliou G., Lindsten T., Yu Q.-C., Yuan J., and Thompson C.B.. 2003. Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J. Cell Biol. 162:59–69. 10.1083/jcb.200302084 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.