Trafficking of integral membrane proteins to cilia is poorly understood. Badgandi et al. show that tubby family proteins TULP3 and TUB act as general adapters for ciliary trafficking of structurally diverse integral membrane cargo like GCPRs and the polycystin 1/2 complex.
Abstract
The primary cilium is a paradigmatic organelle for studying compartmentalized signaling; however, unlike soluble protein trafficking, processes targeting integral membrane proteins to cilia are poorly understood. In this study, we determine that the tubby family protein TULP3 functions as a general adapter for ciliary trafficking of structurally diverse integral membrane cargo, including multiple reported and novel rhodopsin family G protein–coupled receptors (GPCRs) and the polycystic kidney disease–causing polycystin 1/2 complex. The founding tubby family member TUB also localizes to cilia similar to TULP3 and determines trafficking of a subset of these GPCRs to neuronal cilia. Using minimal ciliary localization sequences from GPCRs and fibrocystin (also implicated in polycystic kidney disease), we demonstrate these motifs to be sufficient and TULP3 dependent for ciliary trafficking. We propose a three-step model for TULP3/TUB-mediated ciliary trafficking, including the capture of diverse membrane cargo by the tubby domain in a phosphoinositide 4,5-bisphosphate (PI(4,5)P2)-dependent manner, ciliary delivery by intraflagellar transport complex A binding to the TULP3/TUB N terminus, and subsequent release into PI(4,5)P2-deficient ciliary membrane.
Introduction
The primary cilium is a tiny subcellular compartment comprised of a microtubular axoneme enveloped by a membrane contiguous with the plasma membrane. The ciliary membrane is enriched with multiple integral membrane proteins, including multiple class A (rhodopsin family) G protein–coupled receptors (GPCRs; Hilgendorf et al., 2016), proteins linked to polycystic kidney disease such as the single-pass transmembrane protein fibrocystin (Ward et al., 2003), and the transient receptor potential (TRP) channel family proteins polycystin 1/2 (PC1/2; Pazour et al., 2002; Yoder et al., 2002). Although soluble proteins access cilia by diffusion (Kee et al., 2012; Breslow et al., 2013; Lin et al., 2013) and are further enriched by intraflagellar transport machinery and nuclear targeting-related mechanisms (Qin et al., 2004; Takao et al., 2014), pathways that regulate integral membrane protein trafficking to cilia are poorly understood.
Factors regulating membrane biogenesis with relation to ciliogenesis, such as the Rab8–Rabin8–Rab11 pathway, indirectly impact both ciliary integrity and trafficking of membrane-associated cargo (Moritz et al., 2001; Westlake et al., 2011). The BBSome proteins were initially implicated in GPCR delivery to cilia (Berbari et al., 2008b; Jin et al., 2010; Loktev and Jackson, 2013). However, the BBSome proteins regulate the removal of GPCRs (Domire et al., 2011; Eguether et al., 2014; Liew et al., 2014), polycystins (Xu et al., 2015), and membrane-associated proteins from cilia (Lechtreck et al., 2009, 2013). In addition, membrane composition of cilia is impacted in the BBSome knockouts (Lechtreck et al., 2009, 2013). An increasing number of factors have been reported to be involved in the trafficking of cilia-targeted membrane proteins during transit via the secretory pathway and/or plasma membrane (Mazelova et al., 2009; Follit et al., 2014; Kim et al., 2014; Leaf and Von Zastrow, 2015; Lim and Tang, 2015). In addition, the sequences that determine ciliary localization are diverse and include (a) VXPX or RVXP in the N terminus of PC2 (Geng et al., 2006), the C terminus of rhodopsin (Deretic et al., 1998), and CNGB1 (Jenkins et al., 2006); (b) AX(S/A)XQ in the intracellular loop 3 (IC3) of GPCRs, including somatostatin receptor 3 (Sstr3), melanin-concentrating receptor 1 (Mchr1), and 7-hydroxytryptophan receptor 6 (Htr6; Berbari et al., 2008a,b); (c) (V/I)KARK in the orphan GPCR, Gpr161 IC3 (Mukhopadhyay et al., 2013); (d) (R/K)(I/L)W motif in neuropeptide receptor NPY2R and the orphan GPR83; and (e) the intracellular sequence flanking the transmembrane domain of fibrocystin (Follit et al., 2010). The heterogeneity in localization sequences along with the multiplicity of pathways described for ciliary targeting are confounding, making it difficult to conceptualize the trafficking of different types of integral membrane proteins into a general model.
We previously described the tubby family protein 3 (Tulp3) to be necessary for the trafficking of certain rhodopsin family GPCRs to cilia (Mukhopadhyay et al., 2010). TULP3-dependent GPCRs include Sstr3, Mchr1 (Mukhopadhyay et al., 2010), the neuropeptide receptor 2 (Npy2r; Loktev and Jackson, 2013), and the newly described orphan GPCR, Gpr161, which functions as a negative regulator of the Sonic hedgehog (Shh) pathway (Mukhopadhyay et al., 2013; Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Unlike the ubiquitously expressed Tulp3, the founding tubby family member, Tub, is predominantly expressed only in the brain and retina (Mukhopadhyay et al., 2013). Interestingly, Tub knockouts also exhibit lack of trafficking of a subset of GPCRs to cilia, including Sstr3, Mchr1, and Npy2r (Sun et al., 2012; Loktev and Jackson, 2013). The conserved C-terminal tubby domain in this family of proteins is characterized by its selective and strong association with phosphoinositide 4,5-bisphosphate (PI(4,5)P2; Santagata et al., 2001). In addition, we previously demonstrated that TULP3/TUB binds to the IFT-A core complex through a conserved N-terminal helical domain. TULP3 binds more effectively to IFT-A than TUB and localizes to cilia in an IFT-A core-dependent manner (Mukhopadhyay et al., 2010). TULP3 mutants defective in either PI(4,5)P2 or IFT-A binding have dominant-negative effects in preventing GPCR trafficking to cilia; thus, both domains are critical for TULP3-mediated ciliary trafficking (Mukhopadhyay et al., 2010). However, it is unknown whether TULP3 is a general adapter for trafficking other membrane proteins, whether targeting cargo with diverse ciliary localization sequences (CLSs) by TULP3/TUB is direct or involves additional factors, and whether TUB functions in a capacity similar to TULP3. In this study, we describe TULP3 to be a general adapter for ciliary trafficking of structurally diverse integral membrane cargo and propose a TULP3/TUB-mediated multistep mechanism for delivery to cilia.
Results
TULP3 is required for trafficking of multiple class A GPCRs to cilia
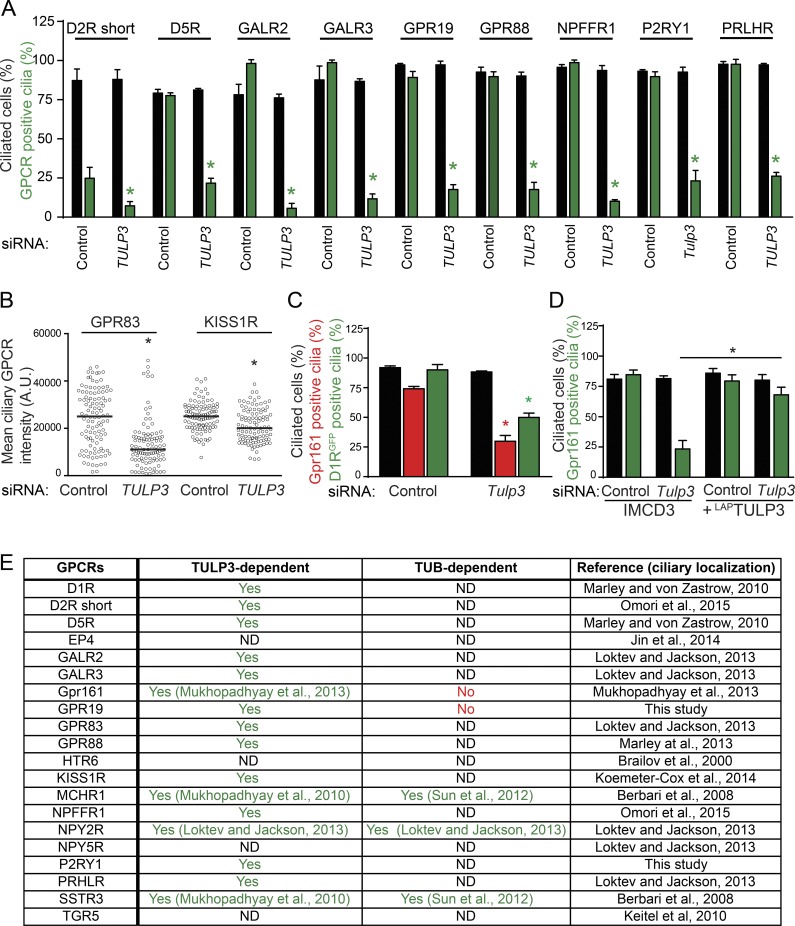
To test whether there is a general adapter for trafficking of ciliary class A GPCRs, we systematically screened cilia-localized GPCRs for dependence on TULP3. To visualize GPCR trafficking to cilia, we generated stable lines of C-terminal GFP-tagged dopamine receptors 2 (short isoform, D2R short) and 5 (D5R), galanin receptors 2 and 3 (GAL2R and GAL3R), orphan receptors GPR83 and GPR88, kisspeptin receptor KISS1R, neuropeptide FF receptor 1 (NPFFR1), and the prolactin-releasing hormone receptor (PRLHR) in retinal pigment epithelial (RPE) hTERT cells (Marley and von Zastrow, 2010; Loktev and Jackson, 2013; Marley et al., 2013; Koemeter-Cox et al., 2014; Omori et al., 2015). In addition, we identified two new GPCRs that localize to cilia: the central nervous system–expressing orphan GPR19 (O’Dowd et al., 1996; Regard et al., 2008) and the ubiquitously expressed purinergic receptor P2RY1 (Regard et al., 2008), and we generated stable cell lines in RPE hTERT and IMCD3 Flp-In cells, respectively. Upon RNAi-mediated knockdown of TULP3, we detected decreased trafficking of these GPCRs to cilia without affecting ciliogenesis (Figs. 1, A and B; and S1, A and B). In most cases, the percentages of cells with GPCR-positive cilia were decreased (Figs. 1 A and S1 A). In two cases, with GPR83 and KISS1R, the steady-state ciliary levels were significantly decreased with respect to the control, although the percentages of cilia positive for the receptors were not reduced, possibly resulting from relatively higher ciliary levels in the overexpression lines (Figs. 1 B and S1 B). The dopamine receptor D1R has been previously reported to localize to cilia in a TULP3-independent manner (Leaf and Von Zastrow, 2015). However, upon generating stable lines in IMCD3 Flp-In cells expressing D1RGFP and RNAi-mediated knockdown of Tulp3, we detected a partial but significant decrease in D1R-positive cilia (Figs. 1 C and S1 C) comparable to a decrease in endogenous Gpr161 trafficking (Fig. 1 C; Mukhopadhyay et al., 2013). Transient overexpression of transfected D1R might have prevented the detection of the role of Tulp3 in the original study. The decreased ciliary localization of endogenous Gpr161 upon siRNA-mediated Tulp3 knockdown was rescued upon stable expression of human LAPTULP3 (LAP, S tag–PreScission-GFP), ruling out nonspecific effects of RNAi (Fig. 1 D). Overall, TULP3 is required for trafficking of at least 16 class A cilia–targeted GPCRs (Fig. 1 E).
Figure 1.
TULP3 determines localization of multiple rhodopsin family GPCRs to primary cilia. (A) Stable RPE hTERT or IMCD3 Flp-In cell lines expressing the indicated GPCRs C-terminally tagged with GFP were transfected with 100 nM TULP3 si #3 siRNA (for RPE, see Materials and methods) or were sequentially transfected with 200 nM Tulp3 siRNA twice (for IMCD3 expressing P2RY1, see Materials and methods) for 72 h and serum starved for the last 24 h before fixation and then were immunostained for GFP, acetylated tubulin, and DNA. GFP-positive cilia were counted from two experiments, and total counted cells are >200 for each condition. Data represent means ± SD. (B) Same as in A, with RPE hTERT stable lines expressing GPR83 and KISS1R, except the ciliary intensity of GFP was quantified. Total counted cells are >100 for each condition. A.U., arbitrary units. (C) Stable IMCD3 Flp-In cells expressing D1R C-terminally tagged with GFP were sequentially transfected with 200 nM Tulp3 siRNA twice for 72 h (see Materials and methods) and serum starved for the last 24 h before fixing and staining for Gpr161, acetylated tubulin, and DNA. D1RGFP/Gpr161-positive cilia were counted from two experiments, and total counted cells are 300–500 for each condition. Data represent means ± SD. (D) IMCD3 Flp-In cells stably expressing LAPTULP3 (LAP; S tag–PreScission-GFP) were sequentially transfected with 200 nM Tulp3 siRNA twice for 72 h (see Materials and methods) and serum starved for the last 24 h before fixing and staining for Gpr161, acetylated tubulin, and DNA. Data represent means ± SD from three experiments, and total counted cells are >400 for each condition. (A–D) *, P < 0.01 (A); *, P < 0.001 (B); *, P < 0.05 (C; A–C, with respect to corresponding siRNA controls); and *, P < 0.001 (D). (E) Table summarizing rhodopsin family GPCRs tested for ciliary localization and the role of TULP3/TUB in trafficking. Unlike the long D2R isoform, the D2R short isoform (NCBI RefSeq database accession no. NP_057658) is ciliary (Omori et al., 2015). ND, not determined or ciliary, but limited stable expression in RPE hTERT cells. Other class A GPCRs reported to be ciliary by transient expression or endogenously in neurons or thyrocytes but not detected in cilia upon stable expression in RPE hTERT cells include the neuromedin receptor NMUR1 (Omori et al., 2015), the orphan PGR15L, the pyroglutamylated RFamide peptide (QRFP) receptor QRFPR (Loktev and Jackson, 2013), and the trace amine receptor TAAR1 (Szumska et al., 2015). Two other cilia-localized seven-transmembrane (7TM) receptors, GPR157 (Takeo et al., 2016) and Gpr175 (TPRA1; Singh et al., 2015), that were not tested are either not readily classifiable or have no homology to known GPCRs, respectively (Foord et al., 2005; Davenport et al., 2013; http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=113). Also see Fig. S1.
GPCR ciliary localization motifs are necessary and sufficient for Tulp3-mediated trafficking to cilia
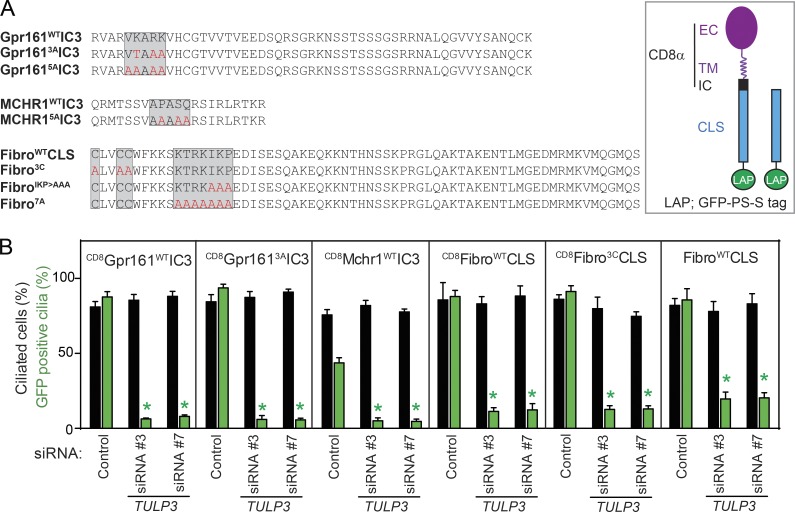
We and others have previously described the (V/I)KARK motif and the AX(S/A)XQ motif in the third intracelullular loop of Gpr161 and Htr6/SStr3/Mchr1, respectively, to be necessary for targeting to cilia (Berbari et al., 2008a,b; Mukhopadhyay et al., 2013). To test if these motifs are sufficient for ciliary targeting, we stably expressed chimeras of the extracellular and transmembrane domains of CD8α (Motley et al., 2003), IC3 of cilia-localized GPCRs (Gpr161 and Mchr1) consisting of CLS motifs and LAP (S tag–PreScission-GFP; hereafter referred to as CD8-CLS-LAP fusions) in ciliated IMCD3 and RPE hTERT cells (Figs. 2 A and S2, A and B). These fusions, including that of a partially mutated Gpr161 IC3 (VKARK>VTAAA; Gpr1613AIC3) were localized to the primary cilia, unlike mutant Gpr161 (VKARK>AAAAA; Gpr1615AIC3; Mukhopadhyay et al., 2013) and MCHR1 (APASQ>AAAAA; MCHR15AIC3) fusions (Figs. 2 A and S2, A and B). Thus, these CLSs are necessary and sufficient to target the chimeras to cilia. As Gpr161 and Mchr1 localization to cilia has been previously demonstrated to be dependent on Tulp3 (Mukhopadhyay et al., 2010, 2013), we determined if these heterologous fusions recapitulated this dependency. Interestingly, upon knockdown of TULP3 in the stably expressing cells, ciliary trafficking of CD8-CLS fusions was completely prevented (Figs. 2 B and S2 B). Localization of the minimal CLS fusions to cilia in a TULP3-dependent manner provided us with an opportunity to study ciliary trafficking of GPCRs using a tractable single transmembrane domain fusion approach.
Figure 2.
CLSs from GPCRs and fibrocystin require Tulp3 for trafficking to cilia. (A) CLS wild-type and mutant sequences used for making CD8-CLS-LAP and CLS-LAP fusions (left), and a diagram representing the CD8-CLS-LAP or CLS-LAP fusions (right). EC, extracellular region; IC, intracellular region; TM, transmembrane domain. (B) Stable RPE lines expressing the indicated CD8-CLS-LAP or CLS-LAP fusions were transfected with 100 nM TULP3 siRNA for 72 h, serum starved for the last 24 h, and processed as in Fig. 1 A. GFP-positive cilia were counted in three experiments, and total counted cells are >300 in each condition. Data represent means ± SD; *, P < 0.001 with respect to corresponding siRNA controls. Also see Fig. S2.
TULP3 determines ciliary trafficking of the minimal fibrocystin CLS
To test if the repertoire of TULP3-regulated ciliary membrane cargo extends beyond GPCRs, we focused on fibrocystin. Fibrocystin is a single-pass transmembrane protein, which is mostly extracellular, except for a short ∼190-residue C terminus (Nagasawa et al., 2002). The fibrocystin CLS has been previously defined to consist of 18 intracellular aa flanking the transmembrane domain, including conserved cysteines, considered to be a palmitoylation motif (Follit et al., 2010). As previous experiments with this CLS were conducted using nontransmembrane fusions (Follit et al., 2010), we wondered whether the conserved cysteines were at all necessary for ciliary localization in the context of a transmembrane domain. The stably expressing CD8-fibrocystin CLS-LAP fusion (CD8-fibrocystinWTCLS-LAP) localized to cilia, and mutating the three cysteines (CLVCC>ALVAA; CD8-fibrocystin3CCLS-LAP) did not affect ciliary localization, suggesting that these residues are not necessary in the context of a transmembrane domain similar to the native protein (Figs. 2 A and S2, C and E). The requirement of the palmitoylation motif in the context of nontransmembrane fusions of fibrocystin CLS is likely a result of their role in targeting the constructs to the membrane. As reported previously (Follit et al., 2010), mutating other motifs downstream of the cysteines (IKP>AAA, CD8fibrocystinIKP>AAACLS-LAP or KTRKIKP>AAAAAAA, CD8fibrocystin7ACLS-LAP) prevented ciliary localization in both CD8α transmembrane and nontransmembrane domain–containing fusions (Figs. 2 A and S2, C and D). Thus, the basic residue stretch consisting of the KTRKIKP motif constitutes the minimal CLS of fibrocystin.
The small G protein Arf4 has been previously reported to delay the trafficking of newly synthesized fibrocystin CLS-SNAP fusions to cilia without affecting steady-state levels (Follit et al., 2014). However, factors regulating steady-state levels of fibrocystin CLS in cilia are not known, except partial reduction upon Rab23 knockdown (Lim and Tang, 2015). Interestingly, upon RNAi-mediated knockdown of TULP3 in ciliated cells stably expressing CD8-fibrocystinWT/3CCLS-LAP or fibrocystinWTCLS-LAP fusions, we noted complete inhibition of steady-state localization to cilia (Figs. 2 B and S2 E). Lack of antibodies that detect endogenous fibrocystin prevented us from determining whether TULP3 is required for trafficking the full-length protein. Thus, TULP3 is required for ciliary trafficking of at least three distinct types of CLSs derived from integral membrane proteins, including fibrocystin and GPCRs.
Proximity biotinylation assays determine PI(4,5)P2-dependent CLS–TULP3 interactions
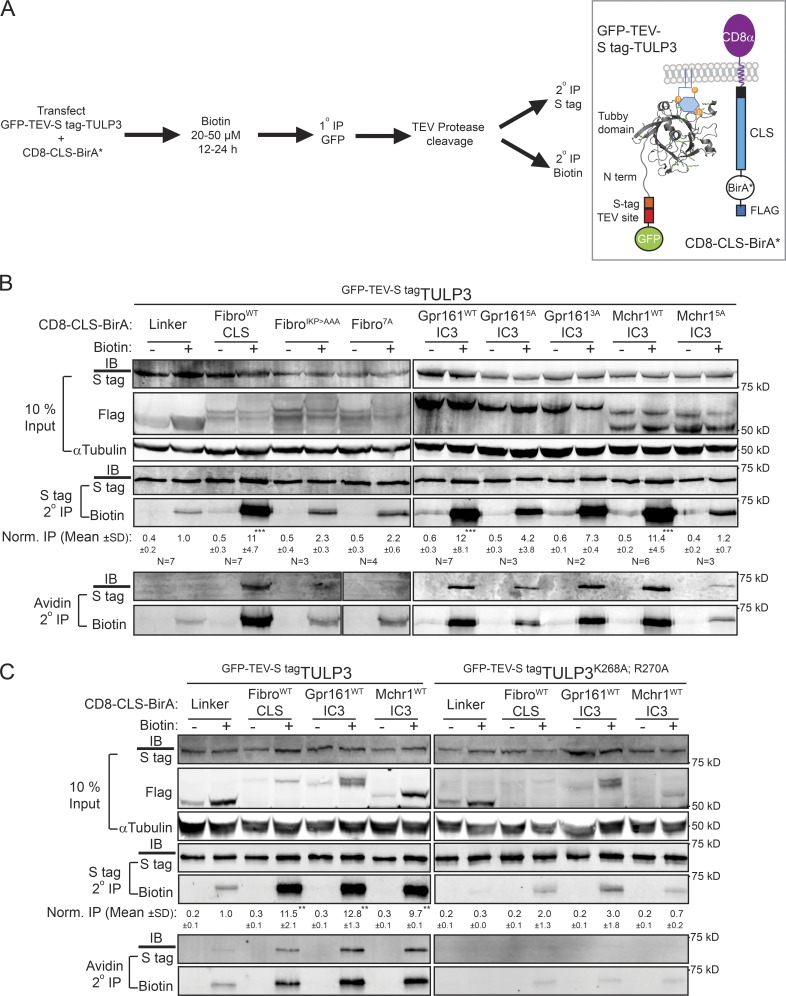
As TULP3 determines the trafficking of CLSs from multiple GPCRs and fibrocystin, we tested for physical interactions between the CLS fusions and TULP3. Typically, weak or transient interactions between adapters and their membrane cargo are difficult to detect by immunoprecipitation (IP), and we were unable to detect binding between CD8-CLSs and TULP3 using conventional coimmunoprecipitation assays. Hence, we used a promiscuous biotin protein ligase approach (biotin protein ligase mutant R118G; BirA*) to test for proximity between TULP3 and CD8-CLS fusions (Roux et al., 2012). Upon cotransfection of CD8-CLS-BirA*-Flag fusions and LAP-TULP3 in nonciliated T-Rex-293 cells and supplementing with biotin, we tested for cumulative biotinylation of LAP-TULP3 (Fig. 3 A). To remove background resulting from nonspecific biotinylation of GFP, we performed tandem affinity purification of LAP-TULP3 (N-terminal LAP, GFP-TEV-S tag) using anti-GFP cross-linked beads, followed by TEV protease digestion (Cheeseman and Desai, 2005). The digested S tag eluates were immunoprecipitated with either S protein agarose to determine biotinylation of total S tag–TULP3 pools using neutravidin dye conjugates (Roux et al., 2012) or with neutravidin agarose to determine S tag–TULP3 in biotinylated immunoprecipitates (Fig. 3 A).
Figure 3.
Proximity biotinylation assays determine CLS–Tulp3 membrane proximity. (A) T-Rex-293 cells were cotransfected with CD8-CLS-BirA* and LAP-TULP3 constructs and processed for a tandem immunopurification (IP) procedure for detecting and quantifying the extent of biotin labeling on LAP-TULP3 as indicated (left). Diagram representing the fusion constructs appears on the right. BirA* will biotinylate primary amine-containing lysine residues in TULP3 fusion, if in close proximity. (B) Western blots of inputs and tandem affinity IPs for the indicated conditions are shown after pretreatment with biotin (50 µM) for 24 h. IB for biotin refers to neutravidin-tagged IRDye 680RD antibody-based detection. Normalized (Norm.) IP values refer to biotin/S tag signal in corresponding S tag secondary IPs, with respect to the control linker biotin-pretreated sample. The number of experiments (N) performed is mentioned beneath each panel. Normalized IP values are reported as means ± SD. (C) T-Rex-293 cells were cotransfected with CD8-CLS-BirA* and the LAP-TULP3 wild type (WT) or PI(4,5)P2 binding-deficient mutant (TULP3K268A;R270A), treated with biotin (20 µM) for 12 h, and processed for tandem IP procedure for detecting and quantifying the extent of biotin labeling as in A. Normalized IP values are reported as means ± SD from two experiments; **, P < 0.01; ***, P < 0.001, with respect to respective control linker biotin-pretreated samples. Also see Figs. S3 and S4.
In contrast to the nonciliary CD8 control linker (GSG(A)5GSG; linker), all three types of CD8-CLS fusions (fibrocystinWTCLS, Gpr161WTIC3, and MCHR1WTIC3) resulted in significant biotinylation of S tag–TULP3 and correspondingly immunoprecipitated S tag–TULP3 in neutravidin agarose pull-downs (Fig. 3 B). In all nonciliary CD8-tagged mutant CLS constructs (Gpr1615AIC3, MCHR15AIC3, fibrocystinIKP>AAACLS, and fibrocystin7ACLS), biotinylation of S tag–TULP3 and IP in neutravidin pull-downs remained unchanged compared with the control linker (Fig. 3 B). Interestingly, the partially mutated CD8-Gpr1613AIC3 fusion that is still ciliary (VKARK>VTAAA) showed significantly more biotinylation and IP of TULP3 in neutravidin pull-downs in contrast to the nonciliary CLS fusion (Gpr1615A; Fig. 3 B). Thus, using a transient cotransfection proximity biotinylation approach, we determined that at least three types of CLS chimeric fusions and TULP3 were in close proximity and maintained specificity in these interactions.
The tubby domain of TULP3 binds to PI(4,5)P2 through a conserved motif similar to TUB (Santagata et al., 2001; Mukhopadhyay et al., 2010). Mutating key residues in the tubby domain of TULP3 that contribute to PI(4,5)P2 binding (TULP3K268A;R270A) completely prevented proximity-induced biotinylation by the CD8-CLS fusions (Fig. 3 C). Thus, plasma membrane association of TULP3 is required for close interactions with all three types of CLS chimeric fusions.
The tubby domain of TULP3 and TUB mediates CLS interactions
To identify the region of TULP3 that determines CLS proximity, we generated mutants of the IFT-A binding predicted helical region without changing lysine content (TULP3–N-terminal helix) and deleted the unstructured linker region between the N terminus and tubby domain of TULP3 (55–183 aa deletion; Fig. S3, A and B). Both of these constructs were effectively biotinylated to the same levels as wild type (Fig. S3, C and D), indicating that neither of these regions is critical for proximity. However, the isolated tubby domain of TULP3 (TULP3–C term; 184–442 aa) was biotinylated upon coexpression with CD8-CLS ciliary fusions and decreased with the nonciliary mutant fusions, indicating that this domain mediates proximity with diverse CLSs (Fig. S4 A). The relative amounts of biotinylation of TULP3 and TUB with homologous tubby domains were similar for all three different CLSs tested (Fig. S4, B and C). Thus, the tubby domain of TULP3 mediates the proximity with CLSs, and TUB interacts similarly to TULP3.
CLS-TULP3 proximity determined using cross-linking assays
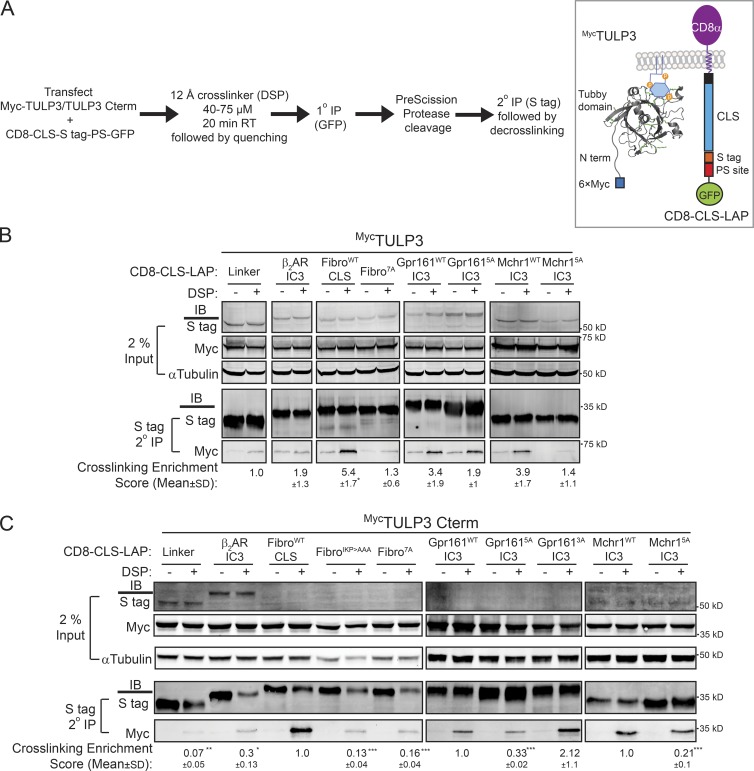
In the proximity biotinylation assays, biotin labeling occurs over a prolonged period of 12–24 h. Hence, we probed for proximity using chemical cross-linking for shorter time periods. Nonciliated T-Rex-293 cells cotransfected with CD8-CLS-LAP fusions and the Myc-TULP3 full-length or C-terminal tubby domain (TULP3–C term) were treated with the 12-Å reversible cell-permeable cross-linker dithiobis(succinimidyl propionate) (DSP) over a short time frame. As before, fusions of control linker (GSG(A)5GSG; linker) and the IC3 of the nonciliary β2 adrenergic receptor (β2AR IC3) were used as negative controls (Marley et al., 2013). The cells were treated with the cross-linker, lysed, and were subjected to tandem affinity purification of the CD8-CLS/mutant/control-LAP fusion using anti-GFP beads followed by S protein agarose and decrosslinked in sample buffer containing reducing agents (see the Chemical cross-linking experiments section of Materials and methods). We next determined levels of Myc-TULP3 in the immunoprecipitates (Fig. 4 A). The tandem affinity purification step removed background resulting from cross-linking of GFP, and the pull-down represents interacting complexes of the CD8-CLS–S tag fusions and Myc-TULP3. We also carefully optimized the cross-linking procedure for overreaction by testing for tubulin levels, taking reduced tubulin band intensities in the cross-linked samples as an indicator of excessive cross-linking.
Figure 4.
CLSs and TULP3 are chemically cross-linked in a ciliary sequence-specific manner. (A–C) T-Rex-293 cells were cotransfected with CD8-CLS-LAP and 6×Myc-TULP3 full-length (B) or 6×Myc–C-terminal tubby domain (C term, 184–442 aa; C) constructs, and processed for cross-linking, quenching, tandem IP procedure, and decross-linking as indicated in the format in the left panel in A. The right panel in A shows a diagram representing the fusion constructs. Western blots of inputs and tandem affinity IPs for the indicated conditions are shown in B and C. Cross-linking enrichment scores shown beneath each respective panel represents (Myc IP/S tag IP)DSP – (Myc IP/S tag IP)NoDSP for each CD8-CLS-LAP–transfected condition normalized with respect to control linker (B) and wild-type CLSs in individual panels in C. The number of lysines in the linker/CLS are as follows: linker GSGAAAAAGSG (0); β2 andrenergic receptor IC3 (β2AR IC3; RVFQEAKRQLQKIDKSEGRFHVQNLSQVEQDGRTGHGLRRSSKFCLKEHKALKT; 5); FibroWTCLS (12); FibroIKPCLS (11); Fibro7ACLS (9); Gpr161WTIC3 (4); Gpr1615AIC3 (2); Gpr1613AIC3 (2); Mchr1WTIC3 (1); and Mchr15AIC3 (1), as shown in Fig. 2 A. In addition, the rest of the fusion in CD8-CLS-LAP, including CD8α, the attB recombinase site, S tag, and the PreScission protease site from LAP, has a total of 11 lysines. All experiments were performed twice, except the last panel in C, which was performed three times. (B and C) Cross-linking enrichment scores are reported as means ± SD; *, P < 0.5 with respect to control linker (B); *, P < 0.05; **, P < 0.01; ***, P < 0.001with respect to wild-type CLS in each panel (C).
The CD8-CLS ciliary fusions of GPCRs and fibrocystin were significantly cross-linked to Myc-tagged full-length TULP3 or tubby domain (TULP3–C term; 184–442 aa), as compared with the nonciliary mutants and negative controls (Fig. 4, B and C). The cross-linker DSP uses N-hydroxysuccinimide chemistry to react with free amines on exposed lysine residues on proteins. The significant difference between ciliary and nonciliary CLSs is not an artifact of the mutational strategy of substituting key CLS lysines with alanines because the MCHR1 CLS is deficient in lysines. Additionally, the fibrocystinIKP>AAA mutant has only one lysine less than the wild-type fibrocystin CLS but was significantly less cross-linked to both full-length TULP3 and to the tubby domain, similar to fibrocystin7A (Fig. 4 C). The Gpr161WT and Gpr1613A mutant fusions were cross-linked significantly more than the nonciliary Gpr1615A mutant to the tubby domain of TULP3 despite having the same number of lysines between Gpr1613A and Gpr1615A (Fig. 4 C). Taking together proximity biotinylation and cross-linking between the CLS and TULP3 wild-type/C-terminal tubby or TUB pairs, we conclude that the tubby domain is responsible for proximity between the CLSs and TULP3.
Differential effects of TUB on ciliary GPCR trafficking
Ciliary trafficking of SSTR3, MCHR1, NPY2R, Gpr161, and GPR19 are all determined by TULP3 alone in cultured RPE hTERT or IMCD3 cell lines (Figs. 1 A and S1 A; Loktev and Jackson, 2013; Mukhopadhyay et al., 2013), which do not express TUB. As opposed to the ubiquitously expressed Tulp3, Tub is predominantly expressed only in brain and retinal photoreceptors, at levels higher than Tulp3 (Fig. S4, C and D; Mukhopadhyay and Jackson, 2011). As multiple GPCRs have been reported to localize to neuronal cilia, it is critical to understand whether TUB affects trafficking of these ciliary GPCRs similar to TULP3. Multiple groups have previously demonstrated Tub to be necessary for the trafficking of certain rhodopsin family GPCRs to neuronal cilia, including Sstr3, Mchr1, and Npy2r (Fig. 1 E; Sun et al., 2012; Loktev and Jackson, 2013). However, it is not known whether TUB localizes to cilia similarly to TULP3 and functions in an identical capacity in trafficking other GPCRs to neuronal cilia.
TUB has been reported to be predominantly nuclear in neurons (He et al., 2000). Similarly, stably expressed LAP-TUB is diffusely cytoplasmic and nuclear in localization without detectable steady-state ciliary localization (Figs. 5 A and S4 E). To further test whether TUB transits through cilia, we performed RNAi-mediated knockdown of Inpp5e. Inpp5e is a 5′-phosphatase that localizes to cilia, and knockouts result in accumulation of ciliary PI(4,5)P2, which increases ciliary pools of IFT-A and Tulp3, possibly resulting from persistent tubby domain–PI(4,5)P2 interactions (Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Interestingly, upon Inpp5e knockdown, we detected clear ciliary accumulation of LAP-TUB in multiple ciliated cell lines (Figs. 5 A and S4 E). Thus, TUB transits through cilia, and the lower steady-state levels in resting cilia in comparison to TULP3 are probably caused by less effective IFT-A binding in vivo, as evident from lower levels of IFT-A complex detected in LAP-TUB tandem affinity purifications (Mukhopadhyay et al., 2010). Using in vitro assays with recombinant MBPTUB and purified IFT-A complex, we detected that TUB bound to the IFT-A complex analogous to MBPTULP3 (Fig. 5 B). An N-terminal fragment of TULP3 (1–183 aa) blocks binding of recombinant TULP3 to purified IFT-A complex (Mukhopadhyay et al., 2010). Similarly, the N-terminal fragment of TULP3 prevented TUB–IFT-A complex binding in vitro (Fig. 5 B). Thus, the N-terminal fragment of TULP3 prevents binding of the IFT-A complex to both TUB and TULP3.
Figure 5.
Differential effects of Tub on ciliary GPCR trafficking. (A) IMCD3 Flp-In cells stably expressing LAP–TUB isoform b (Mukhopadhyay et al., 2010) were sequentially transfected with 200 nM of the indicated siRNAs twice for 72 h (see Materials and methods) and serum starved for the last 24 h before fixation and immunostained for GFP, acetylated tubulin, and DNA. GFP-positive cilia were counted in two experiments, and total counted cells are >400/condition. Data represent means ± SD. Bars, 5 µm. (B) MBPTUB isoform b and MBPTULP3 immobilized on amylose resin were incubated with PreScission eluates from IFT140LAP RPE cells ± HisTULP3 (1–183 aa; see Materials and methods; Mukhopadhyay et al., 2010). LAP, S tag–PreScission-GFP. MBPTUB- and MBPTULP3-bound proteins and corresponding flowthroughs were immunoblotted for S tag (IFT140S tag), maltose-binding protein (MBP), and His-tag as indicated. Data are representative of two experiments. (C and D) Embryonic day 16.5, day in vitro (DIV) 8 hippocampal neurons from wild-type (WT) and Tub mice were immunostained for Gpr161/Gpr19 (green), DyLight594-labeled ACIII (red), and Sstr3 (white; C). Gpr161/Gpr19-positive and -negative cilia are marked by arrows (A and C) and arrowheads (C), respectively. Gpr161/Sstr3 coexpressing cells are marked by yellow arrows. The red dot indicates debris under the coverslip. Data represents means ± SD from cultures of two different embryos belonging to each genotype. Total counted cells are >300 per condition for Gpr161/Sstr3 and >90 per condition for Gpr19. Bars, 5 µm. Ciliary lengths are 3.2 ± 0.1 µm (wild type) and 3.5 ± 0.1 µm (Tub) neurons in experiments with Gpr161 staining and 3.7 ± 0.9 µm (wild type) and 3.5 ± 0.8 µm (Tub) in experiments with Gpr19 staining. Data represent means ± SD. n > 50 each. (E) Embryonic day 16.5 DIV5 hippocampal neurons from wild-type mice were transfected with indicated GFP-tagged N terminus (NT) wild-type or non–IFT-A binding (mut12) TULP3 constructs (Mukhopadhyay et al., 2010), fixed at DIV8, and immunostained for Gpr161 and DyLight594-labeled ACIII. GFP-positive cells were quantified for Gpr161-positive cilia from two different coverslips, and total counted neurons are >24 per condition. (F) Embryonic day 18.5 DIV5 glia from wild-type mice were transfected with constructs as in C, fixed at DIV8, and immunostained for Gpr19 and Arl13b. GFP-positive cells were quantified for Gpr19-positive cilia from cultures from two different mice, and total counted cells are >110 per condition. (A and D–F) *, P < 0.001 with respect to corresponding siRNA control (A); *, P < 0.05 with respect to corresponding wild type; ns, not significant (D); *, P < 0.05 with respect to GFPTULP3-NTmut12 transfected cells (E); and *, P < 0.05 with respect to GFPTULP3-NTmut12-transfected cells. Also see Fig. S4.
To test if other GPCRs are affected by Tub, we determined ciliary trafficking of Gpr161 and Gpr19 in Tub-null mouse brains. We previously reported that Gpr161 localizes to cilia of cultured hippocampal neurons (Figs. 5 C and S4 F; Mukhopadhyay et al., 2013). Similarly, the orphan GPCR, Gpr19, is broadly expressed in the adult brain (O’Dowd et al., 1996). Using a newly developed affinity-purified polyclonal antibody against the C terminus, we detected the endogenous receptor in cilia of both cultured hippocampal neurons and in glia (Figs. 5 C and S4 G). Interestingly, endogenous Gpr161 and Gpr19 trafficking to cilia of cultured hippocampal neurons are not affected in Tub mutants, although Sstr3 trafficking was reduced as previously reported (Fig. 5, C and D; Sun et al., 2012). Expressing the dominant-negative N-terminal fragment of TULP3 that blocks binding of both TUB and TULP3 to IFT-A prevented Gpr161/Gpr19 localization to the cilia of cultured hippocampal neurons/glia (Fig. 5, E and F; and Fig. S4, F and G; Mukhopadhyay et al., 2010). Expression of the IFT-A nonbinding mutant (TULP3NTmut12) did not interfere with Gpr161/Gpr19 localization to cilia (Fig. 5, E and F; and Fig. S4, F and G; Mukhopadhyay et al., 2010). As Tub transits through cilia (Fig. 5 A), is in proximity to the CLSs in biotinylation assays (Fig. S4 B), and binds to the IFT-A core complex (Fig. 5 B; Mukhopadhyay et al., 2010), Tub likely determines ciliary GPCR trafficking in a process similar to Tulp3. Tub exclusively determines trafficking of a subset of GPCRs to neuronal cilia (Sstr3, Mchr1, and Npy2r), and there is a lack of any effect on Gpr161/Gpr19 in Tub knockouts. However, expressing TULP3NT that prevents binding of either TUB or TULP3 to IFT-A inhibits ciliary localization of Gpr161/Gpr19 in neuronal/glial cultures, indicating that both of these proteins possess overlapping functions in ciliary targeting of these receptors in the brain.
Tulp3 determines PC1/2 trafficking to cilia
To test whether the role of Tulp3 in the trafficking of membrane proteins to cilia extends beyond GPCRs and fibrocystin CLS, we further tested its role in the trafficking of polycystins, which belong to the TRP channel family of proteins. The polycystins PC1/2 form a complex in the ER and are trafficked to cilia in an interdependent manner (Cai et al., 2014; Kim et al., 2014; Gainullin et al., 2015). The C terminus cytoplasmic tail of PC2 interacts with a coiled-coil domain at the C terminus of PC1 (Qian et al., 1997). However, factors that mediate ciliary trafficking of polycystins are not known.
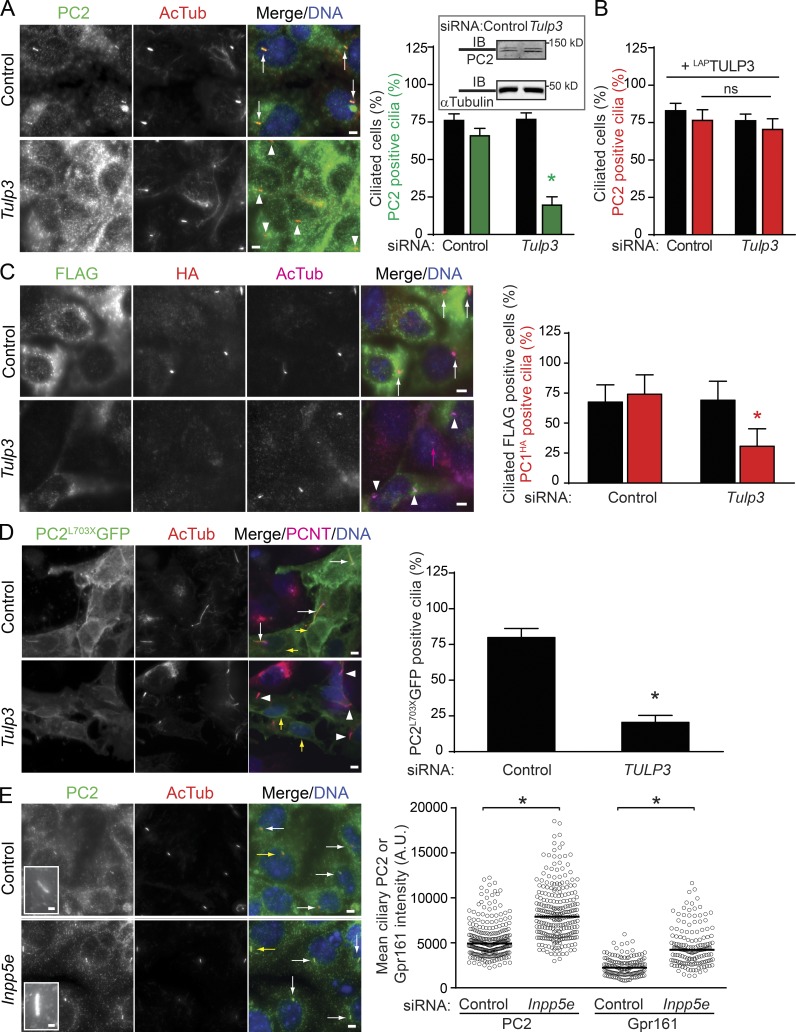
We tested for ciliary localization of polycystins in mIMCD-K2 cells (Kizer et al., 1995), which have been previously reported to localize endogenous PC2 in cilia at high levels (Yoder et al., 2002). Upon RNAi-mediated knockdown of Tulp3, endogenous Gpr161-positive cilia were reduced in mIMCD-K2 cells. Interestingly, the ciliary levels of endogenous PC2 in these cells, detected by two separate polyclonal antibodies, were also sharply reduced, without impacting on total protein levels (Figs. 6 A and S5, A and B). The decreased ciliary localization of endogenous PC2 upon siRNA-mediated Tulp3 knockdown was rescued upon stable expression of human LAPTULP3, ruling out nonspecific effects of RNAi (Fig. 6 B). PC1 traffics to cilia in a PC2-dependent manner, but we were unable to detect endogenous PC1 levels using available antibodies. Hence, we generated stable lines expressing FlagPC1HA in mIMCD-K2 cells (Cai et al., 2004). PC1 is autoproteolytically cleaved to generate extracellular N-terminal and intracellular C-terminal fragments (Ponting et al., 1999). The C-terminal HA-tagged fragment was localized to cilia, whereas the N-terminal Flag tag allowed determination of PC1-expressing cells. Upon RNAi-mediated knockdown of Tulp3, PC1HA-positive cilia were significantly reduced, implicating Tulp3 in trafficking of the PC1/2 complex (Fig. 6 C). We next determined whether PC2 by itself was affected in ciliary trafficking by Tulp3. The PC2703X truncation mutant, which is deleted for the PC1 interaction domain, still traffics to cilia independently of PC1 (Geng et al., 2006). In stable ARPE lines expressing PC2703XGFP, and upon RNAi-based TULP3 knockdown, PC2703XGFP was impaired in ciliary trafficking, although ER localization was not impacted (Fig. 6 D). Interestingly, stable overexpression of PC2703XGFP also resulted in increased ciliary length, as reported previously (Geng et al., 2006; Cai et al., 2014), and lower PC2703XGFP ciliary levels upon TULP3 knockdown coincided with a lack of increase in length (Fig. 6 D). The most parsimonious model would be that Tulp3 regulates the trafficking of the PC1/2 complex to cilia by directly interacting with PC2. Alternatively, Tulp3 could be interacting with both PC1 and PC2 in trafficking the complex to the cilia.
Figure 6.
PC1/2 trafficking to cilia requires Tulp3. (A) mIMCD-K2 cells were sequentially transfected with the indicated 200 nM siRNAs twice for 72 h and serum starved for the last 36 h before fixation and immunostained for PC2 using anti-mouse PC2 rabbit polyclonal serum (from G. Pazour; see the Antibodies section of Materials and methods), acetylated tubulin, and DNA. PC2-positive and -negative cilia are marked by white arrows and arrowheads, respectively. The inset shows PC2 levels in control and Tulp3 siRNA-treated cells by immunoblotting. Data represent means ± SD from three experiments. The total number of cells counted is >800 per condition. Quantification of PC2 using a separate antibody available commercially in Fig. S5 (A and B). (B) mIMCD-K2 cells expressing LAPTULP3 were treated as in A. The total counted cells are >350 for each condition. ns, not significant. (C) mIMCD-K2 cells stably expressing FlagPC1HA were sequentially transfected with the indicated 200-nM siRNAs twice for 72 h (see Materials and methods) and serum starved for the last 36 h before fixation and immunostained for Flag (green), HA (red), AcTub (magenta), and DNA. N-terminal Flag tag allowed determination of PC1-expressing cells. Flag-positive cells were counted for HA-positive cilia from three independent experiments, and total counted cells were >1,000 per condition with ∼10% cells being Flag positive. PC1HA-positive and -negative cilia are marked by white arrows and arrowheads, respectively. The magenta arrow marks a cilium in a cell not expressing Flag. Data represent means ± SD. (D) ARPE cells stably expressing PC2L703XGFP were transfected with the indicated 100 nM siRNAs for 72 h (see Materials and methods) and serum starved for the last 24 h before fixation and immunostained for GFP, acetylated tubulin, pericentrin, and DNA. GFP-positive cilia were counted from two experiments, and the total number of GFP-positive cells counted were >120 per condition. GFP-positive and -negative cilia are marked by white arrows and arrowheads, respectively. Yellow arrows point to perinuclear staining for PC2L703XGFP. Data represent means ± SD. Ciliary lengths of PC2L703XGFP-positive cells treated with control and TULP3 siRNA are 8.5 ± 4.6 and 3.2 ± 0.8 µm, respectively. n = 20 each. (E) mIMCD-K2 cells were transfected with indicated siRNAs as in A and immunostained for PC2 (using anti-mouse PC2 rabbit polyclonal serum) or Gpr161, acetylated tubulin, and DNA. Pixel intensities of PC2- and Gpr161-positive cilia are shown to the right. White arrows mark PC2-positive cilia, and yellow arrows point to cilia shown in insets. Total counted cells were >240 and >140 per condition for PC2 and Gpr161 immunofluorescence, respectively. (A and C–E) *, P < 0.001 with respect to corresponding siRNA control (A); *, P < 0.01 with respect to corresponding siRNA control (C and D); *, P < 0.0001 with respect to controls (E). A.U., arbitrary units. Bars: (A and C–E, main images) 5 µm; (E, insets) 1 µm. Also see Fig. S5.
Inpp5e knockdown increases ciliary pools of endogenous PC2
Depletion of Inpp5e causes accumulation of PI(4,5)P2 in cilia, resulting in increased steady-state levels of Tulp3, IFT-A, and Gpr161 in the compartment (Chávez et al., 2015; Garcia-Gonzalo et al., 2015). In addition, removal of Gpr161 from cilia upon activation of the Shh pathway is impaired in Inpp5e knockouts (Chávez et al., 2015; Garcia-Gonzalo et al., 2015), irrespective of accumulation of the pathway activator Smoothened (Smo) in the compartment (Fig. S5 D). To test whether steady-state levels of other Tulp3-regulated cargo are also impacted upon Inpp5e knockdown, we tested for endogenous PC2 levels in mIMCD-K2 cilia. Interestingly, both PC2 and Gpr161 ciliary pools were increased upon Inpp5e knockdown in these cells (Fig. 6 E). Additionally, in IMCD3 cells, cilia positive for endogenous PC2 increased sharply upon Inpp5e knockdown (Fig. S5 C). Membrane cargoes that are not regulated by Tulp3, such as Smo and adenylyl cyclase III (ACIII), are not affected upon down-regulation of Inpp5e (Fig. S5 D; Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Thus, two different types of Tulp3-regulated cargo accumulate in the abnormally PI(4,5)P2-enriched ciliary membrane upon Inpp5e knockdown.
Tulp3 determines entry of Gpr161 into cilia
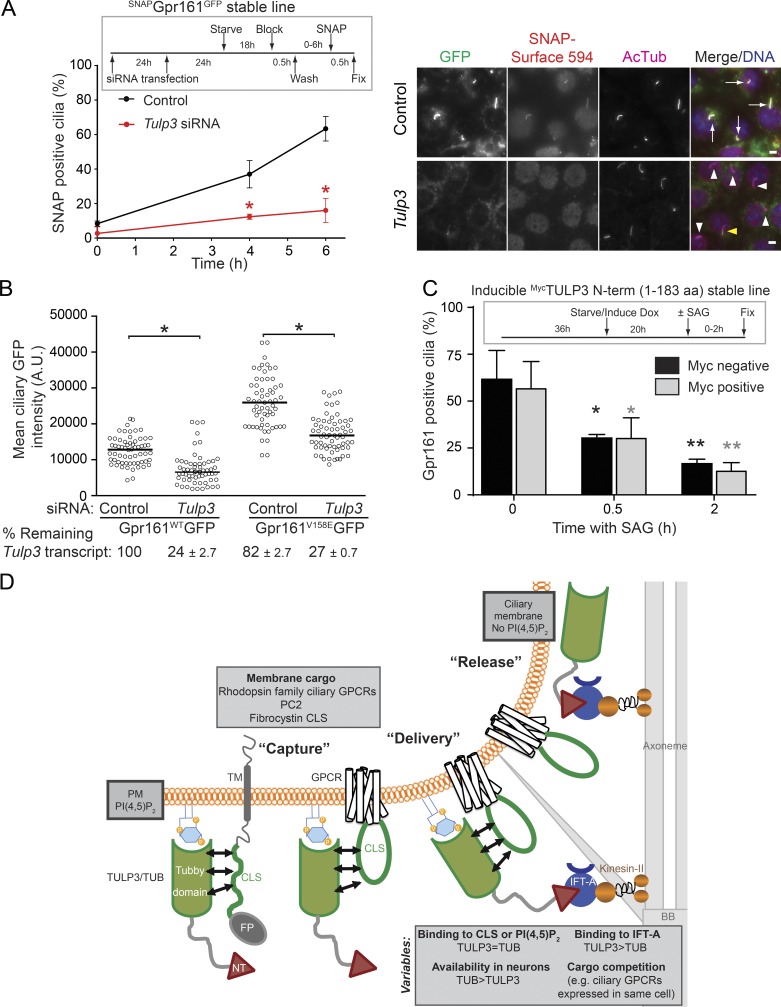
Levels of a protein in ciliary membrane can be regulated at multiple steps, including trafficking to cilia, removal from the compartment, or additional steps in cargo recycling (Pazour and Bloodgood, 2008; Bloodgood, 2012). Therefore, we tested the direct role of Tulp3 in cargo entry into cilia and exit from the compartment. As Gpr161 trafficking in and out of cilia has been well studied, we tested the role of Tulp3 in determining ciliary pools of this particular GPCR (Pal et al., 2016). First, we tested the dynamics of ciliary entry of a stably expressed Gpr161 that was tagged at the extracellular N terminus with SNAP (SNAPGpr161GFP) upon Tulp3 knockdown. After blocking surface pools of SNAPGpr161GFP, we tracked accumulation of newly trafficked proteins in cilia by surface labeling with a non–cell-permeable SNAP substrate (SNAP-surface) at different time points. We detected a sharp decrease of SNAP-surface–labeled cilia upon Tulp3 knockdown, suggesting inhibition of ciliary entry (Fig. 7 A). Next, we tested ciliary pools of a Gpr161 mutant lacking constitutive signaling (Gpr161V158E) that is not removed from cilia (Pal et al., 2016). In stable 3T3 Flp-In cells expressing the Gpr161V158E mutant, we detected a decrease in ciliary pools upon Tulp3 knockdown, suggesting that Tulp3 regulates Gpr161 trafficking into cilia (Figs. 7 B and S5 E). Finally, using an IMCD3 Flp-In stable cell line that inducibly expresses the dominant-negative MycTULP3 N terminus (1–183 aa) upon addition of doxycycline, we tested whether the Shh pathway–dependent removal of endogenous Gpr161 was affected upon induction (Fig. 7 C). We did not detect any differences in removal of Gpr161 from cilia upon addition of the Smo agonist SAG between Myc-positive and -negative cells. Thus, blocking endogenous Tulp3–IFT-A binding by the Myc-TULP3 N terminus does not regulate removal of Gpr161 from cilia (Figs. 7 C and S5 F; Mukhopadhyay et al., 2010). Overall, Tulp3 directly determines entry of Gpr161 into cilia without affecting removal from the compartment.
Figure 7.
TULP3/TUB-mediated trafficking to ciliary membrane. (A) IMCD3 cells stably expressing SNAPGpr161GFP were sequentially transfected with 200 nM of the indicated siRNAs twice as indicated in the experimental format on the top left (see Materials and methods). Starved cells were blocked with SNAP–surface block for 30 min, washed, and then were treated with SNAP–surface 594 at indicated time points. Cells were fixed and immunostained for GFP (green) and acetylated tubulin (magenta). Cilia positive and negative for SNAP–surface 594 are marked by white arrows and arrowheads, respectively. The yellow arrowhead points to cilia faintly stained for SNAP–surface 594. Total counted cells are >150 for control and >200 for Tulp3 siRNA, respectively. Data represent means ± SD from two or more fields from a single experiment. Bars, 5 µm. (B) NIH 3T3 Flp-In cells stably expressing Gpr161-GFP or Gpr161V158E-GFP mutants were sequentially transfected with 200 nM of indicated siRNAs twice for 72 h (see Materials and methods) and serum starved for the last 24 h before fixing and immunostaining for GFP, acetylated tubulin, and DNA. Ciliary pixel intensities for GFP are shown. Total counted cells were >60 per condition from two independent transfections with >30 cells counted per coverslip. A.U., arbitrary units. Also see Fig. S5 E. (C) IMCD3 Flp-In cells stably and inducibly expressing MycTULP3 N terminus (1–183 aa) were starved in the presence of doxycycline (Dox) for 20 h (4 µg/ml). After washing, cells were treated ± SAG (500 nM) for indicated time points in starvation medium before fixing and immunostaining for Gpr161, Myc, acetylated tubulin, and DNA. Myc-positive and -negative cells were scored for Gpr161-positive cilia. Total counted cells are >55 and >180 for Myc-positive and -negative cells, respectively, for each time point. Data represent means ± SD from three coverslips. Also see Fig. S5 F. (A–C) *, P < 0.001 with respect to corresponding siRNA control at each time point measured (A); *, P < 0.0001 with respect to corresponding controls (B); *, P < 0.05; **, P < 0.01 with respect to corresponding 0 h time point (C). (D) Model for TULP3/TUB-mediated trafficking to ciliary membrane. See Discussion. BB, basal body; FP, fusion protein; NT, TULP3/TUB IFT-A binding N-terminus domain. Also see Fig. S5.
Discussion
TULP3 is a general adapter for trafficking integral membrane proteins to cilia
TULP3 functions as a general adapter for ciliary trafficking of structurally diverse integral membrane cargo, including at least 16 out of 23 reported and two novel class A (rhodopsin family) cilia-targeted GPCRs, fibrocystin CLS fusions, and polycystic kidney disease–causing PC1/2 complex. We include two novel cilia-targeted GPCRs, the orphan GPR19 and P2RY1, to the repertoire of TULP3-dependent GPCRs. Notably, other integral membrane proteins, such as the TRP channel family polycystins PC1 and PC2, which traffic to cilia interdependently in a complex, also require Tulp3 for ciliary trafficking. Although TULP3 regulates integral membrane protein cargo, PDE6D and UNC-119b determine trafficking of membrane-associated prenylated (Humbert et al., 2012) and myristoylated cargo (Wright et al., 2011) by binding to the respective lipid moieties. Similarly, Tulp3 does not regulate trafficking of Arl13b, a membrane-associated palmitoylated protein to cilia (Fig. 5 F).
Although TULP3 regulates the trafficking of multiple types of integral membrane proteins to cilia, exceptions to this rule are as follows. First, Smo, a frizzled family seven-transmembrane protein activator of the pathway that is in constant flux through cilia, and is enriched upon transitioning to an "activated" state (Corbit et al., 2005; Rohatgi et al., 2009), is not regulated by Tulp3 (Norman et al., 2009; Mukhopadhyay et al., 2010). Second, and most notably, the cilia-targeted multispan adenylyl cyclases traffic to cilia independent of Tulp3/Tub (Bishop et al., 2007; Mukhopadhyay et al., 2010; Choi et al., 2011; Loktev and Jackson, 2013). In addition, the role of Tulp3 in other ciliary membrane and ciliary pocket–coordinated signaling pathways involving transforming growth factor–β receptors Ι/ΙI, receptor tyrosine kinases, and Notch receptors is presently unknown (Ezratty et al., 2011; Christensen et al., 2012; Pedersen et al., 2016).
A three-step model for TULP3-mediated trafficking to cilia
Mechanistically, we propose that the delivery of integral membrane proteins to cilia by TULP3 involves the following three steps (Fig. 7 D). The first step involves the capture of membrane cargo by TULP3 in the plasma membrane in a PI(4,5)P2-dependent manner. Using proximity biotinylation and cross-linking assays, we show that the tubby domain of TULP3 is in close proximity to at least three different types of CLSs from ciliary GPCRs and fibrocystin. Proximity of the tubby domain to the CLSs requires intact membrane PI(4,5)P2 interactions, suggesting a coincidence detection model, reminiscent of AP2, AP180, and epsin adapter–mediated recruitment of associated cargo during clathrin-mediated endocytosis (Bonifacino and Traub, 2003).
The second step involves the delivery of the CLS–TULP3 complex into the ciliary compartment. The IFT-A complex accumulates at the base of the cilia in the transition zone (Sedmak and Wolfrum, 2010). We previously demonstrated that the ciliary localization of TULP3 absolutely depends on association with the IFT-A core complex (Mukhopadhyay et al., 2010). The CLS–Tulp3 complexes formed at the plasma membrane (step one; Fig. 7 D) likely encounter and bind to the IFT-A complex to form the CLS–Tulp3/IFT-A ternary complex at or near the base of the cilia. Although the PI(4,5)P2-deficient ciliary membrane is unfavorable for either TULP3 to remain membrane associated or for the CLS complex to persist, the IFT-A interaction possibly ensures the integrity of the CLS–TULP3 complex as it moves across the transition zone membrane barrier into cilia.
The third step involves release of cargoes into the ciliary compartment. In line with the cargo–TULP3 interaction being PI(4,5)P2 dependent, lack of PI(4,5)P2 in cilia would result in weakening of cargo–TULP3 interactions upon ciliary delivery and subsequent cargo release. We propose this coordinated release to be crucial for the maintenance of steady-state levels of cargo. Depletion of Inpp5e and the resulting accumulation of PI(4,5)P2 in the ciliary compartment results in increased steady-state ciliary pools of Tulp3/IFT-A and Tulp3-dependent cargo such as Gpr161 and PC2, unlike Tulp3-independent integral membrane proteins (ACIII and Smo; Chávez et al., 2015; Garcia-Gonzalo et al., 2015). In addition, removal of Gpr161 from cilia upon activation of the Shh pathway is impaired in Inpp5e knockouts (Chávez et al., 2015; Garcia-Gonzalo et al., 2015). The reduced off rate of the cargo-Tulp3 interactions in Inpp5e deficient cilia might also impact on TULP3 flux through cilia by IFT trains, resulting in abnormal accumulation of Tulp3 and IFT-A. Accumulation of membrane cargo in cilia can be damaging (e.g., retention of Gpr161 in cilia impacts on Shh signaling and neuronal differentiation; Chávez et al., 2015; Garcia-Gonzalo et al., 2015; Pal et al., 2016).
Differential effects of Tulp3 and Tub on ciliary GPCR trafficking
TUB functions in ciliary GPCR trafficking in a process analogous to TULP3. First, TUB associates with the IFT-A core through the N terminus–conserved helix (Mukhopadhyay et al., 2010), and the TULP3 N terminus fragment prevents this binding. Second, TUB transits through cilia, and although it is not detectable in resting conditions, it accumulates in the compartment upon Inpp5e knockdown, as previously shown for Tulp3 (Chávez et al., 2015; Garcia-Gonzalo et al., 2015). Finally, TUB interacts with three different CLSs in proximity biotinylation assays in a ciliary sequence–specific manner, similar to TULP3.
Interestingly, Tub and Tulp3 demonstrate differential requirements for trafficking of class A GPCRs to neuronal/glial cilia, with some GPCRs requiring Tub and others trafficking independently, but both categories are perturbed by the TULP3 N terminus. Multiple GPCRs such as Gpr161, Sstr3, and Gpr19 are coexpressed in hippocampal neuronal cilia (Fig. 5 C), similar to Caenorhabditis elegans chemosensory neurons (Bargmann, 2006). The differential effects on ciliary localization of GPCRs in Tub mutants could be a consequence of (a) the relative levels of the respective GPCRs that determine their availability for binding to Tub/Tulp3, (b) relative levels of Tub/Tulp3, and (c) differences in binding to the IFT-A core, with TULP3 being more effective than TUB (Fig. 7 D). Thus, less-expressed GPCRs are preferentially trafficked first by being captured by the highly expressed but poor IFT-A–binding Tub. However, highly expressed GPCRs, such as Gpr161 and Gpr19, require Tulp3 and Tub redundantly for trafficking to the compartment.
Tubby domain interactions with diverse cargo
The tubby domain by itself is in close proximity to diverse CLSs from cilia-targeted GPCRs and fibrocystin. Structurally, this process could be analogous to the binding of diverse nuclear recognition sequences by Karyopherin β proteins (Lee et al., 2006) or CRM1-cargo recognition and release in nuclear export pathways (Fung and Chook, 2014). The minimal structural requirement of these diverse sequences might be to generate secondary structure elements that could interact with the tubby domain, similar to the interaction between Gβγ subunit surface and binding peptides (Davis et al., 2005). The IC3 of GPCRs becomes part of the extended fifth transmembrane helix of activated receptors during Gα-protein subunit coupling (Flock et al., 2015), and similar structural elements in this region could mediate interactions with the tubby domain.
Tulp3 and trafficking of polycystins to cilia
In addition to class A GPCRs and fibrocystin CLS, Tulp3 determines ciliary trafficking of the TRP channel family polycystins PC1 and PC2 that traffic to cilia interdependently and the C terminus–deleted PC2L703X mutant that traffics to cilia in a PC1-independent manner. Thus, TULP3 probably regulates PC1/2 complex trafficking by directly regulating PC2. Interestingly, Drosophila melanogaster IFT-A and king tubby (dTulp) mutants have been reported to lack trafficking of the TRPV and TRPN channels to the chordotonal cilia (Lee et al., 2008; Park et al., 2013). Nonciliary mutants of both PC1 and PC2 that are not compromised otherwise cause polycystic disease, suggesting a possible causative role of PC1/2 trafficking to cilia in cyst formation (Cai et al., 2014). Paradoxically, lack of cilia suppresses cyst formation in polycystic kidney disease models (Ma et al., 2013). Tulp3-mediated transport of these integral membrane proteins into cilia possibly prevents a cilia-dependent cystogenic pathway precluding cyst formation (Ma et al., 2013).
In conclusion, our results outline a mechanism inclusive of both Tulp3- and Tub-dependent ciliary trafficking of diverse types of integral membrane proteins. The tripartite steps for capture by Tulp3/Tub, delivery by IFT-A, and release in the PI(4,5)P2-deficient ciliary membrane, with additional layers of complexity in competitive cargo capture between Tub/Tulp3 and delivery mechanisms resulting from differences in IFT-A binding, have important ramifications for ciliary cargo delivery in the brain. By defining Tulp3/Tub as a key factor in trafficking diverse types of membrane cargoes, our study has broader implications in understanding the molecular basis of cilia-generated signaling in ciliopathies, including polycystic kidney disease.
Materials and methods
Antibodies and reagents
Rabbit polyclonal anti-GFP was from J. Seeman (University of Texas Southwestern Medical Center, Dallas, TX), anti-mouse PC2 rabbit polyclonal serum was from G. Pazour (University of Massachusetts Medical School, Worcester, MA; Fig. 6, A and E; and Fig. S5 C), and anti-Smo rabbit polyclonal was from K. Anderson (Memorial Sloan Kettering Cancer Center, New York, NY). Affinity-purified rabbit polyclonal antibody against mouse Gpr161 has been described previously (Pal et al., 2016). An affinity-purified rabbit polyclonal antibody against a C-terminal sequence in mouse Gpr19 (NCBI RefSeq database accession no. NP_001161166; 386–409 aa; Cys-KDSIYDSFDREAREKKLAWPINSN) was generated (Yenzym). Commercial antibodies used were against α-tubulin (rat YL1/2; Santa Cruz Biotechnology, Inc.), acetylated α-tubulin (mAb 6-11B-1; Sigma-Aldrich; and rabbit polyclonal 5335; Cell Signaling Technology), rabbit polyclonal PC2 (H-280; Santa Cruz Biotechnology, Inc.; Fig. S5, A and B), S tag (mouse monoclonal MAC112; EMD Millipore), Myc tag (goat polyclonal ab9132; Abcam [for immunoblotting and immunofluorescence]), Flag (goat polyclonal ab1257; Abcam [for immunoblotting]; and M2 monoclonal F1804; Sigma-Aldrich [for immunofluorescence]), His tag (mouse monoclonal GT359; Sigma-Aldrich), MBP (mouse monoclonal; New England Biolabs, Inc.), Arl13b (mouse monoclonal N295B/66; NeuroMab), Sstr3 (goat polyclonal sc-11617; Santa Cruz Biotechnology, Inc.), and ACIII (rabbit polyclonal sc-588; Santa Cruz Biotechnology, Inc.). DyLight 594–labeled ACIII antibodies were prepared using the DyLight 594 Microscale Antibody Labeling kit (53045; Thermo Fisher Scientific). Fluorescent secondary antibodies for immunofluorescence were from the Jackson ImmunoResearch Laboratories, Inc., and IRDye 680RD, IRDye 800CW secondary antibodies, and neutravidin 680RD for immunoblotting were from LI-COR Biosciences. DSP was from Thermo Fisher Scientific and biotin was from Sigma-Aldrich. SNAP–surface block or SNAP–surface 594 was from New England Biolabs, Inc. Most other reagents were from Sigma-Aldrich.
Plasmids
pG-LAP1 (pCDNA5/FRT/TO-EGFP-TEV-Stag-X) and pG-LAP5 (pEFα-X-Stag-PreScission-EGFP) were from Addgene (Torres et al., 2009). Human TULP3, TUB isoform b, and N- and C-terminal fragments of human TULP3 were generated by Gateway cloning into pG-LAP1 for GFP fusions and into a gatewaytized pCS2-Myc vector for N-terminal 6×Myc fusions (Mukhopadhyay et al., 2010). For inducible expression of the 6×Myc-tagged TULP3 N terminus (1–183 aa), the insert was cloned into pLVX TetOne vector (Takara Bio Inc.) at the BstZ1 site. C-terminal expression constructs for GPCRs were generated by Gateway cloning into pG-LAP5 and for retroviral infections into a gatewaytized LAP5 version of pBABEPuro. Single or multiple amino acid mutations in GPCR CLSs were generated using the Quikchange Site-Directed Mutagenesis kit (Agilent Technologies) or Phusion high-fidelity DNA polymerase (New England Biolabs, Inc.). C-terminal fusions of the CLSs with the extracellular and transmembrane domain of human CD8α (1–206 aa of NCBI RefSeq database accession no. NP_001139345, followed by a KRLK linker) were generated by subcloning the PCR-amplified CLS regions into the AflIII–NotI site of a CD8α/LDLR construct from M. Mettlen (Mettlen et al., 2010). The CD8α-CLS fragments were further subcloned into pG-LAP5 or pBABELAP5 by Gateway cloning to generate CD8-CLS-LAP5 fusions. All constructs were verified by sequencing. All CD8 constructs used in the proximity biotinylation approach were similarly made by Gateway cloning in pDEST-pcDNA5-BirA-FLAG C-term from A.-C. Gingras (The Lunenfeld-Tanenbaum Research Institute at Mount Sinai Hospital, Toronto, Canada). All class A GPCR constructs were obtained from the DNASU Plasmid Repository or the Ultimate ORF Clone Library at the McDermott Center, University of Texas Southwestern Medical Center, unless otherwise mentioned. DNASU clone catalog numbers are as follows: DNASU clones HsCD00040866 (D2R long isoform); HsCD00040882 (D5R); HsCD00718625 (GALR2); HsCD00353856 (GALR3); HsCD00353868 (GPR88); HsCD00515685 (HTR6); HsCD00512792 (NPFFR1); HsCD00288079 (GPR83); HsCD00516096 (KISS1R); HsCD00512758 (NMUR1); HsCD00515567 (P2RY1); MmCD00297454 (Pgr15l); HsCD00353806 (PTGER4); and HsCD00510746 (QRFPR). Invitrogen GPCR clone catalog numbers are as follows: IOH29586 (PRHLR) and IOH28333 (TAAR1). The human C-terminal GFP-tagged GPR19 clone was from OriGene (RG220379). The D1R-eGFP construct was from K. Mykytyn (The Ohio State University, Columbus, OH). D2R short isoform (NCBI RefSeq database accession no. NP_057658) was generated by site-directed mutagenesis using HsCD00040866 (DRD2 long isoform). FlagPC1HA was from S. Somlo (Yale University, New Haven, CT), in which triple HA tag was introduced in-frame immediately before the stop codon, and a triple FLAG tag was introduced in-frame between the leader sequence and leucine-rich repeats (LRR) domain between codons 24 and 25 (Cai et al., 2004). PC2L703XGFP was from G. Pazour. A pENTR plasmid expressing signal peptide–SNAP-Gpr161 (with Gpr161 lacking the first 27 aa from NCBI RefSeq database accession no. NP_001074595) was synthesized by Thermo Fisher Scientific, and C-terminal expression constructs were generated by Gateway cloning into pG-LAP5.
Cell culture and generation of stable cell lines
RPE hTERT and ARPE-19 cells were grown in DMEM F12 media with 10% FBS (Sigma-Aldrich), 0.05 mg/ml penicillin, 0.05 mg/ml streptomycin, and 4.5 mM glutamine. mIMCD-K2 cells were a gift from B. Stanton (Dartmouth College, Hanover, NH). T-Rex-293 (Invitrogen), IMCD3 Flp-In, and Phoenix A (PhA) cells (Indiana University National Gene Vector Biorepository) were cultured in DMEM high glucose (Sigma-Aldrich; supplemented with 10% FBS, 0.05 mg/ml penicillin, 0.05 mg/ml streptomycin, and 4.5 mM glutamine). Transfection of plasmids was done with Polyfect (QIAGEN). Stable cell lines were generated by retroviral infection or transfection. NIH 3T3 Flp-In cell lines stably expressing Gpr161WT/V158E-LAP (Gpr161 followed by Speptide-PrecissionS-EGFP [LAP5]) were generated by retroviral infection and antibiotic selection (Pal et al., 2016). IMCD3 Flp-In cells stably and inducibly expressing 6×Myc-tagged TULP3 N terminus (1–183 aa) were generated by lentiviral infection of the pLVX TetOne vector (Takara Bio Inc.) with the insert. In many cases, stable lines were flow sorted and further selected for GFP.
siRNA transfection
The siRNAs for TULP3/Tulp3 or Inpp5e were predesigned On-target Plus (OTP) siRNA duplexes shown to yield a reduced frequency of off-target effects (GE Healthcare; Jackson et al., 2006). RPE hTERT, IMCD3 Flp-In, mIMCD-K2, and NIH 3T3 cells were passaged on glass coverslips and transfected with siRNA using Lipofectamine RNAiMax (Invitrogen). The OTP siRNA sequences are as follows: TULP3 OTP3 (si #3; J-011415-07), 5′-GAAACAAACGUACUUGGAU-3′; TULP3 OTP7 (si #7; J-011415-11), 5′-GCAGCUAGAAAGCGGAAAA-3′ (Mukhopadhyay et al., 2010); Tulp3 OTP2 (si #2; J-043811-06) 5′-GAACACGAGAGCUUGCUUU-3′ (Mukhopadhyay et al., 2013); INPP5E (J-020852-05) 5′-GGAAUUAAAAGACGGAUUU-3′ (Humbert et al., 2012); and Inpp5e OTP3 (J-041108-07), 5′-GGAAUUAAAAGGCGGAUUU-3′. OTP nontargeting pool (GE Healthcare) was used as control siRNAs in all experiments. Unless otherwise indicated, the siRNA transfection methods were as follows: (a) RPE hTERT and ARPE cells, 100 nM by forward transfection; or (b) IMCD3 Flp-In, mIMCD-K2 cells, NIH 3T3 cells, and IMCD3 SNAPGpr161GFP cells, 200 nM by reverse transfection followed by 200 nM by forward transfection 24 h after plating. Transfection duration is calculated from first transfection.
Immunofluorescence and microscopy
Cells were cultured on coverslips until confluent and starved for indicated periods. Surface labeling of starved cells with SNAP–surface block or SNAP–surface 594 at indicated time points in Fig. 7 A was performed according to the manufacturer’s instructions (New England Biolabs, Inc.). In brief, a 4-mM stock solution of SNAP–surface block in DMSO was added to a final concentration of 20 µM in starving medium, and cells were incubated at 37°C and 5% CO2 for 30 min. Similarly, a 1-mM stock solution of SNAP–surface 594 in DMSO was added to a final concentration of 5 µM in starving medium, and cells were incubated in the dark at 37°C and 5% CO2 for 30 min. Cells were fixed with 4% PFA. When using antibodies against GFP/pericentrin/PC2/HA/FLAG, the cells were postfixed with methanol for 5 min at −20°C. After blocking with 5% normal donkey serum, the cells were incubated with primary antibody solutions for 1 h at room temperature followed by treatment with secondary antibodies for 30 min along with Hoechst 33342 (Invitrogen). For immunofluorescence analysis of hippocampal neurons, the neurons were stained with rabbit anti-Gpr161 and goat anti-Sstr3 antibodies, labeled with secondary antibodies/Hoechst 33342, and washed. This was followed finally by incubation with the DyLight 594–labeled rabbit anti-ACIII antibodies for 30 min and washed. The coverslips were mounted using Fluoromount G (SouthernBiotech). Images were acquired on a microscope (AxioImager.Z1; ZEISS), a sCMOS camera (PCO Edge; BioVision Technologies), and Plan Apochromat objectives (10×/0.45 NA; 20×/0.8 NA; 40×/1.3 NA oil; and 63×/1.4 NA oil) controlled using Micro-Manager software (University of California, San Francisco) at room temperature. Between 8 and 20 z sections at 0.5–0.8-µm intervals were acquired. For quantitative analysis of ciliary localization, stacks of images were acquired from three to eight consecutive fields with confluent cells by looking into the DAPI channel, and percentages of receptor/channel-positive ciliated cells were counted. Maximal projections from images of stacks were exported from ImageJ/Fiji (National Institutes of Health) using a custom-written macro (from M. Mettlen, University of Texas Southwestern Medical Center, Dallas, TX) using similar parameters (image intensity and contrast) for image files from the same experiment. For measuring ciliary pixel intensities, image stacks were acquired with z sections at 0.5-µm intervals. An image interval with maximal intensity was chosen, and cilia were demarcated with a region of interest using fluorescence signal for antiacetylated α-tubulin. The mean pixel intensities for the corresponding surface protein were exported from ImageJ/Fiji.
Proximity biotinylation experiments
T-Rex-293 cells were cotransfected with 5–7.5 µg each of LapN-Tulp3 or LapN-Tubby isoform B or LapN-Tulp3 mutants/fragments and pDEST-pcDNA5-BirA-FLAG C-term expressing the CD8-CLS-BirA fusions. The media was supplemented with 20–50 µM biotin 12–24 h after transfection. Cells were harvested using PBS with 2 mM EDTA and 2 mM EGTA 48 h after transfection. Cells were lysed by resuspending and nutating for 20 min in 50 mM Tris-HCl, pH 7.4, 200 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 1 mM DTT, 0.6% IGEPAL CA-630, 1 mM AEBSF, and 0.01 mg/ml each of leupeptin, pepstatin, and chymostatin. Lysates were centrifuged at 12,000 g for 10 min followed by tandem IPs (Fig. 3). In brief, the GFP immunoprecipitates were first digested with TEV protease for 16 h at 4°C. The supernatants were split to two aliquots and subjected to secondary IPs with S protein agarose and neutravidin, respectively. The resulting secondary IPs were analyzed by Western blotting. Blots were probed with antibodies against S tag (mouse monoclonal MAC112) and Flag (goat polyclonal ab1257) followed by visualization using IRDye-tagged secondary antibodies. IRdye-tagged neutravidin was used for confirming biotinylation signal on the blots.
Chemical cross-linking experiments
T-Rex-293 cells were cotransfected with 5–7.5 µg each of Myc-Tulp3 or Myc-Tulp3 C-terminal fragment and pGLAP5 CD8-CLS. 48 h after transfection, cells were washed three times with PBS. Cells were overlaid with 40–75 µM DSP in a 10% DMSO solution in PBS and rocked gently for 15 min. The DSP cross-linking reaction was quenched, and cells were harvested with 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, and 2 mM EGTA. Care was taken to avoid any contamination with primary amine-containing reagents before the quenching step. The cells were lysed in a buffer containing 50 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 0.6% IGEPAL CA-630, 1 mM AEBSF, and 0.01 mg/ml each of leupeptin, pepstatin, and chymostatin. Lysates were centrifuged at 12,000 g for 10 min followed by tandem IPs (Fig. 5). In brief, the GFP immunoprecipitates were first digested with PreScission protease for 16 h at 4°C. The eluates were subjected to a secondary IP with S protein agarose. The resulting secondary IP consisted of S tag–containing fusion protein cross-linked to proximal proteins via the DSP. Decross-linking was performed by incubating the mixtures with 150 mM DTT and 10 mM β-mercaptoethanol for 2 h at RT, followed by Western blotting. Blots were probed with antibodies against S tag (mouse monoclonal MAC112) and Myc tag (goat polyclonal ab9132) followed by visualization using IRDye-tagged secondary antibodies.
Primary hippocampal neuronal and glial culture
Hippocampi were dissected from embryonic day 16.5–18.5 CD1 mice (Charles River) and incubated in 1× trypsin-EDTA solution (T4174; Sigma-Aldrich) for 15 min at 37°C. Next, cells were dissociated by trituration with PBS containing 10% cosmic calf serum (SH30087.04; Hyclone) and 75 U DNase I (D5025; Sigma-Aldrich). Cells were centrifuged (491 g for 8 min at 4°C), and then neurobasal medium (21103–049; Gibco) was added. 12-mm cover glass (12–545-80; Thermo Fisher Scientific) was coated with poly-d-lysine (P4707; Sigma-Aldrich) and laminin (L2020; Sigma-Aldrich) and placed in a 24-well plate. Neurobasal medium with 1× B27 supplement (127504044; Thermo Fisher Scientific), l-glutamine (G7513; Sigma-Aldrich), and penicillin/streptomycin (P4333; Sigma-Aldrich) was added on the cover glass, and then cells were plated in 50,000 cells/well density. Cells were transfected with DNA constructs expressing GFPTULP3 fragments under the chicken β-actin promoter after 5 d in culture using Lipofectamine 2000 (Invitrogen). Cells were fixed in 4% PFA with 30% sucrose for 15 min and then washed five times with PBS for 10 min each. Cells were then processed for immunofluorescence. GFP-transfected neurons and glia were distinguished based on distinctive morphology; although neurons have thin, long processes (dendrites/axons), glia (astrocytes) typically are flat shaped in morphology with wide and short processes (Beaudoin et al., 2012).
Immunoblotting for PC2 levels
Cell extracts from mIMCD-K2 were prepared by incubating cells in a hypotonic lysis buffer (10 mM Tris, pH 7.0, 1 mM EDTA, 1 mM AEBSF, and 0.01 mg/ml each of leupeptin, pepstatin, and chymostatin) for 30 min at 4°C. Lysate was centrifuged 8,000 g for 15 min and dissolved in Laemmli SDS buffer followed by incubation at 65°C for 5 min (Geng et al., 2006). Blots were probed with antibodies against PC2 (rabbit polyclonal from G. Pazour) and tubulin.
In vitro IFT-A complex binding assays
In vitro IFT-A complex binding assays were performed as described previously (Mukhopadhyay et al., 2010). In short, high-spin lysates from IFT140LAP RPE cell pellets (∼500 µl of packed cell volume) were immunoprecipitated using anti-GFP antibody cross-linked to Affi-Prep Protein A beads (Bio-Rad). The washed beads were treated overnight with PrecissionS, and the digests were eluted in a total of ∼400 µl of LAP150N buffer (50 mM Hepes, pH 7.4, 150 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10% glycerol, and 0.05% NP-40). 30 µl of freshly prepared PrecissionS eluates were added to MBPTULP3 or MBPTUB isoform b immobilized on packed amylose resin in the presence of recombinant HisTULP3 (1–183 aa; 21 µg) in a total volume of 100 µl LAP150N buffer and incubated for 90 min by mixing at 4°C. Flowthrough after 90 min of binding was denatured with 5× sample buffer. After three washes in LAP150N buffer, MBPTUB/TULP3-bound proteins were eluted in 45 µl of 2× sample buffer.
Statistical analyses
Statistical analyses were performed using Student’s t test for comparing two groups or Tukey’s post-hoc multiple comparison tests between all possible pairs using Prism (GraphPad Software). Nonparametric Mann-Whitney U tests were performed for intensity plots in Figs. 1 B, 6 E, and 7 B using Prism.
Online supplemental material
Fig. S1 depicts the role of Tulp3 in localization of GPCRs to cilia. Fig. S2 depicts that CLSs from GPCRs and fibrocystin require Tulp3 for trafficking to cilia. Fig. S3 shows proximity biotinylation assays to characterize CLS–Tulp3 proximity. Fig. S4 (A and B) depicts the proximity of the tubby domain of TULP3 and TUB in biotinylation assays. Fig. S4 (C–G) describes differential effects of Tub on ciliary GPCR trafficking. Fig. S5 (A–D) shows the effects of Tulp3/Inpp5e knockdown on ciliary localization of PC2. Fig. S5 (E and F) demonstrates TULP3/TUB-mediated trafficking of Gpr161 to the ciliary membrane.
Supplementary Material
Acknowledgments
We are thankful for the generous gifts of antibodies from Greg Pazour, Joachim Seeman, and Kathryn Anderson, cell lines from Bruce Stanton, and plasmids from Anne-Claude Gingras, Greg Pazour, Kirk Mykytyn, and Stefan Somlo. We thank Marcel Mettlen for writing the macro for importing images from ImageJ. We thank Peter Jackson, in whose laboratory S. Mukhopadhyay performed the initial experiments with Tub mice as a postdoctoral fellow. We thank Sandra Schmid for comments on the manuscript. Phoenix A cells were provided by the Indiana University National Gene Vector Biorepository.
This project was funded by a recruitment grant from the Cancer Prevention and Research Institute of Texas (R1220 to S. Mukhopadhyay) and a grant from the National Institutes of Health (1R01GM113023-01 to S. Mukhopadhyay).
The authors declare no competing financial interests.
Author contributions: H.B. Badgandi and S. Mukhopadhyay conceived the project, designed experiments, and analyzed data. S.-H. Hwang generated cell lines in Fig. 7 (A and B) and performed experiments described in Figs. 7 B and S5 D. I.S. Shimada contributed to neuronal/glial cultures described in Figs. 5 and S4. E. Loriot contributed to the generation of reagents used in Figs. 1 A and S1 A. H.B. Badgandi performed all other research. H.B Badgandi and S. Mukhopadhyay wrote the manuscript with input from all the authors.
Footnotes
Abbreviations used:
- CLS
- ciliary localization sequence
- DIV
- day in vitro
- DSP
- dithiobis(succinimidyl propionate)
- GPCR
- G protein–coupled receptor
- IP
- immunoprecipitation
- PI(4,5)P2
- phosphoinositide 4,5-bisphosphate
- RPE
- retinal pigment epithelial
- Shh
- Sonic hedgehog
- Smo
- Smoothened
- TRP
- transient receptor potential
References
- Bargmann C.I. 2006. Chemosensation in C. elegans. WormBook. 10.1895/wormbook.1.123.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin G.M. III, Lee S.H., Singh D., Yuan Y., Ng Y.G., Reichardt L.F., and Arikkath J.. 2012. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 7:1741–1754. 10.1038/nprot.2012.099 [DOI] [PubMed] [Google Scholar]
- Berbari N.F., Johnson A.D., Lewis J.S., Askwith C.C., and Mykytyn K.. 2008a Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell. 19:1540–1547. 10.1091/mbc.E07-09-0942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N.F., Lewis J.S., Bishop G.A., Askwith C.C., and Mykytyn K.. 2008b Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA. 105:4242–4246. 10.1073/pnas.0711027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop G.A., Berbari N.F., Lewis J., and Mykytyn K.. 2007. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J. Comp. Neurol. 505:562–571. 10.1002/cne.21510 [DOI] [PubMed] [Google Scholar]
- Bloodgood R.A. 2012. The future of ciliary and flagellar membrane research. Mol. Biol. Cell. 23:2407–2411. 10.1091/mbc.E12-01-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., and Traub L.M.. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447. 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- Breslow D.K., Koslover E.F., Seydel F., Spakowitz A.J., and Nachury M.V.. 2013. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J. Cell Biol. 203:129–147. 10.1083/jcb.201212024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Anyatonwu G., Okuhara D., Lee K.B., Yu Z., Onoe T., Mei C.L., Qian Q., Geng L., Wiztgall R., et al. . 2004. Calcium dependence of polycystin-2 channel activity is modulated by phosphorylation at Ser812. J. Biol. Chem. 279:19987–19995. 10.1074/jbc.M312031200 [DOI] [PubMed] [Google Scholar]
- Cai Y., Fedeles S.V., Dong K., Anyatonwu G., Onoe T., Mitobe M., Gao J.D., Okuhara D., Tian X., Gallagher A.R., et al. . 2014. Altered trafficking and stability of polycystins underlie polycystic kidney disease. J. Clin. Invest. 124:5129–5144. 10.1172/JCI67273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez M., Ena S., Van Sande J., de Kerchove d’Exaerde A., Schurmans S., and Schiffmann S.N.. 2015. Modulation of ciliary phosphoinositide content regulates trafficking and Sonic hedgehog signaling output. Dev. Cell. 34:338–350. 10.1016/j.devcel.2015.06.016 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005. [DOI] [PubMed] [Google Scholar]
- Choi Y.H., Suzuki A., Hajarnis S., Ma Z., Chapin H.C., Caplan M.J., Pontoglio M., Somlo S., and Igarashi P.. 2011. Polycystin-2 and phosphodiesterase 4C are components of a ciliary A-kinase anchoring protein complex that is disrupted in cystic kidney diseases. Proc. Natl. Acad. Sci. USA. 108:10679–10684. 10.1073/pnas.1016214108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.T., Clement C.A., Satir P., and Pedersen L.B.. 2012. Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. J. Pathol. 226:172–184. 10.1002/path.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., and Reiter J.F.. 2005. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Davenport A.P., Alexander S.P., Sharman J.L., Pawson A.J., Benson H.E., Monaghan A.E., Liew W.C., Mpamhanga C.P., Bonner T.I., Neubig R.R., et al. . 2013. International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands. Pharmacol. Rev. 65:967–986. 10.1124/pr.112.007179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T.L., Bonacci T.M., Sprang S.R., and Smrcka A.V.. 2005. Structural and molecular characterization of a preferred protein interaction surface on G protein βγ subunits. Biochemistry. 44:10593–10604. 10.1021/bi050655i [DOI] [PubMed] [Google Scholar]
- Deretic D., Schmerl S., Hargrave P.A., Arendt A., and McDowell J.H.. 1998. Regulation of sorting and post-Golgi trafficking of rhodopsin by its C-terminal sequence QVS(A)PA. Proc. Natl. Acad. Sci. USA. 95:10620–10625. 10.1073/pnas.95.18.10620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domire J.S., Green J.A., Lee K.G., Johnson A.D., Askwith C.C., and Mykytyn K.. 2011. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell. Mol. Life Sci. 68:2951–2960. 10.1007/s00018-010-0603-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T., San Agustin J.T., Keady B.T., Jonassen J.A., Liang Y., Francis R., Tobita K., Johnson C.A., Abdelhamed Z.A., Lo C.W., and Pazour G.J.. 2014. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell. 31:279–290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty E.J., Stokes N., Chai S., Shah A.S., Williams S.E., and Fuchs E.. 2011. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 145:1129–1141. 10.1016/j.cell.2011.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock T., Ravarani C.N., Sun D., Venkatakrishnan A.J., Kayikci M., Tate C.G., Veprintsev D.B., and Babu M.M.. 2015. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 524:173–179. 10.1038/nature14663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., Li L., Vucica Y., and Pazour G.J.. 2010. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J. Cell Biol. 188:21–28. 10.1083/jcb.200910096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., San Agustin J.T., Jonassen J.A., Huang T., Rivera-Perez J.A., Tremblay K.D., and Pazour G.J.. 2014. Arf4 is required for mammalian development but dispensable for ciliary assembly. PLoS Genet. 10 10.1371/journal.pgen.1004170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foord S.M., Bonner T.I., Neubig R.R., Rosser E.M., Pin J.P., Davenport A.P., Spedding M., and Harmar A.J.. 2005. International Union of Pharmacology. XLVI. G protein-coupled receptor list. Pharmacol. Rev. 57:279–288. 10.1124/pr.57.2.5 [DOI] [PubMed] [Google Scholar]
- Fung H.Y., and Chook Y.M.. 2014. Atomic basis of CRM1-cargo recognition, release and inhibition. Semin. Cancer Biol. 27:52–61. 10.1016/j.semcancer.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainullin V.G., Hopp K., Ward C.J., Hommerding C.J., and Harris P.C.. 2015. Polycystin-1 maturation requires polycystin-2 in a dose-dependent manner. J. Clin. Invest. 125:607–620. 10.1172/JCI76972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Phua S.C., Roberson E.C., Garcia G. III, Abedin M., Schurmans S., Inoue T., and Reiter J.F.. 2015. Phosphoinositides regulate ciliary protein trafficking to modulate Hedgehog signaling. Dev. Cell. 34:400–409. 10.1016/j.devcel.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Okuhara D., Yu Z., Tian X., Cai Y., Shibazaki S., and Somlo S.. 2006. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J. Cell Sci. 119:1383–1395. 10.1242/jcs.02818 [DOI] [PubMed] [Google Scholar]
- He W., Ikeda S., Bronson R.T., Yan G., Nishina P.M., North M.A., and Naggert J.K.. 2000. GFP-tagged expression and immunohistochemical studies to determine the subcellular localization of the tubby gene family members. Brain Res. Mol. Brain Res. 81:109–117. 10.1016/S0169-328X(00)00164-9 [DOI] [PubMed] [Google Scholar]
- Hilgendorf K.I., Johnson C.T., and Jackson P.K.. 2016. The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr. Opin. Cell Biol. 39:84–92. 10.1016/j.ceb.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert M.C., Weihbrecht K., Searby C.C., Li Y., Pope R.M., Sheffield V.C., and Seo S.. 2012. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. USA. 109:19691–19696. 10.1073/pnas.1210916109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K., et al. . 2006. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 12:1197–1205. 10.1261/rna.30706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P.M., Hurd T.W., Zhang L., McEwen D.P., Brown R.L., Margolis B., Verhey K.J., and Martens J.R.. 2006. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr. Biol. 16:1211–1216. 10.1016/j.cub.2006.04.034 [DOI] [PubMed] [Google Scholar]
- Jin H., White S.R., Shida T., Schulz S., Aguiar M., Gygi S.P., Bazan J.F., and Nachury M.V.. 2010. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 141:1208–1219. 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee H.L., Dishinger J.F., Blasius T.L., Liu C.J., Margolis B., and Verhey K.J.. 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14:431–437. 10.1038/ncb2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Xu H., Yao Q., Li W., Huang Q., Outeda P., Cebotaru V., Chiaravalli M., Boletta A., Piontek K., et al. . 2014. Ciliary membrane proteins traffic through the Golgi via a Rabep1/GGA1/Arl3-dependent mechanism. Nat. Commun. 5 10.1038/ncomms6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer N.L., Lewis B., and Stanton B.A.. 1995. Electrogenic sodium absorption and chloride secretion by an inner medullary collecting duct cell line (mIMCD-K2). Am. J. Physiol. 268:347–355. [DOI] [PubMed] [Google Scholar]
- Koemeter-Cox A.I., Sherwood T.W., Green J.A., Steiner R.A., Berbari N.F., Yoder B.K., Kauffman A.S., Monsma P.C., Brown A., Askwith C.C., and Mykytyn K.. 2014. Primary cilia enhance kisspeptin receptor signaling on gonadotropin-releasing hormone neurons. Proc. Natl. Acad. Sci. USA. 111:10335–10340. 10.1073/pnas.1403286111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaf A., and Von Zastrow M.. 2015. Dopamine receptors reveal an essential role of IFT-B, KIF17, and Rab23 in delivering specific receptors to primary cilia. eLife. 4 10.7554/eLife.06996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J., Pazour G.J., Ikebe M., and Witman G.B.. 2009. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187:1117–1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F., Brown J.M., Sampaio J.L., Craft J.M., Shevchenko A., Evans J.E., and Witman G.B.. 2013. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J. Cell Biol. 201:249–261. 10.1083/jcb.201207139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.J., Cansizoglu A.E., Süel K.E., Louis T.H., Zhang Z., and Chook Y.M.. 2006. Rules for nuclear localization sequence recognition by karyopherinβ2. Cell. 126:543–558. 10.1016/j.cell.2006.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Sivan-Loukianova E., Eberl D.F., and Kernan M.J.. 2008. An IFT-A protein is required to delimit functionally distinct zones in mechanosensory cilia. Curr. Biol. 18:1899–1906. 10.1016/j.cub.2008.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew G.M., Ye F., Nager A.R., Murphy J.P., Lee J.S., Aguiar M., Breslow D.K., Gygi S.P., and Nachury M.V.. 2014. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev. Cell. 31:265–278. 10.1016/j.devcel.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.S., and Tang B.L.. 2015. A role for Rab23 in the trafficking of Kif17 to the primary cilium. J. Cell Sci. 128:2996–3008. 10.1242/jcs.163964 [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Niewiadomski P., Lin B., Nakamura H., Phua S.C., Jiao J., Levchenko A., Inoue T., Rohatgi R., and Inoue T.. 2013. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat. Chem. Biol. 9:437–443. 10.1038/nchembio.1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loktev A.V., and Jackson P.K.. 2013. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Reports. 5:1316–1329. 10.1016/j.celrep.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Ma M., Tian X., Igarashi P., Pazour G.J., and Somlo S.. 2013. Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat. Genet. 45:1004–1012. 10.1038/ng.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley A., and von Zastrow M.. 2010. DISC1 regulates primary cilia that display specific dopamine receptors. PLoS One. 5 10.1371/journal.pone.0010902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley A., Choy R.W., and von Zastrow M.. 2013. GPR88 reveals a discrete function of primary cilia as selective insulators of GPCR cross-talk. PLoS One. 8 10.1371/journal.pone.0070857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelova J., Astuto-Gribble L., Inoue H., Tam B.M., Schonteich E., Prekeris R., Moritz O.L., Randazzo P.A., and Deretic D.. 2009. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 28:183–192. 10.1038/emboj.2008.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M., Loerke D., Yarar D., Danuser G., and Schmid S.L.. 2010. Cargo- and adaptor-specific mechanisms regulate clathrin-mediated endocytosis. J. Cell Biol. 188:919–933. 10.1083/jcb.200908078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz O.L., Tam B.M., Hurd L.L., Peränen J., Deretic D., and Papermaster D.S.. 2001. Mutant rab8 impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol. Biol. Cell. 12:2341–2351. 10.1091/mbc.12.8.2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A., Bright N.A., Seaman M.N., and Robinson M.S.. 2003. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162:909–918. 10.1083/jcb.200305145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., and Jackson P.K.. 2011. The tubby family proteins. Genome Biol. 12 10.1186/gb-2011-12-6-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Chih B., Nelson C.D., Lane W.S., Scales S.J., and Jackson P.K.. 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24:2180–2193. 10.1101/gad.1966210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Ratti N., Loktev A., Rangell L., Scales S.J., and Jackson P.K.. 2013. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 152:210–223. 10.1016/j.cell.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Nagasawa Y., Matthiesen S., Onuchic L.F., Hou X., Bergmann C., Esquivel E., Senderek J., Ren Z., Zeltner R., Furu L., et al. . 2002. Identification and characterization of Pkhd1, the mouse orthologue of the human ARPKD gene. J. Am. Soc. Nephrol. 13:2246–2258. 10.1097/01.ASN.0000030392.19694.9D [DOI] [PubMed] [Google Scholar]
- Norman R.X., Ko H.W., Huang V., Eun C.M., Abler L.L., Zhang Z., Sun X., and Eggenschwiler J.T.. 2009. Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum. Mol. Genet. 18:1740–1754. 10.1093/hmg/ddp113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dowd B.F., Nguyen T., Lynch K.R., Kolakowski L.F. Jr., Thompson M., Cheng R., Marchese A., Ng G., Heng H.H., and George S.R.. 1996. A novel gene codes for a putative G protein-coupled receptor with an abundant expression in brain. FEBS Lett. 394:325–329. 10.1016/0014-5793(96)00901-5 [DOI] [PubMed] [Google Scholar]
- Omori Y., Chaya T., Yoshida S., Irie S., Tsujii T., and Furukawa T.. 2015. Identification of G protein-coupled receptors (GPCRs) in primary cilia and their possible involvement in body weight control. PLoS One. 10 10.1371/journal.pone.0128422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal K., Hwang S.H., Somatilaka B., Badgandi H., Jackson P.K., DeFea K., and Mukhopadhyay S.. 2016. Smoothened determines β-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J. Cell Biol. 212:861–875. 10.1083/jcb.201506132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee J., Shim J., Han W., Lee J., Bae Y.C., Chung Y.D., Kim C.H., and Moon S.J.. 2013. dTULP, the Drosophila melanogaster homolog of tubby, regulates transient receptor potential channel localization in cilia. PLoS Genet. 9 10.1371/journal.pgen.1003814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., and Bloodgood R.A.. 2008. Targeting proteins to the ciliary membrane. Curr. Top. Dev. Biol. 85:115–149. 10.1016/S0070-2153(08)00805-3 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., San Agustin J.T., Follit J.A., Rosenbaum J.L., and Witman G.B.. 2002. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 12:378–380. 10.1016/S0960-9822(02)00877-1 [DOI] [PubMed] [Google Scholar]
- Pedersen L.B., Mogensen J.B., and Christensen S.T.. 2016. Endocytic control of cellular signaling at the primary cilium. Trends Biochem. Sci. 41:784–797. 10.1016/j.tibs.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Ponting C.P., Hofmann K., and Bork P.. 1999. A latrophilin/CL-1-like GPS domain in polycystin-1. Curr. Biol. 9:585–588. 10.1016/S0960-9822(99)80379-0 [DOI] [PubMed] [Google Scholar]
- Qian F., Germino F.J., Cai Y., Zhang X., Somlo S., and Germino G.G.. 1997. PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat. Genet. 16:179–183. 10.1038/ng0697-179 [DOI] [PubMed] [Google Scholar]
- Qin H., Diener D.R., Geimer S., Cole D.G., and Rosenbaum J.L.. 2004. Intraflagellar transport (IFT) cargo. J. Cell Biol. 164:255–266. (published erratum appears in J. Cell Biol. 2004. 164:943) 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard J.B., Sato I.T., and Coughlin S.R.. 2008. Anatomical profiling of G protein-coupled receptor expression. Cell. 135:561–571. 10.1016/j.cell.2008.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., Corcoran R.B., and Scott M.P.. 2009. Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc. Natl. Acad. Sci. USA. 106:3196–3201. 10.1073/pnas.0813373106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., and Burke B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196:801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S., Boggon T.J., Baird C.L., Gomez C.A., Zhao J., Shan W.S., Myszka D.G., and Shapiro L.. 2001. G-protein signaling through tubby proteins. Science. 292:2041–2050. 10.1126/science.1061233 [DOI] [PubMed] [Google Scholar]
- Sedmak T., and Wolfrum U.. 2010. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J. Cell Biol. 189:171–186. 10.1083/jcb.200911095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Wen X., and Scales S.J.. 2015. The orphan G protein-coupled receptor Gpr175 (Tpra40) enhances hedgehog signaling by modulating cAMP levels. J. Biol. Chem. 290:29663–29675. 10.1074/jbc.M115.665810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Haley J., Bulgakov O.V., Cai X., McGinnis J., and Li T.. 2012. Tubby is required for trafficking G protein-coupled receptors to neuronal cilia. Cilia. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumska J., Qatato M., Rehders M., Führer D., Biebermann H., Grandy D.K., Köhrle J., and Brix K.. 2015. Trace amine-associated receptor 1 localization at the apical plasma membrane domain of fisher rat thyroid epithelial cells is confined to cilia. Eur. Thyroid J. 4(Suppl 1):30–41. 10.1159/000434717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao D., Dishinger J.F., Kee H.L., Pinskey J.M., Allen B.L., and Verhey K.J.. 2014. An assay for clogging the ciliary pore complex distinguishes mechanisms of cytosolic and membrane protein entry. Curr. Biol. 24:2288–2294. 10.1016/j.cub.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo Y., Kurabayashi N., Nguyen M.D., and Sanada K.. 2016. The G protein-coupled receptor GPR157 regulates neuronal differentiation of radial glial progenitors through the Gq-IP3 pathway. Sci. Rep. 6 10.1038/srep25180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J.Z., Miller J.J., and Jackson P.K.. 2009. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics. 9:2888–2891. 10.1002/pmic.200800873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.J., Yuan D., Masyuk T.V., Wang X., Punyashthiti R., Whelan S., Bacallao R., Torra R., LaRusso N.F., Torres V.E., and Harris P.C.. 2003. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum. Mol. Genet. 12:2703–2710. 10.1093/hmg/ddg274 [DOI] [PubMed] [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. . 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764. 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K.J., Baye L.M., Olivier-Mason A., Mukhopadhyay S., Sang L., Kwong M., Wang W., Pretorius P.R., Sheffield V.C., Sengupta P., et al. . 2011. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 25:2347–2360. 10.1101/gad.173443.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Zhang Y., Wei Q., Huang Y., Li Y., Ling K., and Hu J.. 2015. BBS4 and BBS5 show functional redundancy in the BBSome to regulate the degradative sorting of ciliary sensory receptors. Sci. Rep. 5 10.1038/srep11855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder B.K., Hou X., and Guay-Woodford L.M.. 2002. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 13:2508–2516. 10.1097/01.ASN.0000029587.47950.25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.