Starr and Rose discuss work by Saunders et al. demonstrating that torsinA and LAP1 regulate nuclear movement during fibroblast polarization.
Abstract
How LINC complexes are regulated to connect nuclei to the cytoskeleton during nuclear migration is unknown. Saunders et al. (2017. J. Cell Biol. https://doi.org/10.1083/jcb.201507113) show that the AAA+ ATPase torsinA and its partner LAP1 are required for nuclear migration during fibroblast polarization by mediating the dynamics of LINC complexes.
Nuclear migration is central to several cellular and developmental processes including fertilization, muscle development, neuronal development, and cellular polarization (Bone and Starr, 2016). Many mechanisms to move nuclei exist; the best understood use the LINC complex, which consists of SUN proteins in the inner nuclear membrane connected to KASH proteins in the outer nuclear membrane. LINC complexes bridge the nuclear envelope to connect the cytoskeleton to the nucleoskeleton (Gundersen and Worman, 2013; Bone and Starr, 2016). A variety of LINC complexes and partners have been described, yet how LINC complex assembly is regulated to move specific nuclei at the proper time is poorly understood. The AAA+ ATPase torsinA has been proposed to regulate LINC complexes (Gerace, 2004; Nery et al., 2008; Saunders and Luxton, 2016). In this issue Saunders el al. show that torsinA localizes to specific structures that move nuclei and functions to regulate LINC complex formation during nuclear movement in polarizing fibroblasts.
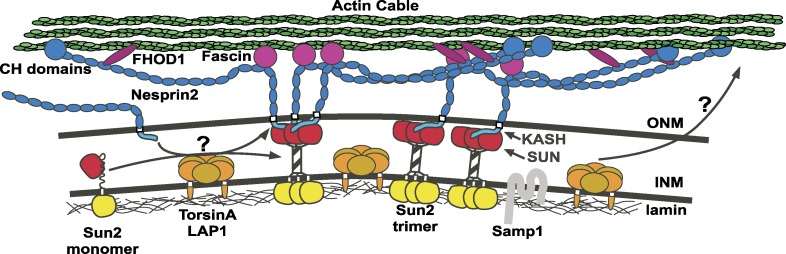
Nuclear migration is often studied in developing myotubes, migrating neurons, or various cells in model organisms. Because these take place in developing tissues, their usefulness for mechanistic studies are limited (Bone and Starr, 2016). To more easily characterize mechanisms of nuclear migration, an inducible nuclear migration model in wounded fibroblast monolayers was developed in which centrosomes orient toward the wound edge and nuclei move away from the wound edge (Gomes et al., 2005). Nuclei migrate using linear arrays of LINC complexes to hold onto apical actin cables undergoing retrograde flow (Luxton et al., 2010). LINC complexes connect to actin cables through FHOD1, Fascin, and the calponin domains of the KASH protein nesprin-2, whereas in the nucleoplasm, LINC complexes use laminA, emerin, and Samp1 to anchor to the nucleoskeleton (Gundersen and Worman, 2013; Bone and Starr, 2016; Fig. 1). Together, linear arrays of LINC complexes connected to actin cables are called transmembrane actin-associated nuclear (TAN) lines and they move away from the wound as a unit (Luxton et al., 2010). Although the existence of TAN lines in three-dimensional, in vivo tissues remains to be shown, this system has been fruitful in characterizing mechanisms involved in connecting nuclei to moving actin cables. However, the regulation and assembly of TAN lines is not well described. Specifically, it is poorly understood how SUN proteins are targeted to the inner nuclear membrane, how trimerization of SUN proteins is controlled, how SUN proteins choose or switch their KASH partners, or how the actin cables are assembled to then organize TAN lines (Luxton et al., 2010). TorsinA, which binds KASH proteins in vitro and is involved in the localization of KASH proteins, has long been thought to be a candidate to mediate many of these processes (Saunders and Luxton, 2016).
Figure 1.
TAN lines. TAN lines include LINC complexes made of the inner nuclear membrane (INM) protein Sun2 and the outer nuclear membrane (ONM) protein nesprin-2. LINC complexes connect to lamins and other INM proteins, including Samp1. Nesprin-2 mediates interactions with actin cables through its calponin homology (CH) domains, FHOD1 and Fascin. TorsinA and LAP1 form a heterocomplex at the INM; arrows and question marks indicate their possible targets. (left) TorsinA/LAP1 could mediate LINC complex formation by freeing nesprin-2 to associate in TAN lines or allowing Sun2 monomers to form trimers. (right) TorsinA/LAP1 could function through unknown targets independently of the LINC to regulate actin assembly.
TorsinA is a member of the AAA+ ATPase superfamily of proteins. ATPase proteins typically use energy generated from ATP hydrolysis to cause conformational changes in their substrates, thereby regulating diverse cellular functions (Cascalho et al., 2016; Laudermilch and Schlieker, 2016). TorsinA was first identified as the causative mutation of the DYT1 form of dystonia, a neuromuscular disease that causes abnormal twisting, or torsion, of the limbs. Although mutations in the Tor1A gene act in a dominant fashion to cause dystonia, several studies indicate that the most common causative mutation, an in-frame deletion that results in the loss of a single glutamate residue (ΔE), results in a loss of torsinA function (Cascalho et al., 2016; Laudermilch and Schlieker, 2016).
TorsinA was originally characterized as a luminal ER protein and many studies have identified connections between torsins and ER homeostasis (Cascalho et al., 2016). However, the role of torsins in the nuclear envelope, which is contiguous with the ER, has become a major focus in the field (Cascalho et al., 2016; Laudermilch and Schlieker, 2016; Saunders and Luxton, 2016). Significantly, both torsinA(ΔE) and the ATP-locked form, torsinA(E171Q), are abnormally concentrated in the nuclear envelope, suggesting that torsinA function in the perinuclear space could be relevant to disease. A screen for proteins that bind the ATP-locked form of torsinA identified the inner nuclear membrane component lamin-associated polypeptide 1 (LAP1) and the ER resident membrane protein luminal domain like LAP1 (LULL1). LAP1 and LULL1 share homology in their luminal domains, which directly interact with torsinA. Further evidence of a role for torsinA in the nuclear envelope came from studies showing that Tor1A and LAP1 knockout mice, and Tor1A(ΔE) mutant mice, have severe defects in nuclear envelope architecture where large out-pocketings of the envelope are filled with vesicle-like structures. Similar defects are observed in DYT1 patient cells as well as Drosophila melanogster and nematode torsin mutants (Cascalho et al., 2016; Laudermilch and Schlieker, 2016).
Several connections between torsinA and LINC proteins have been reported. SUN proteins are involved in localization of torsinA to the nuclear envelope and torsinA interacts with multiple KASH domains. Additionally, knockdown of Tor1A disrupts localization of Nesprin-3, the KASH protein that connects nuclei to intermediate filaments (Saunders and Luxton, 2016). Morever, torsinA was implicated in nuclear polarization in migrating fibroblasts (Nery et al., 2008), but the mechanisms were not explored. These findings led to the working model that torsinA mediates nuclear migration by regulating interactions between SUN and KASH proteins in the perinuclear space.
To test the plausibility of this model, Saunders et al. (2017) examined nuclear migration in mouse NIH3T3 fibroblasts after Tor1A siRNA and in mouse embryonic fibroblasts (MEFs) from Tor1A−/− mice. Cultured fibroblasts at the edge of a scratch-induced wound normally polarize in response to growth factors by moving their nuclei away from the wound so that centrosomes are oriented toward the wound, in front of nuclei (Gomes et al., 2005). Saunders et al. (2017) found that knockdown of torsinA affected centrosome polarization by blocking rearward nuclear movement. Those few nuclei that did move migrated at significantly reduced speeds. In control cells, torsinA strongly colocalized with nesprin-2 and actin cables in TAN lines. Importantly, expressing a wild-type copy of torsinA rescued nuclear movement and centrosome orientation in Tor1A siRNA and MEFs. Thus, torsinA both localizes to TAN lines and is necessary for TAN line–mediated nuclear movements in polarizing fibroblasts.
Saunders et al. (2017) next demonstrated that Tor1A mutants defective in either ATP binding or hydrolysis blocked rearward nuclear movements. Furthermore, the torsinA(E171Q) ATPase-deficient form of the protein failed to rescue nuclear migration and cell polarization when introduced in to Tor1A−/− MEFs. Interestingly, the torsinA(E171Q) protein still accumulated in TAN lines. These data imply that localization of torsinA to TAN lines is independent of nucleotide state, but torsinA function during nuclear movement requires its ATPase activity.
The inner nuclear membrane protein LAP1 and the ER resident protein LULL1 were recently shown to be stimulators of torsinA ATPase activity. AAA+ ATPases generally act as hexameric rings in which the ATPase activity of one subunit is stimulated by the adjacent subunit via a conserved arginine residue. This arginine is missing in torsins and torsinA lacks ATPase activity unless copurified with either LAP1 or LULL1 (Cascalho et al., 2016; Laudermilch and Schlieker, 2016). Structural and cross-linking studies suggest that torsinA forms an alternating torsinA–LAP1/LULL1 heterohexamer where the LAP/LULL1 subunits provide the catalytic arginine for torsinA ATPase activity. Consistent with this model, torsinA copurified with LAP1/LULL1 (R>A) mutant proteins have a dramatic reduction of ATPase activity in vitro (Laudermilch and Schlieker, 2016). Saunders et al. (2017) therefore tested whether LAP1 or LULL1 are required for rearward nuclear movements in polarizing fibroblasts, and found that only LAP1 was required. When the luminal domain of LULL1 was targeted to the inner nuclear membrane using a LAP1/LULL1 chimera, it was not sufficient to move nuclei in LAP1 knockdown cells. Surprisingly, however, LAP1(R>A) was able to rescue polarization in LAP1 siRNA cells. Although this result could be interpreted to mean that ATPase stimulation is not required for the role of LAP1 in nuclear polarization, this at odds with the observation that torsinA(E171Q) is not functional for nuclear polarization. Thus it seems more likely that the reduction of ATPase activity exhibited by the LAP1(R>A) mutant in vivo is weaker than shown in vitro. Nonetheless, these results, together with the chimera data, suggest that that the luminal domain of LAP1 has a role in addition to tethering torsinA at the inner nuclear membrane.
Saunders et al. (2017) next addressed the mechanisms of the involvement of torsinA and LAP1 in TAN line assembly. siRNAs against either Tor1A or LAP1 significantly reduced the number of TAN lines as assayed by actin cables. TAN lines that did form were less stable than normal and moved rearward at about half the speed of controls. One model for torsinA and LAP1 function in TAN line assembly is that they act to remodel SUN2–nesprin-2 interactions in the perinculear space. To test this hypothesis the dynamics of LINC components within the nuclear envelope were examined by FRAP. Sun2 recovered normally in TorA−/− MEFs; however, nesprin-2 had a significantly increased recovery time, suggesting that torsinA and LAP1 normally function to free nesprin-2 to assemble in TAN lines. Interestingly, the effect on nesprin-2 mobility is not general to all KASH proteins, because nesprin-3 recovered normally after FRAP in TorA−/− MEFs. Therefore, Saunders et al. (2017) propose that torsinA and LAP1 act to mobilize nesprin-2 in the nuclear envelope, which results in TAN line assembly.
In summary, Saunders et al. (2017) significantly advance our mechanistic understanding of the role of torsinA in fibroblast polarization. They demonstrate that torsinA works with LAP1 to promote rearward nuclear movement by regulating TAN line assembly and stability as well as the retrograde flow of dorsal perinuclear actin cables. Furthermore, torsinA/LAP1 likely function independently of the microtubule-based mechanism that keeps the centrosome in the center of the cell, as disruptions of torsinA/LAP1 do not cause centrosome positioning defects. The effects on TAN line assembly and retrograde flow in torsinA/LAP1-depleted cells are consistent with a model where torsinA/LAP1 heterohexamers directly regulate Sun2–nesprin-2 interactions. In support of this, nesprin-2 is significantly less mobile in TorA−/− MEFs, suggesting that torsinA frees up nesprin-2 from another binding partner to allow it to assemble into TAN lines. The in vivo relevance of the ability of torsinA to bind KASH peptides in vitro is unclear, as torsinA/LAP1 is likely tethered to the inner nuclear membrane where it would be difficult to bind directly to KASH peptides (Fig. 1, left). Another model is that torsinA/LAP1 could regulate the monomer-to-trimer equilibrium of Sun2 to facilitate nesprin-2 binding, as KASH proteins bind SUN trimers. The advantage of this model is that torsinA/LAP1 could function at TAN lines to promote Sun2 trimerization, which could then interact with nesprin-2 to assemble TAN lines. However, torsinA/LAP1 heterohexamers could also function independently of LINC to remodel other complexes in the nuclear envelope (Fig. 1, right). To better understand the mechanisms of torsinA/LAP1 regulation of TAN lines and nuclear movement, the field must focus on identifying the cellular substrates of torsinA/LAP1 AAA+ ATPases.
TorsinA has been implicated in several different processes including cell polarity, ER protein quality control, nuclear membrane modeling, lipid metabolism, and LINC protein localization (Cascalho et al., 2016; Laudermilch and Schlieker, 2016; Saunders and Luxton, 2016). Is this new role of torsinA in regulating actin-based nuclear migration in fibroblasts relevant to the neuromuscular disorder dystonia? The mechanisms of TAN line–mediated nuclear migration in fibroblasts are quite different than those in migrating neurons or developing skeletal muscles, which either use LINC to connect to microtubule motors or use LINC-independent mechanisms to move nuclei (Bone and Starr, 2016). Nonetheless, the results of Saunders et al. (2017) provide the most direct evidence yet for a role for torsinA and its activator LAP1 in remodeling LINC protein interactions. Furthermore, they found that the disease-associated form of torsinA, Tor1A(ΔE), which fails to bind LAP1, shows the same defects in TAN line assembly. The fibroblast polarization model should prove useful in future studies to identify the direct substrates of torsinA/LAP1 and help unravel the details of the mechanism of torsinA action.
Acknowledgments
We apologize that because of space constraints we were unable to directly cite many excellent primary articles.
Research in the laboratory of D.A. Starr is funded by National Institutes of Health grant R01GM073874. Research in the laboratory of L.S. Rose is funded by National Institutes of Health grant R01GM68744 and National Institute of Food and Agriculture CA-D*-MCB-6239-H.
The authors declare no competing financial interests.
References
- Bone C.R., and Starr D.A.. 2016. Nuclear migration events throughout development. J. Cell Sci. 129:1951–1961. 10.1242/jcs.179788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascalho A., Jacquemyn J., and Goodchild R.E. Membrane defects and genetic redundancy: Are we at a turning point for DYT1 dystonia? Mov. Disord. 2016 doi: 10.1002/mds.26880. [DOI] [PubMed] [Google Scholar]
- Gerace L. 2004. TorsinA and torsion dystonia: Unraveling the architecture of the nuclear envelope. Proc. Natl. Acad. Sci. USA. 101:8839–8840. 10.1073/pnas.0402441101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., and Gundersen G.G.. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463. 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Gundersen G.G., and Worman H.J.. 2013. Nuclear positioning. Cell. 152:1376–1389. 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudermilch E., and Schlieker C.. 2016. Torsin ATPases: Structural insights and functional perspectives. Curr. Opin. Cell Biol. 40:1–7. 10.1016/j.ceb.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W.G., Gomes E.R., Folker E.S., Vintinner E., and Gundersen G.G.. 2010. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 329:956–959. 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery F.C., Zeng J., Niland B.P., Hewett J., Farley J., Irimia D., Li Y., Wiche G., Sonnenberg A., and Breakefield X.O.. 2008. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J. Cell Sci. 121:3476–3486. 10.1242/jcs.029454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C.A., and Luxton G.W.G.. 2016. LINCing defective nuclear-cytoskeletal coupling and DYT1 dystonia. Cell. Mol. Bioeng. 9:207–216. 10.1007/s12195-016-0432-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C.A., Harris N.J., Wiley P.W., Woolums B.M., Wang Y., McQuown A.J., Worman H.J., Dauer W.T., Gundersen G.G., and Luxton G.W.G.. 2017. TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J. Cell Biol. 10.1083/jcb.201507113 [DOI] [PMC free article] [PubMed] [Google Scholar]