Abstract
Catalase is well-known as an antioxidant dismutating H2O2 to O2 and H2O. However, catalases evolved when metabolism was largely sulfur-based, long before O2 and reactive oxygen species (ROS) became abundant, suggesting catalase metabolizes reactive sulfide species (RSS). Here we examine catalase metabolism of H2Sn, the sulfur analog of H2O2, hydrogen sulfide (H2S) and other sulfur-bearing molecules using H2S-specific amperometric electrodes and fluorophores to measure polysulfides (H2Sn; SSP4) and ROS (dichlorofluorescein, DCF). Catalase eliminated H2Sn, but did not anaerobically generate H2S, the expected product of dismutation. Instead, catalase concentration- and oxygen-dependently metabolized H2S and in so doing acted as a sulfide oxidase with a P50 of 20 mmHg. H2O2 had little effect on catalase-mediated H2S metabolism but in the presence of the catalase inhibitor, sodium azide (Az), H2O2 rapidly and efficiently expedited H2S metabolism in both normoxia and hypoxia suggesting H2O2 is an effective electron acceptor in this reaction. Unexpectedly, catalase concentration-dependently generated H2S from dithiothreitol (DTT) in both normoxia and hypoxia, concomitantly oxidizing H2S in the presence of O2. H2S production from DTT was inhibited by carbon monoxide and augmented by NADPH suggesting that catalase heme-iron is the catalytic site and that NADPH provides reducing equivalents. Catalase also generated H2S from garlic oil, diallyltrisulfide, thioredoxin and sulfur dioxide, but not from sulfite, metabisulfite, carbonyl sulfide, cysteine, cystine, glutathione or oxidized glutathione. Oxidase activity was also present in catalase from Aspergillus niger. These results show that catalase can act as either a sulfide oxidase or sulfur reductase and they suggest that these activities likely played a prominent role in sulfur metabolism during evolution and may continue do so in modern cells as well. This also appears to be the first observation of catalase reductase activity independent of peroxide dismutation.
Keywords: Hydrogen sulfide, Polysulfide, Garlic, Reactive oxygen species, Thioredoxin, Evolution, Aspergillus niger
Graphical abstract
Highlights
-
•
Reactive sulfide species (RSS) and reactive oxygen species (ROS) are biochemically similar.
-
•
RSS predate ROS in evolution and RSS metabolism likely persists in extant organisms.
-
•
Catalase is a multifunctional sulfur oxidoreductase capable of oxidizing H2S or generating H2S
-
•
Catalase appears to be important in oxygen sensing and response to hypoxia.
-
•
Catalase oxidization of DCF suggests that ROS measurements may be overestimated.
1. Introduction
Reactive oxygen species (ROS) are produced from one-electron reductions of oxygen that sequentially form superoxide (O2•-), hydrogen peroxide (H2O2), hydroxyl radical (HO•) and ultimately terminate in water;
| O2 (-e-) --> O2•- (-e-) --> H2O2 (-e-) --> HO• (-e-) --> H2O | (1) |
Reactive sulfide species (RSS) are chemically, biochemically and physiologically similar to ROS [1] and can be produced from sequential one-electron oxidations of hydrogen sulfide (H2S) to form a thiyl radical (HS•), hydrogen persulfide (H2S2) and persulfide radical (HS2•-) before terminating in elemental sulfur (S2); the latter usually cyclizing to S8;.
| H2S (+e-) --> HS• (+e-) --> H2S2 (+e-) --> HS2•- (+e-) --> S2 | (2) |
While ROS have pathophysiological consequences when in excess, there is considerable evidence that H2O2, and perhaps O2•- are important homeostatic signaling entities under normal circumstances [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18]. As chalcogens with six valence electrons, oxygen and sulfur would be expected to exhibit some commonalities in their biological actions and this has become quite apparent in regard to signaling via cysteine sulfur (Cys-S) in regulatory proteins. Peroxidation of Cys-S produces the sulfenyl, Cys-SOH and persulfidation (a.k.a. sulfhydration) produces a cysteine persulfide (Cys-S-SH; [19]). In the few regulatory systems where both peroxidation and persulfidation have been examined in detail the effector responses appear to be identical [20], [21], [22], [23], [24], [25] with the added caveat that, unlike H2O, H2S can also reduce protein disulfide bonds and effect enzyme activity [26]. In addition, we have shown that many of the methods used to measure ROS are sensitive to RSS and often more so [27]. This further confounds issues of the relative biological importance of ROS versus RSS in terms of tissue production, metabolism and intracellular signaling.
Cells have purportedly developed a number of “antioxidant” mechanisms to regulate ROS and guard against their toxicity. Catalase is one of the earliest known and best characterized of the antioxidant enzymes catalyzing the dismutation of peroxide to water and oxygen;
| 2H2O2 –> 2H2O + O2 | (3) |
However, because catalase appears to have appeared in evolution long before oxygen was present and at a time when RSS were more likely to be involved in cellular metabolism [1], (also see discussion), we wondered if catalase could also dismutate persulfide, i.e.;
| 2H2S2 –> 2H2S + S2 | (4) |
While we observed that catalase did indeed remove persulfides from solution we also observed, unexpectedly, that catalase also removed H2S from solution. Because sulfur in H2S is in its most reduced state (−2), H2S dismutation is impossible and the most logical scenario is that H2S is oxidized. We also observed that under certain conditions catalase generated H2S from other sulfur-bearing molecules. Thus catalase appears to be a “primordial” sulfur oxidoreductase. In the present study we examine these aspects of catalase-mediated sulfur metabolism and attempt to place them into an evolutionary perspective where these functions most likely evolved and suggest how they may still play a homeostatic role in modern animals.
2. Materials and methods
2.1. Chemicals
SSP4 (3',6'-Di(O-thiosalicyl)fluorescein), Na2S2, Na2S3 and Na2S4 were purchased from Dojindo molecular Technologies Inc. (Rockville, MD). Thioredoxin was purchased from ThermoFisher Scientific (Grand Island, NY). Carbon monoxide (CO, 1 mM), carbonyl sulfide (COS, 20 mM) and sulfur dioxide (SO2, 1.4 M) solutions were prepared by bubbling pure gas through a sintered glass aerator into buffer for 20–30 min. All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Phosphate buffer (in mM): 137 NaCl, 2.7, KCl, 8 Na2HPO4, 2 NaH2PO4, pH 7.4.
Sorensen's buffer in (mM): 200 Na2HPO4, 200 NaH2PO4, ratio sdjusted to pH 6, 7 or 8.
2.2. Polysulfide measurement
The polysulfide-specific fluorophore, SSP4 was used to measure polysulfides. Samples and test compounds were aliquoted into black 96 well plates in a darkened room and fluorescence was measured on a SpectraMax M5e plate reader (Molecular Devices, Sunnyvale, CA). Typically, fluorescence was measured every 10 min over 90 min. In order to reduce the potential loss of H2S due to volatilization in these and other experiments the cover of the well plate was lined with parafilm in an attempt to seal off the wells.
2.3. Amperometric measurement of O2, H2O2 and H2S
Amperometric O2 and H2O2 sensors, ISO-OXY-2 and ISO-HPO-2, respectively, were purchased from WPI (World Precision Instruments, Sarasota, FL). They are designed for tissue culture with 2 mm dia replaceable membrane sleeves and a reported detection limit of 0.1% (ISO-OXY-2) and <100 nM (ISO-HPO-2). It should be noted that the ISO-HPO-2H2O2 sensor cannot be used when H2S is present as it is 24 times more sensitive to H2S than it is to H2O2 [27].
H2S amperometric sensors with a sensitivity of 14 nM H2S gas (~100 nM total sulfide) were constructed in-house as described previously [28]. The sensors were connected to WPI TBR 4100 Free Radical Analyzers and data was archived on a laptop PC with software provided by the manufacturer and exported into Microsoft Excel. The H2S sensor was calibrated periodically throughout each day with fresh standards made up in anoxic phosphate buffer (pH 7.4). This sensor does not respond to polysulfides or other oxidized forms of sulfur.
A reaction chamber with a side ports for the H2S and O2 sensors and a 1-cm wide by 2 cm deep central well was purchased from WPI (NOCHM-4). A polycarbonate stopper with a hole in the stopper permitted venting the head space air when the stopper was lowered into the chamber and provided an access port for sample injection with a Hamilton microliter syringe. The chamber was placed on a magnetic stirrer and stirred with a Teflon micro stir bar. Compounds of interest were injected through the stopper and the reactions monitored for 10–30 min or longer if necessary.
2.4. Oxygen sensitivity of H2S oxidation by catalase
To determine if catalase-mediated inactivation of H2S was an oxidative process buffer containing catalase was deoxygenated by passing 100% N2 into the chamber via a 21 ga needle inserted into the stopper until O2 was removed as indicated by the O2 electrode. This decreased the rate of H2S consumption confirming that this was an oxidative process. Preliminary experiments showed that H2S oxidation was not affected by a 6% O2 balance N2 mixture. In order to examine O2 tensions below this the 6% O2/bal N2 gas was mixed with 100% N2 using a Wösthoff Digamix gas mixing pump (H. Wösthoff Messtechnik GmbH, Bochum, Germany). Samples were gassed as above and Po2 was continuously monitored. The partial pressure of O2 at which catalase oxidation was halved (P50) was determined from the graph of percent H2S consumption vs percent O2 in chamber. Oxygen concentration in μM was determined from the prevailing barometric pressure (PB) measured in the laboratory with a mercury barometer, water vapor pressure (PH2O, 17.5 mmHg at 20 °C) and the oxygen solubility coefficient (α, at 300 mosm L-1 and 20 °C =1.7196 μmol L-1 mmHg-1; [29]; O2 (μM) =α•0.209•(PB-PH2O)).
2.5. pH sensitivity of catalase-mediated H2S oxidation
In order to determine if the catalase preferentially reacts with dissolved H2S or the hydrosulfide anion (HS-) the rate of H2S oxidation in Sorensen's buffer at pH 6, 7 and 8 was monitored with the amperometric H2S sensor. As the pKa1 of the reaction, H2S <–> HS- is 6.98 at 20 °C [30], this allowed us to adjust the H2S:HS- ratio from 90:10 to 10:90.
Vetrano et al. [31] have shown that catalase oxidizes dichlorofluorescein (DCF) and we observed DCF oxidation competes with H2S oxidation (Olson unpublished). In these experiments we first measured DCF oxidation by catalase at pH 6,7 and 8 in 96 well plates in the absence of H2S to determine the pH sensitivity of catalase. By repeating these experiments in the presence of H2S it was then possible to identify the sulfide species that competes with DCF in the catalase-mediated oxidation process.
2.6. Sodium azide
In preliminary experiments we monitored H2O2 concentration with the H2O2 sensor and found that catalase dismutation of 10 μM H2O2 could be completely inhibited by 50 mM sodium azide (NaN3; not shown). This concentration of azide was used in all further experiments. The effects of azide on H2S metabolism were measured by adding 10 μM H2S to catalase in the presence or absence of 50 mM azide. As this appeared to decrease the rate of catalase-mediated H2S oxidation we then examined the possibility that another electron acceptor, H2O2, might take the place of O2. In these experiments 10 or 100 μM H2O2 was added 2–5 min after H2S in the presence of catalase with or without azide.
2.7. Aspergillus niger catalase
Catalase from the fungus Aspergillus niger was used to investigate limited aspects of sulfide:sulfur metabolism. This enzyme does not contain or readily utilize NADPH cofactors and only forms Compound I.
2.8. Data Analysis
Data was analyzed and graphed using QuatroPro (Corel Corporation, Ottawa Ont, Canada) and SigmaPlot 13.0 (Systat Software, Inc., San Jose, CA). Statistical significance was determined using one-way ANOVA and the Holm-Sidak test (SigmaPlot 13.0). Results are given as mean + or±SE; significance was assumed when p≤0.05.
3. Results
3.1. Catalase as a sulfide oxidase
3.1.1. Effects of catalase on mixed polysulfide
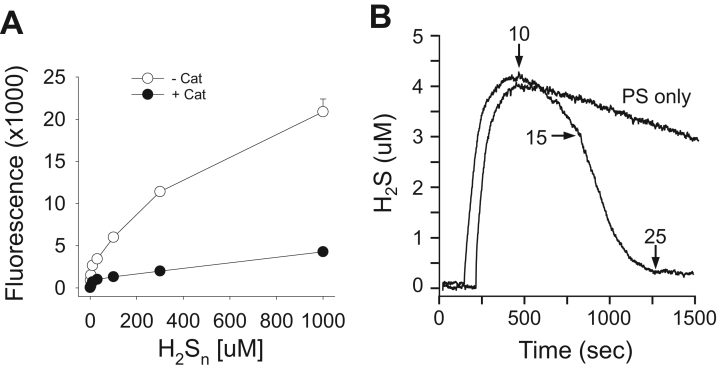
When dissolved, K2Sn forms a mixture of polysulfides, H2Sn, where n=1–8, i.e., H2S, H2S2, H2S3… H2S8. As shown in Fig. 1A, 20 μM H2Sn alone in solution concentration-dependently increased SSP4 fluorescence and this was almost completely prevented by adding 25 μM catalase. As we presumed this was due to catalase dismutation of the polysulfide to H2S (Eq. (4)) we then measured H2S directly and in real time with the amperometric sensor. As shown in Fig. 1B, dissolving K2Sn produced H2S which slowly out-gassed through the hole in the stopper. However, when catalase was added the H2S concentration began to decrease more rapidly and increasing catalase concentration from 10 to 25 μM decreased the H2S concentration from ~4 μM to essentially nil in approximately 7 min. Addition of a second 25 μM catalase (50 μM total) did not affect H2S concentration confirming that H2S was not produced from the remaining polysulfides. These results clearly show that under aerobic conditions catalase metabolizes polysulfides. They also suggest that polysulfide metabolism is either not a dismutative process (in that no H2S is produced), or if H2S is produced it is also metabolized by catalase or tightly bound to it.
Fig. 1.
(A) Concentration-dependent polysulfide (H2Sn) mediated SSP4 fluorescence in the presence or absence of 25 μM catalase. Catalase essentially inhibits fluorescence over a wide range of polysulfide concentrations. Mean +SE, n =4, many error bars are within symbols. (B) Amperometric measurement of H2S concentration in 20 μM polysulfide in the absence of catalase (PS only) or after additions of 10, 15 and 25 μM catalase (total catalase, 10, 25 and 50 μM). In the absence of catalase H2S slowly out-gases from the chamber. Addition of 10 μM catalase increases the rate of H2S disappearance and this is further increased by a second addition of 15 μM catalase which completely removes all H2S. Doubling the total catalase concentration to 50 μM by a third addition of catalase did not affect H2S concentration.
It should be noted that the exact ratio of polysulfide species produced when K2Sn is dissolved is not known. We show in Fig. 1B that dissolving 20 μM of the salt produced ~4 μM H2S. If there is equal distribution of the remaining 16 μM of S2-S7 sulfur species there would be 72 μM sulfur. If catalase dismutates these polysulfides there would be enough catalase at 25 μM (100 μM heme) to bind one sulfide per heme. Subsequent studies were aimed at resolving this issue.
3.1.2. Catalase reactions with H2S
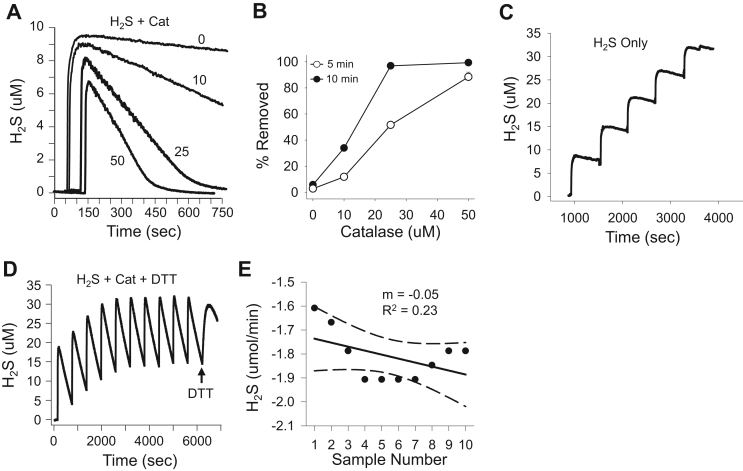
Catalase from 0 to 50 μM concentration-dependently increased the rate of 10 μM H2S removal from buffer when measured amperometrically (Fig. 2A, B). Under aerobic conditions administering H2S in five consecutive 10 μM doses cumulatively increased H2S concentration (Fig. 2C), whereas in the presence of 25 μM catalase, ten consecutive 20 μM H2S injections were continuously removed and did not accumulate (Fig. 2D). The average rate of removal of these 10H2S injections in the presence of catalase (−1.81±0.03 μmoles H2S/min) remained relatively constant over the course of the experiment (Fig. 2D, E). This suggests that H2S does not remain bound to catalase thereby affecting its activity. This is supported by the fact that at the end of the experiment the H2S concentration (200 μM) was twice as much as the four heme groups in 25 μM catalase (100 μM). In another experiment, 13 consecutive H2S additions were applied with the same results (not shown).
Fig. 2.
Amperometric measurements of H2S metabolism by catalase. (A, B) Catalase from 0 to 50 μM concentration dependently increased the rate of disappearance of 10 μM H2S; (A) representative traces, (B) average percent removed at 5 (open circles) and 10 (black circles) min. Mean +SE (n =3), all values with catalase >0 are significantly different from each other (p<0.001). Repetitive injections of 10 μM H2S in the absence of catalase cumulatively increase H2S concentration (C), whereas in the presence of 25 μM catalase ten consecutive 20 μM H2S injections are efficiently removed (D). Addition of 1 mm DTT after H2S injections (D, arrow) further increases H2S concentration. (E) Plot of the decay slope of H2S removal in (from D) as a function of injection number showing random variation in slope.
3.1.3. O2 sensitivity of catalase-mediated H2S oxidation
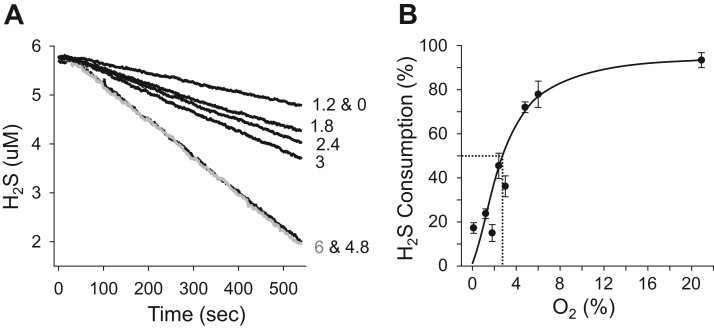
Because H2S sulfur is in its most reduced form (−2), catalase metabolism of H2S is likely an oxidative process. This possibility was examined by measuring H2S metabolism at different oxygen tensions. The oxygen sensitivity of 25 μM catalase-mediated metabolism of H2S (measured amperometrically) is shown in Fig. 3A. There was a progressive decrease in the rate of consumption of 10 μM H2S as the percent O2 fell below 4.8%. The rate of H2S consumption was halved at ~2.7% O2 (Fig. 3B) which at average barometric pressure (745 mmHg) and 100% humidity (PH2O =17.5 mmHg) resulted in an apparent P50 of ~20 mmHg.
Fig. 3.
Oxygen sensitivity of H2S metabolism by 25 μM catalase. A) Decrease in H2S concentration at various % O2. (B) Rate of H2S consumption as a function of % O2. The rate of H2S consumption is halved at ~2.7% O2, which at average barometric pressure (745 mmHg) and 100% humidity (PH2O =17.5 mmHg) results in an apparent P50 of ~20 mmHg. (A) Representative traces from single amperometric measurements. (B) Average H2S consumption; mean±SE (n=3), line fit by eye.
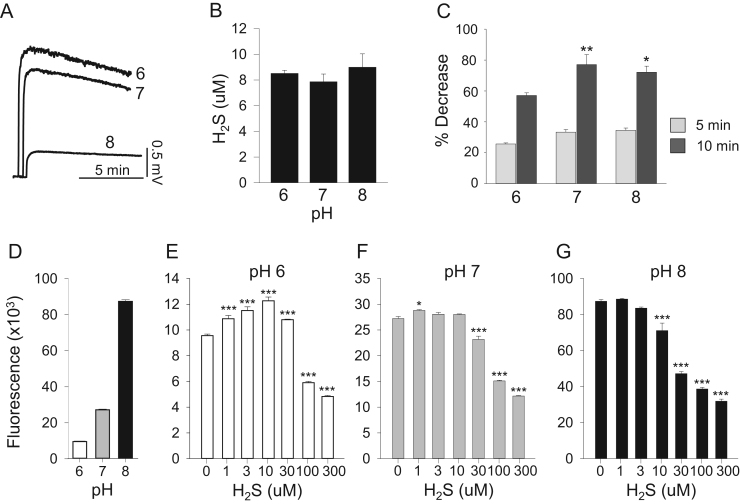
3.1.4. pH sensitivity of catalase-mediated H2S and DCF oxidation
Because the amperometric sensor only measures dissolved H2S gas and this decreases as pH increases the sensor was calibrated at pH 6, 7 and 8 and H2S consumption was corrected accordingly (Fig. 4A, B). Metabolism of 10 μM H2S by 25 μM catalase at 5 and 10 min after H2S injection, measured amperometrically and corrected for pH, increased from pH 6 to pH 7 but did not change from pH 7–8 even though the H2S/HS- ratio decreased another 10 fold (Fig. 4C).
Fig. 4.
Effect of pH on catalase-mediated H2S oxidation measured amperometrically (A-C) and on interactions with DCF oxidation (D–G). (A) Sensor response to 10 μM H2S standard (output in mV) decreases as pH increases due to pH-dependent decrease in dissolved H2S gas. (B) Sensor response to 10 μM H2S in presence of 25 μM catalase corrected for pH effect on dissolved H2S. Peak H2S is similar after correction but less than 10 μM due to initial H2S consumption. (C) Percent H2S consumption by 25 μM catalase at 5 and 10 min after H2S injection. D. Catalase-mediated oxidation of 10 μM DCF increased over nine-fold as pH was increased from 6.0 to 8.0. E-G. Effect of H2S on catalase-mediated DCF oxidation at pH 6.0, 7.0 and 8.0. At pH 6.0, low H2S concentrations increased DCF oxidation and higher concentrations inhibited it. As pH increased, the stimulatory effect was lost and H2S became solely inhibitory. Mean±SE (n=3, B, C; n=4, D-G); *, p<0.5; **, p<0.01; ***, p<0.001.
The pH sensitivity of H2S metabolism by catalase cannot be determined solely from amperometric measurements because both catalase activity and the H2S:HS- ratio can be affected by pH. As a surrogate for the effect of pH on catalase during H2S metabolism, we measured pH sensitivity of catalase-mediated oxidation of DCF. As shown in Fig. 4D, DCF fluorescence increased nearly 3 fold as pH was increased from 6 to 7 and another 3 fold from pH 7–8. Background DCF fluorescence also increased as a function of pH (291, 357, 533 at pH 6, 7 and 8, arbitrary fluorescence units) but this 1.8-fold increase was far less than the 9.1-fold increase in the presence of catalase. The failure of H2S metabolism to increase between pH 7 and 8, commensurate with increased catalase activity, suggests that dissolved H2S is preferred over HS- as a substrate for catalase.
Knowing the effect of pH on DCF oxidation (Fig. 4D) and the fact that H2S appears to be a competitive inhibitor of DCF oxidation (Olson, unpublished observation), we then examined the interaction between H2S and DCF at different pH in order to confirm if dissolved H2S was more reactive than HS-. At pH 6.0, H2S from 1 to 30 μM concentration-dependently increased catalase-mediated DCF oxidation and decreased it at 100 and 300 μM (Fig. 4E). At pH 7.0, 1 μM H2S slightly, but significantly increased DCF oxidation, whereas oxidation was concentration-dependently decreased from 30 to 300 μM H2S (Fig. 4F). At pH 8.0, 10–300 μM H2S concentration-dependently inhibited DCF oxidation and there was no stimulatory effect of H2S (Fig. 4G). These results suggest that H2S is metabolized by catalase and that HS- might inhibit this process.
3.1.5. Effects of sodium azide on H2S metabolism
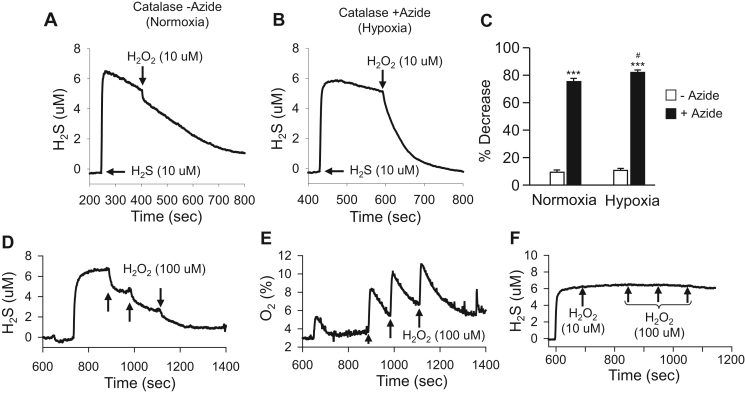
As shown in Fig. 5A and C, addition of 10 μM H2O2 produced a slight transient decrease in 10 μM H2S concentration in the presence of 25 μM catalase in both normoxia and hypoxia. However, when catalase was inhibited with sodium azide, 10 μM H2O2 removed approximately 80% of the 10 μM H2S in less than 1 min (Fig. 5B, C). Hypoxia in combination with azide inhibition slightly, but significantly (p<0.05), enhanced the effect of H2O2 (Fig. 5C). By comparison, three consecutive 100 μM injections of H2O2 did not completely remove 10 μM H2S from solution when catalase was not inhibited with azide in hypoxia (Fig. 5D), although each H2O2 injection reduced the H2S concentration by approximately 25%. Without azide catalase dismutation of H2O2 was clearly evident in this experiment as O2 production increased with each addition of H2O2 followed by a decrease in oxygen presumably due to catalase-induced consumption of oxygen by sulfide (Fig. 5E). H2O2 did not affect H2S concentration in the presence of azide, but without catalase, (Fig. 5F). In the absence of H2O2 azide also decreased the rate of catalase oxidation of H2S (not shown).
Fig. 5.
Sodium azide (50 mM) inhibition of catalase promotes H2S oxidation by H2O2. (A) In normoxia with 25 μM catalase but without sodium azide, 10 μM H2S (measured amperometrically) rapidly decreased while addition of 10 μM H2O2 produced a slight, rapid decrease bur did not change slope. (B) In hypoxia, azide and catalase, 10 μM H2O2 nearly completely removed 10 μM H2S within 1 min. (C) Summary of azide effects. H2O2 (10 μM) had little effect on H2S concentration in the presence of catalase without azide (white bars) in either normoxia or hypoxia but it decreased H2S concentration by around 80% when catalase was inhibited by azide and this was slightly augmented in hypoxia. (D) In hypoxia, each of three 100 μM H2O2 injections reduced H2S by ~20% in the presence of catalase without azide but increased O2 (E). (F) With azide but without catalase H2O2 did not affect H2S concentration. Mean +SE (n=3); *** effect of azide inhibition at same O2(p<0.001), # effect of O2(p<0.05).
3.1.6. Catalase-mediated polysulfide production from H2S and catalase-mediated H2S metabolism
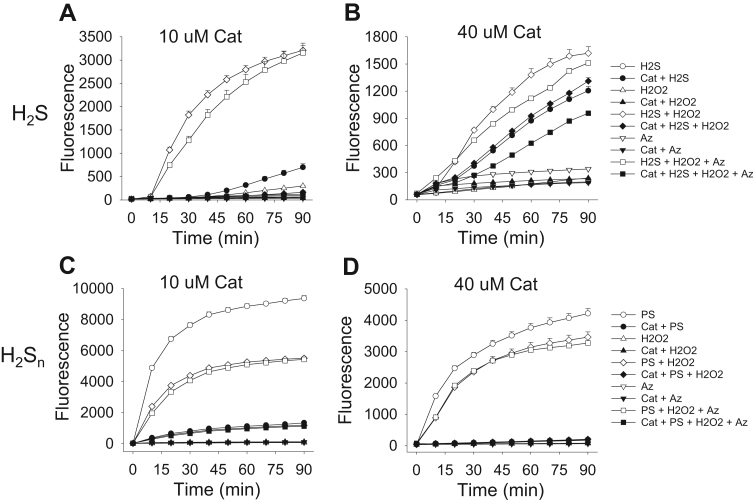
Although catalase metabolized polysulfides from the K2Sn salt (Fig. 1), it is possible that catalase also generated polysulfide intermediates during H2S oxidation and that these were different from those in H2Sn or metabolized by catalase as they were produced. In order to examine these possibilities we compared the production and metabolism of putative polysulfides and potential polysulfide oxides from H2S and H2Sn with SSP4 fluorescence. As shown in Fig. 6A and B, 300 μM H2S in combination with either 300 μM H2O2 or H2O2 +50 mM azide produced the greatest increase in SSP4 fluorescence in the absence of catalase, the effects of azide appeared to be negligible. There was little polysulfide in H2S. With low catalase there was a small increase in fluorescence from H2S that was approximately 20% of that produced from H2S plus H2O2, but other combinations of compounds did not increase fluorescence. In the presence of 40 μM catalase, fluorescence from H2S was increased to 70% of that produced from H2S plus H2O2 in the absence of catalase. Fluorescence from H2S with 40 μM catalase was not significantly increased by adding H2O2 but it was decreased by nearly 25% by azide. Catalase alone did not affect SSP4 fluorescence.
Fig. 6.
(A, B) Effects of combinations of H2S (300 μM), H2O2 (300 μM) and sodium azide (Az, 50 mM) with or without 10 μM (A) or 40 μM (B) catalase (Cat) on putative polysulfide production measured by SSP4 fluorescence. H2S in combination with either H2O2 or H2O2 + azide produced the greatest increase in fluorescence. Low (10 μM) catalase produced a small amount of fluorescence from H2S. Conversely, substantial fluorescence was produced by H2S and 40 μM catalase. H2O2 did not significantly increase fluorescence production from H2S and 40 μM catalase, whereas fluorescence was decreased by azide. Catalase alone did not affect SSP4 fluorescence. (C, D) Effects of combinations of the mixed polysulfide H2Sn (PS; 300 μM), H2O2 (300 μM) and sodium azide (50 mM) with or without 10 μM (C) or 40 μM (D) catalase on SSP4 fluorescence. In the absence of catalase, polysulfide alone produced the greatest increase in fluorescence and fluorescence was decreased by H2O2 alone and to the same extent by H2O2 plus azide. Polysulfide fluorescence was reduced by more than 80% by 10 μM catalase and completely eliminated by 40 μM catalase. The inhibitory effect of catalase was unaffected by addition of H2O2, azide or H2O2 plus azide. Traces of SSP4 or SSP4 plus catalase did not affect fluorescence and have been omitted for clarity. Mean +SE, n =4, many error bars are within symbols. The difference in total fluorescence in A, B or C, D was due to different lot numbers of SSP4.
The mixed polysulfide (H2Sn, where n=1–8; 300 μM) produce the greatest increase in SSP4 fluorescence and this was partially inhibited to the same extent by either 300 μM H2O2 or 300 μM H2O2 +50 mM azide, again suggesting effect was due to H2O2 not azide (Fig. 6C, D). Polysulfide fluorescence was reduced by more than 80% by 10 μM catalase and completely eliminated by 40 μM catalase. The inhibitory effect of catalase was unaffected by addition of H2O2, azide or H2O2 plus azide.
Collectively, the above results suggest that while catalase interacts with H2S to increase SSP4 fluorescence (Fig. 6A, B), this fluorescence is not due to generation of a “classical” polysulfide because catalase efficiently prevented fluorescence from the mixed polysulfide H2Sn (Fig. 6C, D). These results are consistent with those of Fig. 1 and confirm that catalase potently metabolizes polysulfides.
3.2. Catalase as a sulfur reductase
3.2.1. Catalase releases H2S from dithiothreitol (DTT)
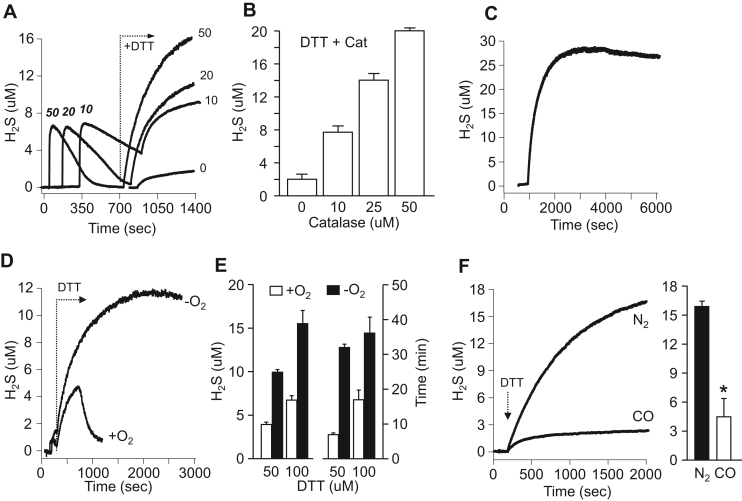
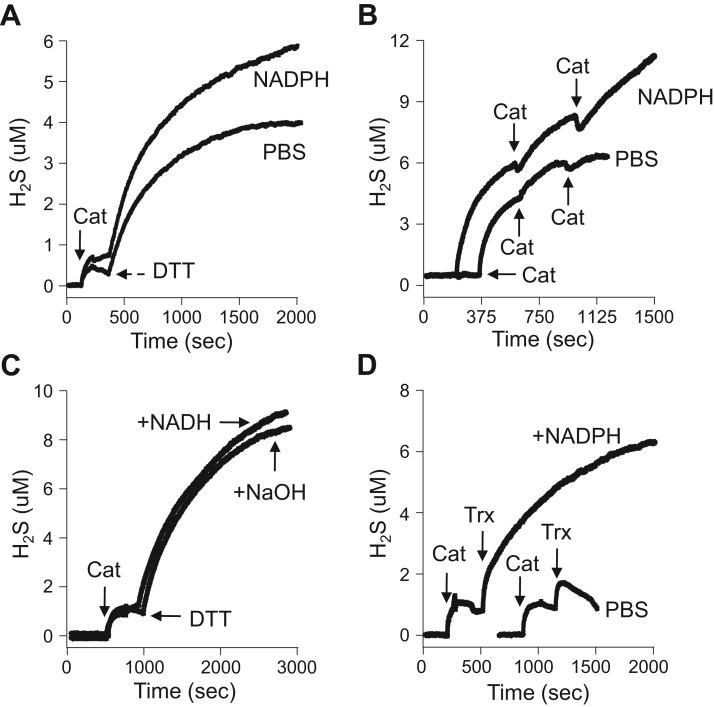
Although we observed that catalase efficiently removed polysulfides (Fig. 1), we also noticed that after exposing 25 μM catalase to ten consecutive 20 μM H2S injections (200 μM total) subsequent addition of 1 mM DTT produced an additional increase in H2S (Fig. 2D). This suggests that either DTT released H2S from polysulfides that were formed by catalase-mediated oxidation of H2S, which seems unlikely given that catalase rapidly metabolizes polysulfides (Fig. 1, Fig. 6), or that catalase directly generates H2S from DTT. In order to evaluate these possibilities 1 mM DTT was added 10 min after H2S was metabolized by increasing concentrations of catalase. As shown in Fig. 7A, the amount of H2S released appeared to be correlated with the catalase concentration as both 20 and 50 μM catalase removed all of the H2S in this time period. To directly examine the role of catalase in H2S release from DTT, these experiments were repeated without pre-exposure to H2S. This also produced a catalase concentration-dependent increase in H2S release (Fig. 7B) confirming that catalase reacts directly with DTT to generate H2S.
Fig. 7.
Catalase directly releases H2S from 1 mM DTT and this is inhibited by 1 mM carbon monoxide (CO). (A) Representative amperometric traces of H2S release from DTT alone (0) or when 1 mM DTT was added 10 min after 10 μM H2S was metabolized by 10, 20 or 50 μM catalase. Numbers in bold/italic indicate catalase concentration after H2S addition; plain numbers are from same curve as above showing H2S generated 10 min after DTT addition in same experiment. (B) Effect of catalase concentration on H2S release from 1 mM DTT without prior addition of H2S. All values are significantly (p<0.001) different from each other (mean +SE, n =3 replicates). (C) H2S released from 50 μM catalase metabolism of 1 mM DTT remains relatively constant for nearly 90 min. (D) Amperometric traces of H2S released from 50 μM catalase metabolism of 100 μM DTT in normoxia and hypoxia. In normoxia, H2S concentration increases then there is a relatively abrupt decrease in concentration. In hypoxia H2S concentration continues to increase until it is twice that in normoxia and it does not abruptly decrease. (E) H2S released from 50 μM catalase metabolism of 50 and 100 μM DTT in normoxia and hypoxia and duration of H2S response which is shorter in normoxia (mean +SE, n =3 replicates; all hypoxia are significantly (p<0.001) different from respective normoxia and all 50 μM DTT are significantly (p<0.001)different from 100 μM DTT). (F) Production of H2S from 100 μM DTT by 50 μM catalase in hypoxia (N2) and in hypoxia with 1 mM carbon monoxide (CO). Left, typical amperometric traces; right, average H2S production (mean +SE, n=3; *p=0.006).
Although 50 μM catalase metabolizes nearly 90% of a 10 uM H2S injection in 5 min (Fig. 1A), the H2S released from the interaction of 50 μM catalase with 1 mM DTT remained in the reaction chamber essentially unchanged even after nearly 90 min (Fig. 7C). This poses a conundrum, does DTT and H2S compete for the same site on the enzyme, or is there so much H2S released it overwhelms the capacity of catalase to remove this H2S in the observation interval? In order to examine these possibilities, lower DTT concentrations were employed. As shown in Fig. 7D, H2S released by 50 μM catalase metabolism of 100 μM DTT in normoxia initially increased then abruptly decreased. However, in the absence of oxygen H2S concentration increased to more than twice as much as that produced in normoxia and H2S remained elevated for the duration of the experiment. These effects were observed when either 50 or 100 μM of DTT was employed, although the magnitude and duration of the response was greater with the latter (Fig. 7E). These studies show that catalase directly releases H2S from DTT independent of the presence of oxygen and that in the presence of oxygen H2S production and metabolism appear to occur concurrently.
In order to verify that DTT could nevertheless release H2S from polysulfides, 1 mM DTT was added to dissolved polysulfide salts, Na2S2, Na2S3 Na2S4. As shown in Table 1, H2S was spontaneously released from these polysulfides; the amount of H2S released essentially doubled with each additional sulfur indicating that DTT quantitatively reduced these polysulfides back to H2S.
Table 1.
H2S (μM) released spontaneously (spont) upon dissolving 10 μM H2S2, H2S3 or H2S4 standards in normoxia and after subsequent addition of 1 mM DTT (+DTT).
| H2S2 | H2S3 | H2S4 | |
|---|---|---|---|
| H2S (spont) | 9.1 ±0.36* | 5.9 ±0.51 | 5.7 ±0.62 |
| + DTT | 3.3 ±1.12a,b, | 7.7 ±0.84a,c | 16.8 ±1.28b,c |
| Total H2S from polysulfide | 12.4 | 13.6 | 22.5 |
Measured with amperometric H2S sensor. Mean±SE; n =10–12 (H2S), 5–6 (DTT). * significantly different from either H2S3 or H2S4(p<0.001). a; significantly different from same letter at (p=0.015); b, c, significantly different from same letter at (p<0.001). +DTT corrected for H2S released by DTT alone (3.9 μM).
3.2.2. CO inhibition of H2S production from DTT
In order to determine if catalase-mediated H2S production from DTT involves the catalase heme iron the buffer was sparged with CO gas for 20 min (which also removed O2) then the buffer was added to the chamber followed by 50 μM catalase then 100 μM DTT. H2S production was measured amperometrically and compared to catalase-mediated H2S production in N2 sparged buffer. As shown in Fig. 7F, 1 mM CO reduced H2S production by 70% demonstrating that the heme iron in catalase is also involved in the reductive component of catalase metabolism of sulfur-bearing molecules.
3.2.3. Catalase-mediated H2S production from other sulfur compounds
The ability of catalase to release H2S from DTT prompted an inquiry into the effects of catalase on other sulfur-bearing compounds. In these experiments, buffer in the reaction chamber was sparged with 100% N2 for 20 min, non gaseous sulfur compounds were then added and sparged another 10 min with N2, the stopper was then lowered to eliminate headspace and catalase was added. H2S release was measured amperometrically for at least 10 min after the catalase was added. This procedure allowed us to determine if any H2S was released from the sulfur-bearing compound in the absence of catalase. With gaseous sulfur compounds, COS and SO2, the buffer was sparged with N2 as above, the stopper was then lowered to eliminate headspace and minimize volatilization of COS or SO2, and the dissolved gas was added. After the response had stabilized (within a few min) catalase was added and H2S release measured as above. The effects of SO2 on catalase oxidation of DCF were also examined.
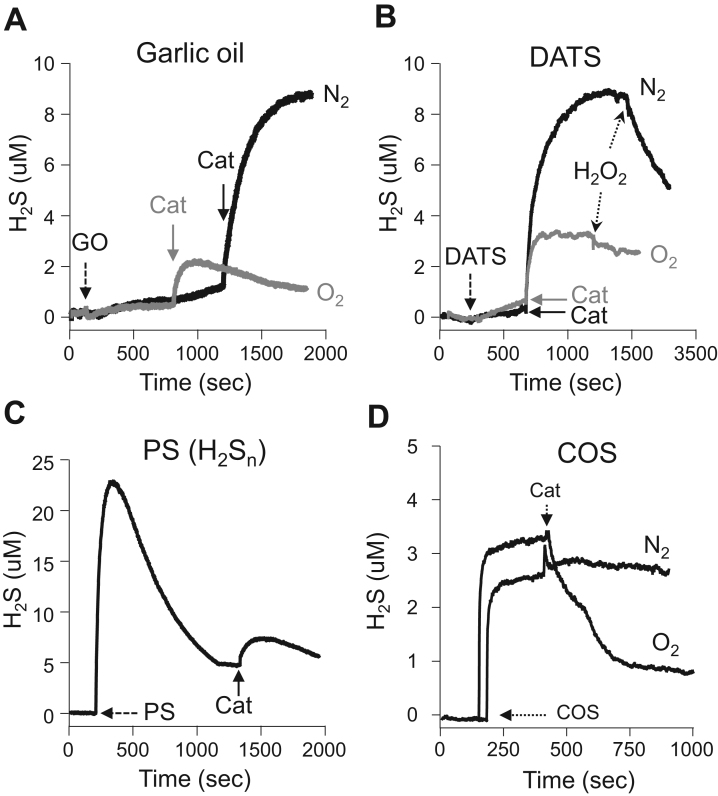
As shown in Fig. 8, 50 μM catalase produced H2S from 100 μM garlic oil and 100 μM diallyl disulfide (DATS) in hypoxia, whereas in normoxia considerably less H2S was produced. Addition of 100 μM H2O2 to H2S released by DATS plus catalase produced a rapid decrease in H2S concentration in hypoxia but not in normoxia (Fig. 8B).
Fig. 8.
Amperometric traces showing catalase-mediated H2S production from sulfur-bearing molecules. Under hypoxic (N2) conditions catalase (Cat, 50 μM) produced H2S from (A) 100 μM garlic oil (GO) or (B) 100 μM diallyl disulfide (DATS). H2S production from garlic oil and DATS was considerably reduced in normoxia (O2). Peroxide (H2O2; 100 μM) rapidly reduced H2S generated from DATS in hypoxia. (C) Considerable H2S is produced when 100 μM of the polysulfide (PS) salt K2Sn is dissolved (producing the mixed polysulfide H2Sn); whereas relatively little H2S is recovered after subsequent addition of 50 μM catalase in hypoxia. (D) H2S was released from the spontaneous decomposition of carbonyl sulfide (COS) and metabolized by catalase in normoxia but not hypoxia. Catalase did not increase H2S production from COS in either normoxia or hypoxia.
A considerable amount of H2S was released when 100 μM polysulfide (K2Sn) was dissolved but only several μM more were released when 50 μM catalase was added (Fig. 8C). This suggests that catalase does not release H2S from polysulfides or of it does the H2S is removed by a non-oxidative process, which seems unlikely. H2S was also released from the spontaneous decomposition of 30 μM COS, however, when catalase was added the H2S concentration decreased in normoxia but did not change in hypoxia (Fig. 8D). These results suggest that catalase oxidized H2S that was formed from COS decomposition but catalase did not react directly with COS to produce H2S. Catalase did not produce H2S when incubated with 100 μM cysteine, cystine, glutathione, oxidized glutathione, thiosulfate (S2O32-) or metabisulfite (S2O52-) in either normoxia or hypoxia (not shown).
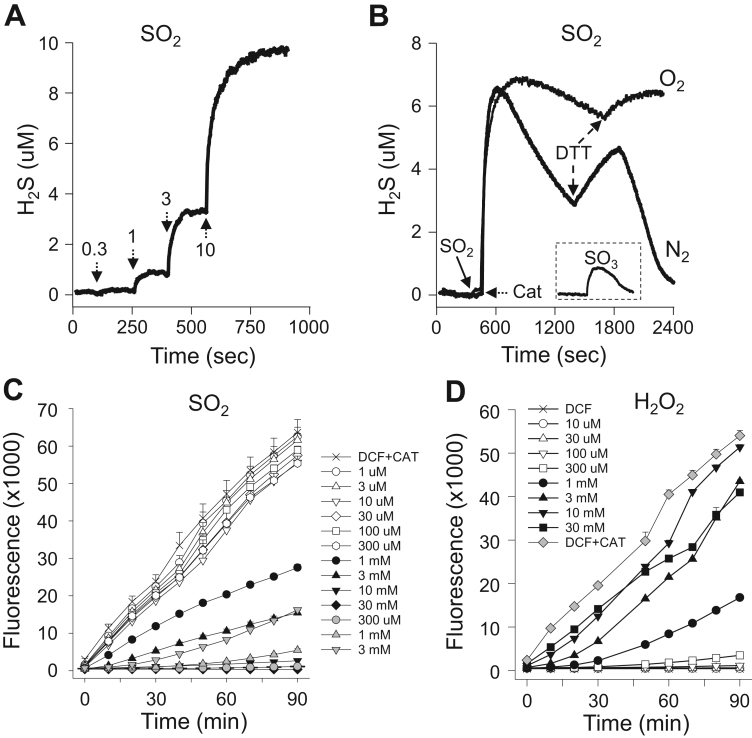
Sulfur dioxide (SO2) did not spontaneously produce appreciable amounts of H2S, whereas there was a concentration-dependent increase in H2S release in the presence of catalase (Fig. 9A) with a threshold SO2 less than 1 mM. The release of H2S from SO2 was unaffected by the presence or absence of oxygen, although H2S concentration appeared to decrease more rapidly in hypoxia (Fig. 9B). Addition of DTT produced a further increase in H2S. Sulfite (SO32-), supposedly in equilibrium with dissolved SO2, produced a minimal amount of H2S in the presence of catalase in hypoxia (inset, Fig. 9B). These results suggest that SO2 gas, not the cognate anion, SO3-2, reacts with catalase.
Fig. 9.
(A) Amperometric traces showing H2S was SO2 concentration (arrows)- and catalase (50 μM)-dependently released in hypoxia or (B) from 7 mM SO2 in both normoxia and hypoxia; H2S release appeared to be somewhat prolonged in normoxia. Addition of 1 mM DTT produced a further increase in H2S in both conditions. Inset in (B) shows that only a small amount of H2S was released from 7 mM sodium sulfite (SO3, shown in same concentration-time scale. (C) effects of SO2 on DCF (20 μM) oxidation in the absence (gray symbols) or presence of 20 μM catalase (open or solid symbols at low and high SO2 concentrations, respectively). SO2 concentration-dependently oxidized DCF between 300 μM and 3 mM SO2 in the absence of catalase, whereas SO2 inhibited catalase oxidation of DCF at concentrations >~300 μM and completely inhibited oxidation at 10 mM. (D) Comparison of DCF oxidation by increasing concentrations of H2O2 (open and solid symbols) or 20 μM catalase (gray diamonds). More DCF is oxidized by 20 μM catalase than by 30 mM H2O2.
SO2 alone concentration-dependently oxidized 20 μM DCF, between 300 μM and 3 mM (Fig. 9C) but not at higher or lower concentrations (not shown). SO2 concentration-dependently inhibited catalase oxidation of DCF with an EC50 between 300 μM and 1 mM and DCF oxidation was essentially completely inhibited by SO2 >3 mM (Fig. 9C).
DCF is commonly used an an indicator of ROS and DCF oxidation is often assumed to be an indicator of H2O2 [32]. DCF is oxidized by catalase alone and as shown in Fig. 9D, 20 μM DCF is concentration-dependently oxidized by H2O2 with an EC50 of approximately 1.5 mM and maximal oxidation at 10 mM H2O2. This is slightly less than the amount of oxidation produced by 20 μM catalase alone. This suggests that even a small amount of active catalase could give the impression of significant ROS production.
These results show that in the absence of oxygen, catalase acts as a selective sulfur reductase capable of directly producing reduced sulfur in the form of H2S from a variety of oxidized sulfides. They also suggest that in the presence of oxygen or another electrophile, such as H2O2, catalase oxidizes H2S concurrently with its formation.
3.2.4. Contribution of NADPH and NADH to H2S production from DTT and thioredoxin
NADPH is a well-known co-factor for bovine catalase that has been proposed to prevent formation of inactive compound II and this effect can be enhanced by addition of non-bound NADPH [33]. In these studies we examined the possibility that exogenous NADPH can provide a pathway for the reaction of DTT and the endogenous thiol, thioredoxin with catalase. As shown in Fig. 10A, the addition of NADPH increased H2S release from DTT and catalase by ~50%. NADPH nearly doubled H2S release from DTT after three consecutive additions of catalase (Fig. 10B). H2S was not released from DTT and NADPH in the absence of catalase (not shown). Conversely, NADH did not augment H2S release from DTT in the presence of catalase compared to the alkaline vehicle that was necessary to dissolve NADPH (Fig. 10C). The alkaline-induced increase in catalase activity is consistent with our observations that alkalinity increases catalase oxidase activity (Fig. 4) indicating that both the oxidase and reductase activities of catalase are pH dependent.
Fig. 10.
Amperometric measurements illustrating the effect of NADPH and NADH on catalase-mediated H2S release from DTT and thioredoxin (Trx) in hypoxia. (A) Catalase (50 μM) releases more H2S from 50 μM DTT in the presence of 400 μM NADPH than in phosphate buffer (PBS). (B) Release of H2S following multiple additions of 50 μM catalase is sustained in the presence of 400 μM NADPH but not in PBS. (C) NADH (400 μM) does not augment H2S release from 50 μM catalase and 50 μM DTT compared to alkaline vehicle (NaOH). (D) H2S release from 50 μM catalase and 10 μM thioredoxin is greatly increased by 400 μM NADPH compared to PBS.
In addition, as shown in Fig. 10D, approximately 1 μM of H2S is transiently produced when 10 μM Trx is added to 50 μM catalase in hypoxia. Reversing the order of addition of Trx and catalase produced a similar amount of H2S that also was not sustained (not shown). We next examined the contribution of NADPH to this reaction and in the presence of NADPH nearly 7 μM H2S were produced from the same reactants (Fig. 10C). Accounting for some H2S volatility or degradation, the amount of H2S released in the presence of NADPH is nearly equivalent to the amount of Trx used. It is clear that Trx is an effective endogenous substrate for catalase and that NADPH is required. Conversely, NADH did not augment H2S release from thioredoxin (not shown) which is similar to its lack of effect on DTT.
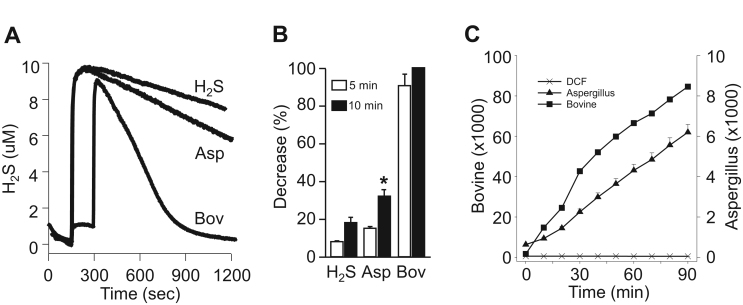
3.2.5. Sulfur metabolism by catalase from Aspergillus niger
Because NADPH is not a cofactor in catalase from the fungus Aspergillus niger and it does not form compound II [34], [35] we conducted several studies to determine if this enzyme is capable of oxidizing H2S or DCF and/or if it can generate H2S from Trx. As shown in Fig. 11A and B, 20 μM of A. Niger catalase enhanced the rate of H2S disappearance compared to H2S alone indicative of an oxidative capability, although this was only about 20–30% of that produced by bovine catalase. A. Niger catalase also oxidized DCF and again its catalytic activity was only about 20% of that of bovine catalase (Fig. 11C). However, when 400 μM NADPH and 10 μM Trx were added to 20 μM A. Niger catalase there was no H2S production (not shown). These experiments show that catalase from A. niger possesses oxidative activity and they also suggest that the lack of the NADPH binding site prevents the transfer of reducing equivalents and subsequent formation of H2S from sulfur-donating compounds.
Fig. 11.
Oxidase function of catalase from Aspergillus niger. (A) Amperometric measurements of a 10 μM H2S standard and 10 μM H2S with 20 μM A. niger catalase (Asp) and 20 μM bovine catalase (Bov) in hypoxia and (B) average decrease in H2S at 5 (open bars) and 10 (solid bars) min (mean +SD, n=3, H2S and Bov, n=2 Asp; * significantly greater than respective control (p=0.006); all Bov are greater than H2S or Asp (p<0.001). (C) Oxidation of 20 μM DCF A. Niger or bovine catalase. DCF is readily oxidized by A. Niger catalase compared to DCF alone, however, bovine catalase is 10-fold more efficacious (mean 3 replicates +SE).
4. Discussion
Vetrano et al., [31] provided the first evidence for an oxidase function of catalase, other than peroxide, using the xenobiotic 10-acetyl-3,7-dihydrophenoxazine (ADP). They also showed that catalase could oxidize DCF and the endogenous compounds, indole and β-phenethylamine. Furthermore, they and others have suggested catalase may not be a major contributor to H2O2 metabolism in that the H2O2 turnover rate is extremely high, the affinity of catalase for H2O2 is very low (Km ≥10 mM) and at low and presumably “physiological” H2O2 concentrations peroxidatic activity would be expected to predominate [31], [36], [37], [38]. This raises the question of the actual catalytic role of catalases under normal conditions, are they dismutative, peroxidatic or oxidative? Furthermore, are there other endogenous compounds that are preferred, or more physiologically relevant catalase substrates?
Our experiments show that catalase can act as a sulfide/sulfur oxidase and that this is not accomplished by a “classical” dismutation reaction. We also show that catalase can act as a reductase and generate H2S from a variety of sulfur-bearing molecules. This to our knowledge is the first demonstration of catalase as a peroxide-independent reductase. Collectively, our results suggest that catalase can play a role in sulfur metabolism, sulfur signaling, and through oxidation of DCF, may inadvertently led to misinterpretation of redox stress.
4.1. Physiological functions of catalase-mediated sulfur metabolism
4.1.1. Detoxification
The toxicity of H2S and other sulfur compounds are well known [39] and sulfur toxicity may have been an even greater problem early in evolution when these sulfur molecules were more prevalent [1]. In fact, the ability of catalase to metabolize H2S and polysulfides may have taken its origin in these sulfidic environments. The identity of potential electron acceptors in these early anoxic environments is unclear, although the ability of the modern bovine catalase to utilize H2O2 suggests some flexibility, perhaps ancient enzymes used other sulfur or nitrogen oxides that were present.
4.1.2. Response to hypoxia
However, the catalase-mediated production of H2S from sulfur-bearing molecules under hypoxic conditions versus oxidation of H2S in normoxia is suggestive of a regulatory system that is responsive to oxygen availability. The two most logical candidates that would benefit from this catalase activity are tissue (especially vascular) oxygen sensing and protection from ischemia and reperfusion injury.
4.1.2.1. Oxygen sensing
There is growing evidence that H2S serves as an O2 sensor in a variety of tissues including vascular and non-vascular smooth muscles, chemoreceptors, and airway epithelia. Here H2S transduces the hypoxic response by the oxygen-dependent balance between constitutive H2S production and H2S oxidation. Thus hypoxia increases H2S and this in turn activates a variety of downstream H2S-mediated, homeostatic effector responses [40].
Prior work from our lab has provided substantial evidence that during hypoxia tissue H2S production increases and this constitutes an important mechanism in vascular oxygen sensing [40]. While the hypoxia-induced increase in H2S is believed to result from decreased oxidative metabolism in tissue, decreased uptake and metabolism of H2S by RBC could amplify the response as RBCs avidly remove H2S from the circulation under normoxic conditions [28]. Although, catalase, which is abundant in RBC is thought to be the main mechanism of H2O2 metabolism [41], this process is not oxygen sensitive, whereas H2S metabolism is. In fact, the P50 for the oxyhemoglobin dissociation, typically ~25 mmHg, is strikingly similar to the 20 mmHg P50 for catalase oxidation of H2S (Fig. 3). Thus, hypoxia would be expected to nearly simultaneously increase unloading of O2 and inhibit H2S oxidation. As RBCs are also capable of generating H2S [42], catalase would directly affect H2S levels in these cells in a Po2-sensitive manner. The increase in H2S would not only convey its cytoprotective effects to the tissues but promote vasodilation to help restore O2 delivery. Furthermore, the tissue acidosis that usually accompanies ischemia would also decrease catalase oxidase activity (Fig. 4) and further decrease H2S oxidation.
4.1.2.2. Protection from ischemia and reperfusion injury
The location of catalase in peroxisomes in most cells except RBC may also provide anti-ischemic regulatory opportunities and the kidney may especially benefit from this. H2S and sulfides are now well recognized for their role in protecting kidneys from ischemic reperfusion injury and in ischemic conditioning [43], [44]. Unlike most tissues, from 7 to 80 times more H2S is derived from d-cysteine than l-cysteine in the kidney and brain and in these tissues uptake of d-cysteine and conversion to bound sulfane sulfur is faster than that of l-cysteine. It has also been demonstrated that d-cysteine, but not l-cysteine, protects the kidney from ischemia-reperfusion injury [45].
Although it is not clear how hypoxia is coupled to H2S formation, peroxisomal d-cysteine appears to be the primary source of H2S production [45]. Here d-cysteine is metabolized to 3-mercaptopyruvate (3-MP) by d-amino acid oxidase. It has been proposed that 3-MP is then transferred to the mitochondrion via peroxisome-mitochondrial vesicular trafficking and the mitochondrial enzyme 3-mercaptopyruvate sulfur transferase (3-MST) accepts the sulfur thereby forming a sulfane sulfur (3-MST-S) on the enzyme. This sulfur can be released as H2S by intracellular antioxidants and diffuse back to the peroxisome, or it can be transferred to other intracellular thiols as a polysulfide. Assuming catalase is involved in peroxisomal H2S and polysulfide metabolism, the oxygen sensitivity of this enzyme would provide the oxygen “sensing” transducer that couples ischemic insult to the appropriate homeostatic responses. Catalase may also generate H2S directly during hypoxia from other sulfur-bearing sources (see below).
4.1.3. Physiological functions of catalase-mediated H2S production from sulfur-bearing compounds
Catalase-mediated generation of H2S from a variety of exogenous and endogenous sulfur-bearing molecules suggests an important, and heretofore unrecognized function of this enzyme. This has both physiological implications and the potential for creating experimental artefacts.
4.1.3.1. DTT
DTT is a commonly used reductant in many experimental situations. Its ability to reduce poly- and persulfides is well known [46], [47], [48], [49] and we clearly show its efficacy in generating H2S from polysulfides in buffer (Table 1). However, we also show that catalase generates significant quantities of H2S directly from DTT (Fig. 7) and that it is likely that this is also an enzymatic process involving the heme center as it is inhibited by CO (Fig. 7F). Inadvertent H2S production from DTT could have profound effects on experiments in which DTT is added to cells to serve as a thiol reductant. This could be further confounded in experiments where oxygen tension is varied. Furthermore, even the general interpretation of ROS production in cells could be an artifact of DCF oxidation by catalase.
4.1.3.2. Garlic compounds
Garlic oil (GO) and its main active ingredient, diallyl trisulfide (DATS) are well known for their garlic-related health benefits and this has been attributed to their ability to release H2S. In buffer H2S can be released non-enzymatically from GO and DATS by relatively high concentrations of cysteine or glutathione (GSH; [50], [51]). Benavides et al. [50] have also shown that addition of garlic or DATS to RBCs releases H2S and this is augmented by glucose. They propose that garlic reacts with exofacial thiols on the RBC membrane to cross the membrane and then react with GSH to produce H2S and oxidized GSH (GSSG). Glucose metabolism by the RBC provides NADPH to reduce GSSG to GSH and sustain the reaction. Our experiments suggest an alternative mechanism catalyzed by catalase. It is expected that this would also be sustained by NADPH which is consistent with our results showing NADPH increases catalase-mediated H2S release from DTT and thioredoxin (Fig. 10).
4.1.3.3. SO2
SO2 is released in large quantities from volcanoes [52], [53] and early organisms could have exploited this in catalase-mediated redox reactions possibly setting the precedence for this enzyme (see below). SO2 is also endogenously generated in vertebrate tissues including the vasculature and its production appears to be physiologically regulated [54], [55], [56], [57]. Like H2S, SO2 is toxic in excess [58], [59], but at lower concentrations it vasodilates arteries and protects tissues against ischemic conditions, reperfusion injury and oxidative stress [57], [60], [61], [62], [63], [64], [65], [66], [67]. SO2 is reportedly rapidly hydrated in the lung and with a pKa of ~9 form sulfite (SO3-2) and bisulfite (HSO3-1) in a 3:1 Molar ratio [68]. However, SO2 gas is a far stronger vasodilator than either sulfite or bisulfite [57] suggesting that the gaseous form is the biologically active moiety. Furthermore, only about 10% of the dissolved SO2 is ionized in aqueous solution [69].
We show that H2S can be released from SO2 by catalase (Fig. 9A,B) which suggests that perhaps the ultimate biologically active “form” is actually H2S, or that H2S at least contributes to the biological activity of SO2. This is further supported by our observation that H2S is not released from sulfite (Fig. 9B) which could explain why sulfite is not as efficacious a vasodilator as gaseous SO2 [57].
While significant amounts of H2S are released from 7 mM SO2 by catalase, 1 mM SO2 halves the ability of catalase to oxidize DCF (Fig. 9C). We did not examine the concentration-dependency of catalase-mediated H2S production from SO2 but it would be interesting if there was a regulatory feedback inhibition on H2S production. The production of H2S from SO2 may involve a dismutation reaction of SO2 such as the following over-all reaction;
| 2SO2 + 2H2O –> H2S + O2 + H2SO4 | (5) |
4.1.3.4. Thioredoxin
Thioredoxin (Trx) is an evolutionarily conserved antioxidant present in essentially all living organisms [70], [71]. The redox-active site of Trx contains two cysteines in a Cys-X-X-Cys sequence where X=amino acids [72] which is similar to DTT (S-C-C-C-C-S, where C = carbon). The ability of catalase to release H2S from Trx (Fig. 10D) suggests that Trx is an endogenous substrate of catalase and that the mechanism of H2S generation is similar to that of DTT. This is further supported by the increased release of H2S from both DTT and Trx in the presence of NADPH. The nearly stoichiometric production of H2S from Trx in the presence of NADPH suggests that Trx could be a significant source of H2S production. This could provide another link to hypoxic responses where NADPH would be expected to increase and thereby increase H2S.
4.1.4. Mechanism of catalase-mediated H2S release from DTT and Trx
It is not clear how catalase mediates the release of H2S from DTT. H2S can be released from thiols by elimination reactions catalyzed by cystathionine β synthase or cystathionine γ lyase, e.g., RSH + R'SH –> R-S-R’ + H2S, or RSH + H2O –> ROH + H2S. To our knowledge no such function for catalase has been described. If H2S is released by a redox reaction an additional oxidant would be required as the formal oxidation state of sulfur in DTT and H2S (and reduced thioredoxin) is −2. The oxidant does not appear to be oxygen as H2S release is enhanced in hypoxia (Fig. 7D). However, the ability of CO to reduce H2S production from DTT by nearly 80% (Fig. 7F) suggests that the reaction does occur at the heme iron, which supports a redox mechanism. Clearly additional studies are necessary to identify the process involved.
4.1.5. Role of NADPH
Bovine catalase is an evolutionarily recent mono-functional clade 3 catalase with NADPH tightly bound to each of its four subunits [35], [73]. The NADPH is believed to prevent formation of inactive compound II from compound I when H2O2 concentrations are low (reviewed in [74]). Because bound NADPH is too large to enter the catalytic pore (which restricts access to molecules larger than H2O2) it is believed that electrons are transferred from unbound NADPH to bound NADPH and then tunneled to the heme [75].
Our experiments showing that NADPH is required for H2S generation from molecules as large as DTT, DATS and Trx raises the question of how (or where) do the reactants access the catalytic site. Vetrano et al. [31] examined the crystal structure of bovine catalase and identified a potential large binding pocket adjacent to the β-barrel region abutting the heme environments that would accommodate and orient large and diverse electronegative substrates such as DCF through interaction with Arg74, Arg111, Arg364 and Phe131. This would appear to be the likely site of sulfur metabolism by bovine catalase as well as provide access for two-electron donation by unbound NADPH. SO2 which is only weakly bound to water may find its way directly to the heme by the channel used by H2O2.
Catalase from A. niger is a large subunit mono-functional clade 2 enzyme that does not have the narrow catalytic pore nor does it bind NADPH or form compound II during low concentrations of H2O2 [73], [74]. We did not find evidence that A. niger catalase could generate H2S from Trx even in the presence of NADPH. This does not completely rule out the possibility of A. niger catalase as a sulfur reductase as other substrates and electron donors may be involved. This could not be pursued in the present studies due to the limited availability of this catalase. However, it is evident that A. niger catalase, is similar to bovine catalase in its ability to oxidize H2S and DCF, albeit at a slower rate (Fig. 11). This is not surprising as the rate of H2O2 dismutation is also slower for A. niger catalase [76].
4.1.6. Product(s) of H2S oxidation
Our studies suggest that the majority of the reaction products of H2S oxidation are most likely elemental sulfur or sulfur oxides (SOn or SnOn) because there was minimal recovery of H2S by DDT after catalase-mediated H2S oxidation when H2S was measured amperometrically and because catalase readily consumes polysulfides. DTT, which unfortunately also liberates a small amount of H2S when dissolved (Table 1; [47]) clearly liberated H2S from polysulfide standards (H2Sn, n=2–4; Table 1) but was unable to do so after H2S reaction with catalase (Fig. 2).
In an attempt to ‘trap’ polysulfides as they are formed, we added combinations of H2S, H2O2 and azide with or without catalase to SSP4, a fluorophore that irreversibly reacts with polysulfides (Fig. 6) and other sulfanes. The greatest SSP4 fluorescence was produced by the combination of H2S and H2O2 in the absence of catalase. Catalase plus H2S also increased SSP4 fluorescence and this was slightly increased by addition of H2O2 in the presence of 40 μM catalase and slightly decreased with H2O2 and 10 μM catalase. Overall, H2O2 appeared to have little consistent effect on catalase mediated formation of polysulfide from H2S suggesting that H2O2 was dismutated so rapidly that it did not have time to react with H2S. Azide inhibited fluorescence produced by H2S in the presence of catalase but not fluorescence produced by H2S and H2O2 in the absence of catalase. This is consistent with azide inhibition of catalase-mediated H2S consumption measured amperometrically and suggests that this is operating at the site of O2 consumption. However, when catalase was added to mixed polysulfides derived from K2Sn, it completely inhibited SSP4 fluorescence irrespective of the presence of H2O2 or azide (Fig. 6C). This suggests that the ‘polysulfides’ produced by catalase and H2S were not of the form H2Sn, even though they remained reactive with SSP4 in the absence of catalase. Thus, if polysulfides are formed they must either rapidly decompose under aerobic conditions, as suggested by Wedmann et al. [77], or they are further metabolized by catalase.
4.1.7. Limitations to the above studies
While these studies point out the potential for catalase to greatly affect sulfur metabolism in cells a few limitations to the study need to be identified. First, the concentrations of H2S and polysulfides that we used likely exceed intracellular levels. This is especially the case for the plate reader experiments where the sensitivity of the fluorophores predetermined the lower limit of concentration. Furthermore the actual concentrations of these sulfur moieties in cells and their distribution has not been resolved. Second, the purity of polysulfides is an issue especially once in solution and exposed to oxygen. Therefore, the measurements of H2S produced from polysulfides (Table 1) are only estimates, although there are clear differences that correlate with the number of sulfur atoms. Finally, there is always some uncertainty regarding the specificity of the fluorophores.
4.1.8. Catalase and evolution
Life began around 3.8 billion years ago (bya) and was chemolithotrophic; the energy that sustained and drove early evolution was obtained from inorganic reducing compounds in the environment. Although still somewhat controversial, a strong case can be made that H2S emanating from hydrothermal vents provided this energy as well as creating the chemical backbone for the earliest catalysts and cell walls [1]. Escape from the chemotrophic existence probably came within the next few hundred million years (~3.6 bya) in the form of anoxigenic photosynthesis [78], [79]. It is also possible that H2S would have been the likely substrate in this reaction [1];
| H2S + CO2 + hv –> (CH2O)n + H2O + S(n) | (6) |
thereby generating a variety of reactive sulfide species that would need metabolic attention. It took over a billion years for organisms (namely cyanobacteria) to develop sufficiently efficient light gathering antennae to replace H2S with the far more abundant H2O and its oxidization product became O2 [79]. Although this “great oxidation event” (GOE) may have transiently increased atmospheric O2 up to 1%, the oceans remained anoxic and became more sulfidic [80]. Eukaryotes appeared around 1.5 bya and upon engulfing cyanobacteria, created the first “plants.” The combined efforts of cyanobacteria and plants accelerated O2 production but it took nearly another billion years, to 0.6 bya before the vast quantities of ferrous iron and sulfide, which previously “mopped up” O2 were oxidized. Only then did O2 begin to accumulate in the oceans and rise to present day levels in the atmosphere. It is generally thought that during this period of Earth's oxygenation organisms either retreated to anoxic environments, died or developed strategies to remove excess O2. To deal with the latter, the “ox-tox” hypothesis, posits that this led to the appearance of the classical antioxidant mechanisms including SOD, catalase, glutathione reductase, peroxiredoxins and thioredoxins, molecules that were designed to rapidly dispose of ROS [81].
We propose an alternate scenario; the metabolic “machinery” that was necessary to deal with RSS had to have developed early in evolution commensurate with H2S-based anoxigenic photosynthesis. These pathways were then fine-tuned over a billion years of evolution before organisms began producing oxygen. Just as the transition from H2S- to H2O-based photosynthesis merely required better light-gathering capabilities, the transition from RSS to ROS based metabolism probably required only slight enzymatic modifications. It is not surprising, then, that many (if not all) of the “antioxidant” mechanisms, including superoxide dismutases, thioredoxins, peroxiredoxins and catalases appeared early in evolution [70], [72], [82], [83], [84], [85], [86], [87], [88], probably well before either oxygen or ROS were a serious threat.
Manganese catalase, most likely the ancestral catalase, appeared at least 3 billion years ago, nearly three-quarters of a billion years before the GOE. It has been proposed that the function of Mn catalase was to generate oxygen from peroxide dismutation and thereby provide oxygen to early aerobes [79], [89]. We have questioned this hypothesis based on the perceived difficulty of finding a sufficient and reliable source of peroxide to sustain life compared to the ready availability of RSS [1]. Our identification of the ability of catalase to generate H2S from sulfur-bearing molecules under reducing (hypoxic) conditions and to paradoxically oxidize them under oxidizing conditions bespeaks of an enzyme that initially dealt with RSS. It also supports the hypothesis that some of these “primordial” functions have been retained in present-day organisms to metabolize and regulate RSS.
Collectively, our results suggest that our view of catalase as just an antioxidant enzyme may be too myopic. The next challenge will be in identifying additional endogenous sulfur substrates and products that may be under homeostatic control by this and other “redox-regulatory” enzymes.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors (http://Www.Nature.Com/Nature).
Author contributions
K.R.O. designed the study, M.A. F.A. N.A, E.R.D. and Y.G. performed the experiments. K.R.O., E.R.D and K.D.S. analyzed the data interpreted the results. K.R.O. wrote the manuscript and all authors discussed the results and commented on the manuscript.
Acknowledgments
This research was supported by National Science Foundation Grant No. IOS 1446310.
References
- 1.Olson K.R., Straub K.D. The role of hydrogen sulfide in evolution and the evolution of hydrogen sulfide in metabolism and signaling. Physiology. 2016;31(1):60–72. doi: 10.1152/physiol.00024.2015. [DOI] [PubMed] [Google Scholar]
- 2.Brandes N., Tienson H., Lindemann A., Vitvitsky V., Reichmann D., Banerjee R., Jakob U. Time line of redox events in aging postmitotic cells. Elife. 2013;2:e00306. doi: 10.7554/eLife.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown D.I., Griendling K.K. Regulation of signal transduction by reactive oxygen species in the cardiovascular system. Circ. Res. 2015;116(3):531–549. doi: 10.1161/CIRCRESAHA.116.303584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y.R., Zweier J.L. Cardiac mitochondria and reactive oxygen species generation. Circ. Res. 2014;114(3):524–537. doi: 10.1161/CIRCRESAHA.114.300559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chouchani E.T., Pell V.R., James A.M., Work L.M., Saeb-Parsy K., Frezza C., Krieg T., Murphy M.P. A unifying mechanism for mitochondrial superoxide production during ischemia-reperfusion injury. Cell Metab. 2016;23(2):254–263. doi: 10.1016/j.cmet.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Circu M.L., Aw T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010;48(6):749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Circu M.L., Aw T.Y. Intestinal redox biology and oxidative stress. Semin. Cell Dev. Biol. 2012;23(7):729–737. doi: 10.1016/j.semcdb.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson S.B. Investigating the role of reactive oxygen species in regulating autophagy. Methods Enzym. 2013;528:217–235. doi: 10.1016/B978-0-12-405881-1.00013-6. [DOI] [PubMed] [Google Scholar]
- 9.Goncalves R.L., Quinlan C.L., Perevoshchikova I.V., Hey-Mogensen M., Brand M.D. Sites of superoxide and hydrogen peroxide production by muscle mitochondria assessed ex vivo under conditions mimicking rest and exercise. J. Biol. Chem. 2015;290(1):209–227. doi: 10.1074/jbc.M114.619072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambeth J.D., Neish A.S. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu. Rev. Pathol. 2014;9:119–145. doi: 10.1146/annurev-pathol-012513-104651. [DOI] [PubMed] [Google Scholar]
- 11.Li X., Fang P., Mai J., Choi E.T., Wang H., Yang X.F. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013;6:19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 13.Olschewski A., Weir E.K. Redox regulation of ion channels in the pulmonary circulation. Antioxid. Redox Signal. 2014 doi: 10.1089/ars.2014.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radi E., Formichi P., Battisti C., Federico A. Vol. 42. 2014. Apoptosis and oxidative stress in neurodegenerative diseases; pp. S125–S152. (J. Alzheimer's Dis.). [DOI] [PubMed] [Google Scholar]
- 15.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24(10):R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz E., Wenzel P., Munzel T., Daiber A. Mitochondrial redox signaling: interaction of mitochondrial reactive oxygen species with other sources of oxidative stress. Antioxid. Redox Signal. 2014;20(2):308–324. doi: 10.1089/ars.2012.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma K. Obesity and diabetic kidney disease: role of oxidant stress and redox balance. Antioxid. Redox Signal. 2016;25(4):208–216. doi: 10.1089/ars.2016.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winterbourn C.C. Are free radicals involved in thiol-based redox signaling? Free Radic. Biol. Med. 2014 doi: 10.1016/j.freeradbiomed.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Lo C.M., Carroll K.S. The redox biochemistry of protein sulfenylation and sulfinylation. J. Biol. Chem. 2013;288(37):26480–26488. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filipovic M.R. Persulfidation (S-sulfhydration) and H2S. Handb. Exp. Pharmacol. 2015;230:29–59. doi: 10.1007/978-3-319-18144-8_2. [DOI] [PubMed] [Google Scholar]
- 21.Kimura H. Signaling molecules: hydrogen sulfide and polysulfide. Antioxid. Redox Signal. 2015;22:362–376. doi: 10.1089/ars.2014.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy P. Mechanistic chemical perspective of hydrogen sulfide signaling. Methods Enzymol. 2015;554:3–29. doi: 10.1016/bs.mie.2014.11.036. [DOI] [PubMed] [Google Scholar]
- 23.Ono K., Akaike T., Sawa T., Kumagai Y., Wink D.A., Tantillo D.J., Hobbs A.J., Nagy P., Xian M., Lin J., Fukuto J.M. Redox chemistry and chemical biology of HS, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic. Biol. Med. 2014 doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul B.D., Snyder S.H. Protein sulfhydration. Methods Enzym. 2015;555:79–90. doi: 10.1016/bs.mie.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Toohey J.I., Cooper A.J. Thiosulfoxide (sulfane) sulfur: new chemistry and new regulatory roles in biology. Molecules. 2014;19(8):12789–12813. doi: 10.3390/molecules190812789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palinkas Z., Furtmuller P.G., Nagy A., Jakopitsch C., Pirker K.F., Magierowski M., Jasnos K., Wallace J.L., Obinger C., Nagy P. Interactions of hydrogen sulfide with myeloperoxidase. Br. J. Pharmacol. 2015;172(6):1516–1532. doi: 10.1111/bph.12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLeon E.R., Gao Y., Huang E., Arif M., Arora N., Divietro A., Patel S., Olson K.R. A case of mistaken identity: are reactive oxygen species actually reactive sulfide species? Am. J. Physiol Regul. Integr. Comp. Physiol. 2016;310(7) doi: 10.1152/ajpregu.00455.2015. (R549-560) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield N.L., Kreimier E.L., Verdial F.C., Skovgaard N., Olson K.R. Reappraisal of H2S/sulfide concentration in vertebrate blood and its potential significance in ischemic preconditioning and vascular signaling. Am. J. Physiol Regul. Integr. Comp. Physiol. 2008;294(6):R1930–R1937. doi: 10.1152/ajpregu.00025.2008. [DOI] [PubMed] [Google Scholar]
- 29.Boutilier R.G., Heming T.A., Iwama G.K. Academic Press; 1984. Physiochemical Parameters for Use in Fish Respiratory Physiology. (City) [Google Scholar]
- 30.Olson K.R. A practical look at the chemistry and biology of hydrogen sulfide. Antioxid. Redox Signal. 2012;17(1):32–44. doi: 10.1089/ars.2011.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vetrano A.M., Heck D.E., Mariano T.M., Mishin V., Laskin D.L., Laskin J.D. Characterization of the oxidase activity in mammalian catalase. J. Biol. Chem. 2005;280(42):35372–35381. doi: 10.1074/jbc.M503991200. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson M., Kurz T., Brunk U.T., Nilsson S.E., Frennesson C.I. What does the commonly used DCF test for oxidative stress really show? Biochem. J. 2010;428(2):183–190. doi: 10.1042/BJ20100208. [DOI] [PubMed] [Google Scholar]
- 33.Gaetani G.F., Ferraris A.M., Sanna P., Kirkman H.N. A novel NADPH:(bound) NADP+ reductase and NADH:(bound) NADP+ transhydrogenase function in bovine liver catalase. Biochem. J. 2005;385(Pt 3):763–768. doi: 10.1042/BJ20041495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuchi-Torii K., Hayashi S., Nakamoto H., Nakamura S. Properties of Aspergillus niger catalase. J. Biochem. 1982;92(5):1449–1456. doi: 10.1093/oxfordjournals.jbchem.a134069. [DOI] [PubMed] [Google Scholar]
- 35.Kirkman H.N., Gaetani G.F. Catalase: a tetrameric enzyme with four tightly bound molecules of NADPH. Proc. Natl. Acad. Sci. USA. 1984;81(14):4343–4347. doi: 10.1073/pnas.81.14.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen G., Hochstein P. Generation of hydrogen peroxide in erythrocytes by hemolytic agents. Biochemistry. 1964;3:895–900. doi: 10.1021/bi00895a006. [DOI] [PubMed] [Google Scholar]
- 37.Heck D.E., Shakarjian M., Kim H.D., Laskin J.D., Vetrano A.M. Mechanisms of oxidant generation by catalase. Ann. N.Y. Acad. Sci. 2010;1203:120–125. doi: 10.1111/j.1749-6632.2010.05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacob H.S., Ingbar S.H., Jandl J.H. Oxidative hemolysis and erythrocyte metabolism in hereditary acatalasia. J. Clin. Investig. 1965;44:1187–1199. doi: 10.1172/JCI105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Truong D.H., Eghbal M.A., Hindmarsh W., Roth S.H., O'Brien P.J. Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab. Rev. 2006;38(4):733–744. doi: 10.1080/03602530600959607. [DOI] [PubMed] [Google Scholar]
- 40.Olson K.R. Hydrogen sulfide as an oxygen sensor. Antioxid. Redox Signal. 2014 doi: 10.1089/ars.2014.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller S., Riedel H.D., Stremmel W. Direct evidence for catalase as the predominant H2O2 -removing enzyme in human erythrocytes. Blood. 1997;90(12):4973–4978. [PubMed] [Google Scholar]
- 42.Searcy D.G., Lee S.H. Sulfur reduction by human erythrocytes. J. Exp. Zool. 1998;282(3):310–322. doi: 10.1002/(sici)1097-010x(19981015)282:3<310::aid-jez4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 43.Bos E.M., Leuvenink H.G., Snijder P.M., Kloosterhuis N.J., Hillebrands J.L., Leemans J.C., Florquin S., van G.H. Hydrogen sulfide-induced hypometabolism prevents renal ischemia/reperfusion injury. J. Am. Soc. Nephrol. 2009;20(9):1901–1905. doi: 10.1681/ASN.2008121269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bos E.M., Snijder P.M., Jekel H., Weij M., Leemans J.C., van Dijk M.C., Hillebrands J.L., Lisman T., van G.H., Leuvenink H.G. Beneficial effects of gaseous hydrogen sulfide in hepatic ischemia/reperfusion injury. Transpl. Int. 2012;25(8):897–908. doi: 10.1111/j.1432-2277.2012.01514.x. [DOI] [PubMed] [Google Scholar]
- 45.Shibuya N., Koike S., Tanaka M., Ishigami-Yuasa M., Kimura Y., Ogasawara Y., Fukui K., Nagahara N., Kimura H. A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat. Commun. 2013;4:1366. doi: 10.1038/ncomms2371. [DOI] [PubMed] [Google Scholar]
- 46.Miyamoto R., Otsuguro K.I., Yamaguchi S., Ito S. Contribution of cysteine aminotransferase and mercaptopyruvate sulfurtransferase to hydrogen sulfide production in peripheral neurons. J. Neurochem. 2014 doi: 10.1111/jnc.12698. [DOI] [PubMed] [Google Scholar]
- 47.Olson K.R., DeLeon E.R., Gao Y., Hurley K., Sadauskas V., Batz C., Stoy G.F. Thiosulfate: a readily accessible source of hydrogen sulfide in oxygen sensing. Am. J. Physiol Regul. Integr. Comp. Physiol. 2013;305:R592–R603. doi: 10.1152/ajpregu.00421.2012. [DOI] [PubMed] [Google Scholar]
- 48.Shibuya N., Tanaka M., Yoshida M., Ogasawara Y., Togawa T., Ishii K., Kimura H. 3-Mercaptopyruvate sulfurtransferase produces hydrogen sulfide and bound sulfane sulfur in the brain. Antioxid. Redox Signal. 2009;11:703–714. doi: 10.1089/ars.2008.2253. [DOI] [PubMed] [Google Scholar]
- 49.Tanabe S. Development of assay methods for endogenous inorganic sulfur compounds and sulfurtransferases and evaluation of the physiological functions of bound sulfur. Yakugaku Zasshi. 2008;128(6):881–900. doi: 10.1248/yakushi.128.881. [DOI] [PubMed] [Google Scholar]
- 50.Benavides G.A., Squadrito G.L., Mills R.W., Patel H.D., Isbell T.S., Patel R.P., Darley-Usmar V.M., Doeller J.E., Kraus D.W. Hydrogen sulfide mediates the vasoactivity of garlic. Proc. Natl. Acad. Sci. USA. 2007;104(46):17977–17982. doi: 10.1073/pnas.0705710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLeon E.R., Gao Y., Huang E., Olson K.R. Garlic oil polysulfides: H2s- and O2-independent prooxidants in buffer and antioxidants in cells. Am. J. Physiol Regul. Integr. Comp. Physiol. 2016;310(11):R1212–1225. doi: 10.1152/ajpregu.00061.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bekker A., Barley M.E., Fiorentini M.L., Rouxel O.J., Rumble D., Beresford S.W. Atmospheric sulfur in Archean komatiite-hosted nickel deposits. Science. 2009 2009;326(5956):1086–1089. doi: 10.1126/science.1177742. [DOI] [PubMed] [Google Scholar]
- 53.Keppler H. Experimental evidence for the source of excess sulfur in explosive volcanic eruptions. Science. 1999;284(5420):1652–1654. doi: 10.1126/science.284.5420.1652. [DOI] [PubMed] [Google Scholar]
- 54.Balazy M., bu-Yousef I.A., Harpp D.N., Park J. Identification of carbonyl sulfide and sulfur dioxide in porcine coronary artery by gas chromatography/mass spectrometry, possible relevance to EDHF. Biochem. Biophys. Res. Commun. 2003;311(3):728–734. doi: 10.1016/j.bbrc.2003.10.055. [DOI] [PubMed] [Google Scholar]
- 55.Du S.X., Jin H.F., Bu D.F., Zhao X., Geng B., Tang C.S., Du J.B. Endogenously generated sulfur dioxide and its vasorelaxant effect in rats. Acta Pharmacol. Sin. 2008;29(8):923–930. doi: 10.1111/j.1745-7254.2008.00845.x. [DOI] [PubMed] [Google Scholar]
- 56.Luo L., Chen S., Jin H., Tang C., Du J. Endogenous generation of sulfur dioxide in rat tissues. Biochem. Biophys. Res. Commun. 2011;415(1):61–67. doi: 10.1016/j.bbrc.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 57.Meng Z., Li J., Zhang Q., Bai W., Yang Z., Zhao Y., Wang F. Vasodilator effect of gaseous sulfur dioxide and regulation of its level by Ach in rat vascular tissues. Inhal. Toxicol. 2009;21(14):1223–1228. doi: 10.3109/08958370902798463. [DOI] [PubMed] [Google Scholar]
- 58.Hansell A., Oppenheimer C. Health hazards from volcanic gases: a systematic literature review. Arch. Environ. Health. 2004;59(12):628–639. doi: 10.1080/00039890409602947. [DOI] [PubMed] [Google Scholar]
- 59.Wang X.B., Du J.B., Cui H. Sulfur dioxide, a double-faced molecule in mammals. Life Sci. 2014;98(2):63–67. doi: 10.1016/j.lfs.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 60.Chen S., Du J., Liang Y., Ochs T., Liu D., Zhu L., Tang X., Tang C., Jin H. Sulfur dioxide inhibits excessively activated endoplasmic reticulum stress in rats with myocardial injury. Heart Vessels. 2011 doi: 10.1007/s00380-011-0192-7. [DOI] [PubMed] [Google Scholar]
- 61.Chen S., Du J., Liang Y., Zhang R., Tang C., Jin H. Sulfur dioxide restores calcium homeostasis disturbance in rat with isoproterenol-induced myocardial injury. Histol. Histopathol. 2012;27(9):1219–1226. doi: 10.14670/HH-27.1219. [DOI] [PubMed] [Google Scholar]
- 62.Chen S., Zheng S., Liu Z., Tang C., Zhao B., Du J., Jin H. Endogeous sulfur dioxide protects against oleic acid-induced acute lung injury in association with inhibition of oxidative stress in rats. Lab. Investig. 2015;95(2):142–156. doi: 10.1038/labinvest.2014.147. [DOI] [PubMed] [Google Scholar]
- 63.Du S.X., Jin H.F., Bu D.F., Zhao X., Geng B., Tang C.S., Du J.B. Endogenously generated sulfur dioxide and its vasorelaxant effect in rats. Acta Pharmacol. Sin. 2008;29(8):923–930. doi: 10.1111/j.1745-7254.2008.00845.x. [DOI] [PubMed] [Google Scholar]
- 64.Huang P., Sun Y., Yang J., Chen S., Liu A.D., Holmberg L., Huang X., Tang C., Du J., Jin H. The ERK1/2 signaling pathway is involved in sulfur dioxide preconditioning-induced protection against cardiac dysfunction in isolated perfused rat heart subjected to myocardial ischemia/reperfusion. Int. J. Mol. Sci. 2013;14(11):22190–22201. doi: 10.3390/ijms141122190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin H., Wang Y., Wang X., Sun Y., Tang C., Du J. Sulfur dioxide preconditioning increases antioxidative capacity in rat with myocardial ischemia reperfusion (I/R) injury. Nitric Oxide. 2013;32:56–61. doi: 10.1016/j.niox.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 66.Liang Y., Liu D., Ochs T., Tang C., Chen S., Zhang S., Geng B., Jin H., Du J. Endogenous sulfur dioxide protects against isoproterenol-induced myocardial injury and increases myocardial antioxidant capacity in rats. Lab. Investig. 2011;91(1):12–23. doi: 10.1038/labinvest.2010.156. [DOI] [PubMed] [Google Scholar]
- 67.Wang X.B., Huang X.M., Ochs T., Li X.Y., Jin H.F., Tang C.S., Du J.B. Effect of sulfur dioxide preconditioning on rat myocardial ischemia/reperfusion injury by inducing endoplasmic reticulum stress. Basic Res. Cardiol. 2011;106(5):865–878. doi: 10.1007/s00395-011-0176-x. [DOI] [PubMed] [Google Scholar]
- 68.Meng Z., Li Y., Li J. Vasodilatation of sulfur dioxide derivatives and signal transduction. Arch. Biochem. Biophys. 2007;467(2):291–296. doi: 10.1016/j.abb.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 69.Falk M., Giguere P.A. On the nature of sulphurous acid. Can. J. Chem. 1958;36(7):1121–1125. [Google Scholar]
- 70.Novoselov S.V., Gladyshev V.N. Non-animal origin of animal thioredoxin reductases: implications for selenocysteine evolution and evolution of protein function through carboxy-terminal extensions. Protein Sci. 2003;12(2):372–378. doi: 10.1110/ps.0226503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams C.H., Arscott L.D., Muller S., Lennon B.W., Ludwig M.L., Wang P.F., Veine D.M., Becker K., Schirmer R.H. Thioredoxin reductase two modes of catalysis have evolved. Eur. J. Biochem. 2000;267(20):6110–6117. doi: 10.1046/j.1432-1327.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 72.Lu J., Holmgren A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 73.Zamocky M., Gasselhuber B., Furtmuller P.G., Obinger C. Molecular evolution of hydrogen peroxide degrading enzymes. Arch. Biochem. Biophys. 2012;525(2):131–144. doi: 10.1016/j.abb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirkman H.N., Gaetani G.F. Mammalian catalase: a venerable enzyme with new mysteries. Trends Biochem. Sci. 2007;32(1):44–50. doi: 10.1016/j.tibs.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 75.Olson L.P., Bruice T.C. Electron tunneling and ab initio calculations related to the one-electron oxidation of NAD(P)H bound to catalase. Biochemistry. 1995;34(22):7335–7347. doi: 10.1021/bi00022a006. [DOI] [PubMed] [Google Scholar]
- 76.Switala J., Loewen P.C. Diversity of properties among catalases. Arch. Biochem. Biophys. 2002;401(2):145–154. doi: 10.1016/S0003-9861(02)00049-8. [DOI] [PubMed] [Google Scholar]
- 77.Wedmann R., Bertlein S., Macinkovic I., Boltz S., Miljkovic J.L., Munoz L.E., Herrmann M., Filipovic M.R. Working with "HS": facts and apparent artifacts. Nitric Oxide. 2014 doi: 10.1016/j.niox.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 78.Hohmann-Marriott M.F., Blankenship R.E. Evolution of photosynthesis. Annu. Rev. Plant Biol. 2011;62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- 79.Raymond J., Blankenship R.E. The origin of the oxygen-evolving complex. Coord. Chem. Rev. 2008;252:377–383. [Google Scholar]
- 80.Planavsky N.J., Reinhard C.T., Wang X., Thomson D., McGoldrick P., Rainbird R.H., Johnson T., Fischer W.W., Lyons T.W. Earth history. low mid-Proterozoic atmospheric oxygen levels and the delayed rise of animals. Science. 2014;346(6209):635–638. doi: 10.1126/science.1258410. [DOI] [PubMed] [Google Scholar]
- 81.Kurland C.G., Andersson S.G. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 2000;64(4):786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dibrova D.V., Cherepanov D.A., Galperin M.Y., Skulachev V.P., Mulkidjanian A.Y. Evolution of cytochrome bc complexes: from membrane-anchored dehydrogenases of ancient bacteria to triggers of apoptosis in vertebrates. Biochim. Biophys. Acta. 2013;1827(11–12):1407–1427. doi: 10.1016/j.bbabio.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grace S.C. Phylogenetic distribution of superoxide dismutase supports an endosymbiotic origin for chloroplasts and mitochondria. Life Sci. 1990;47(21):1875–1886. doi: 10.1016/0024-3205(90)90399-c. [DOI] [PubMed] [Google Scholar]
- 84.Hanschmann E.M., Godoy J.R., Berndt C., Hudemann C., Lillig C.H. Thioredoxins, glutaredoxins, and peroxiredoxins--molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid. Redox Signal. 2013;19(13):1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inupakutika M.A., Sengupta S., Devireddy A.R., Azad R.K., Mittler R. The evolution of reactive oxygen species metabolism. J. Exp. Bot. 2016;67(21):5933–5943. doi: 10.1093/jxb/erw382. [DOI] [PubMed] [Google Scholar]
- 86.Knoops B., Loumaye E., Van Der Eecken V. Evolution of the peroxiredoxins. Subcell. Biochem. 2007;44:27–40. doi: 10.1007/978-1-4020-6051-9_2. [DOI] [PubMed] [Google Scholar]
- 87.Miller A.F. Superoxide dismutases: ancient enzymes and new insights. FEBS Lett. 2012;586(5):585–595. doi: 10.1016/j.febslet.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang X.H., Weissbach H. Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases. Biol. Rev. Camb. Philos. Soc. 2008;83(3):249–257. doi: 10.1111/j.1469-185X.2008.00042.x. [DOI] [PubMed] [Google Scholar]
- 89.Kim K.M., Qin T., Jiang Y.Y., Chen L.L., Xiong M., Caetano-Anolles D., Zhang H.Y., Caetano-Anolles G. Protein domain structure uncovers the origin of aerobic metabolism and the rise of planetary oxygen. Structure. 2012;20(1):67–76. doi: 10.1016/j.str.2011.11.003. [DOI] [PubMed] [Google Scholar]