Abstract
Background
The BioBank Japan (BBJ) Project was launched in 2003 with the aim of providing evidence for the implementation of personalized medicine by constructing a large, patient-based biobank (BBJ). This report describes the study design and profile of BBJ participants who were registered during the first 5-year period of the project.
Methods
The BBJ is a registry of patients diagnosed with any of 47 target common diseases. Patients were enrolled at 12 cooperative medical institutes all over Japan from June 2003 to March 2008. Clinical information was collected annually via interviews and medical record reviews until 2013. We collected DNA from all participants at baseline and collected annual serum samples until 2013. In addition, we followed patients who reported a history of 32 of the 47 target diseases to collect survival data, including cause of death.
Results
During the 5-year period, 200,000 participants were registered in the study. The total number of cases was 291,274 at baseline. Baseline data for 199,982 participants (53.1% male) were available for analysis. The average age at entry was 62.7 years for men and 61.5 years for women. Follow-up surveys were performed for participants with any of 32 diseases, and survival time data for 141,612 participants were available for analysis.
Conclusions
The BBJ Project has constructed the infrastructure for genomic research for various common diseases. This clinical information, coupled with genomic data, will provide important clues for the implementation of personalized medicine.
Keywords: BioBank Japan Project, Biobank, Personalized medicine, Genomic research
Highlights
-
•
The BioBank Japan Project (BBJ) enrolled 200,000 patients with 47 target diseases.
-
•
The BBJ is one of the largest patient-based biobanks in the world.
-
•
The BBJ may allow for personalized medicine in the future.
Introduction
With the rapid advancement of human genetic research, researchers have found many genetic variations that contribute to disease susceptibility and drug responses.1, 2, 3 Based on these achievements, personalized medicine is expected to provide optimal medical treatment by taking into account individual genetic makeup.4, 5
To better achieve this goal, the Japanese Ministry of Education, Culture, Sports, Science and Technology launched the BioBank Japan (BBJ) Project in 2003. This project is being conducted in three 5-year periods. In the first period (April 2003 to March 2008), the project constructed a large, patient-based biobank, referred to as the BBJ, as the basic infrastructure for genetic research for common diseases; this was accomplished through the collection of clinical information and biological samples related to 47 target diseases and with the cooperation of 12 Japanese medical institutes. In the second period (April 2008 to March 2013), the project conducted various genomic investigations and identified genetic variations related to disease susceptibility and drug responses using a genome-wide association study approach.6 In the third period (April 2013 to March 2018), the project will expand the BBJ by enrolling new participants with 38 target diseases, starting sequence-based genomic analyses, and constructing a tissue bank.
Ultimately, the BBJ aims to (1) discover the genes that contribute to various diseases and those related to the efficacy or adverse reactions of drugs, (2) to provide useful information regarding molecular targets for the evidence-based development of new drugs and/or diagnostic tools, (3) to examine interactions between genetic and treatment factors related to the progression of diseases, and (4) to provide important medical information that can be applied to the implementation of personalized medicine.7 This project is jointly managed by The Institute of Medicine of The University of Tokyo and the RIKEN Center for Integrative Medical Sciences.
This paper provides an overview of the BBJ Project as well as a profile of the BBJ participants who were registered in the first 5-year period of the project.
Methods
Study participants
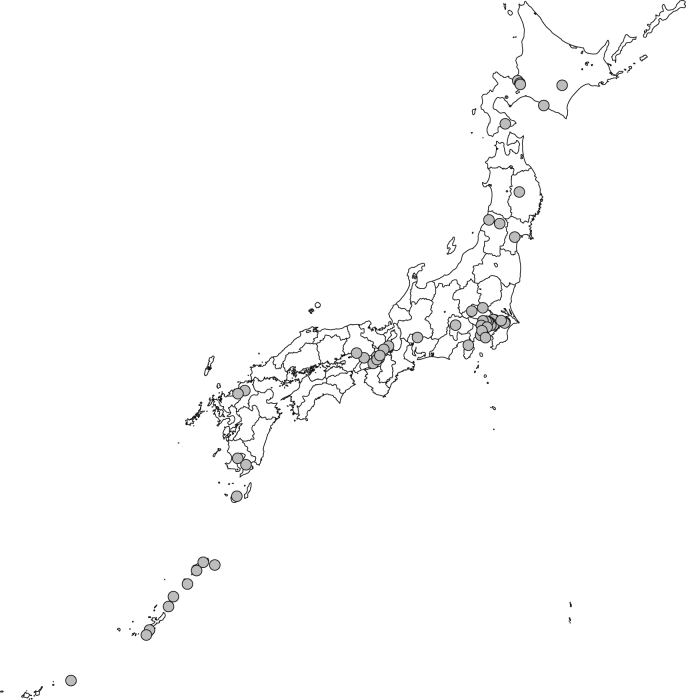
The BBJ is a multi-institutional hospital-based registry that was initially designed to focus on the use of human genetic research. Therefore, this project registered not only patients with newly developed diseases (incident cases) but also patients who were diagnosed and treated before starting the project (prevalent cases). Project participants were recruited between June 2003 and March 2008 from 66 hospitals, which consisted of 12 cooperating medical institutions located throughout Japan (Fig. 1). Attending physicians identified patients with any of 47 target diseases. Before enrollment, patients received a detailed explanation regarding the project from expert, independent medical coordinators who were specifically trained for this project. Biological samples and clinical information were collected and anonymized on-site at the cooperating hospitals. All study participants had been diagnosed with one or more of 47 target diseases, which are listed in Table 2. These target diseases were selected by clinical importance based on morbidity or mortality in Japan through discussion with cooperating medical institutions. Diagnoses of these diseases were based on the physicians' diagnoses at cooperating hospitals. We excluded patients who had received a bone marrow transplant and those who were not of East Asian descent.
Fig. 1.
Geographical distribution of cooperating hospitals.
Table 2.
Number of cases of 47 diseases and disease durations at the time of entry in the BioBank Japan Project.
| Disease group | Disease name | Number of cases (with serum) | Median disease duration at entry (interquartile range, years) | ||
|---|---|---|---|---|---|
| Malignant tumors | Lung cancer | 3779 | (3777) | 0.7 | (0.2–2.9) |
| Esophageal cancer | 1291 | (1290) | 0.4 | (0.2–2.8) | |
| Gastric cancer | 6322 | (6316) | 1.0 | (0.2–4.0) | |
| Colorectal cancer | 6759 | (6754) | 1.0 | (0.2–3.4) | |
| Liver cancer | 1924 | (1922) | 1.2 | (0.3–3.3) | |
| Pancreas cancer | 392 | (392) | 0.4 | (0.1–1.5) | |
| Gallbladder/Cholangiocarcinoma | 392 | (393) | 0.5 | (0.2–2.1) | |
| Prostate cancer | 5066 | (5065) | 1.0 | (0.3–2.8) | |
| Breast cancer | 6336 | (6336) | 0.8 | (0.2–3.6) | |
| Cervical cancer | 1218 | (1218) | 1.8 | (0.4–5.3) | |
| Uterine cancer | 1026 | (1026) | 1.5 | (0.4–4.4) | |
| Ovarian cancer | 888 | (888) | 1.8 | (0.5–4.9) | |
| Hematopoietic tumor | 1307 | (1303) | 2.4 | (0.9–5.1) | |
| Cerebral diseases | Cerebral infarction | 16,534 | (16,522) | 2.2 | (0.4–5.6) |
| Cerebral aneurysm | 2710 | (2709) | N/A | ||
| Epilepsy | 2303 | (2275) | 5.6 | (1.7–13.4) | |
| Respiratory diseases | Bronchial asthma | 8700 | (8642) | 9.0 | (3.0–20.0) |
| Pulmonary tuberculosis | 863 | (862) | N/A | ||
| Chronic obstructive pulmonary diseasea | 2774 | (2771) | 3.0 | (1.0–8.0) | |
| Interstitial lung disease/Pulmonary fibrosis | 808 | (804) | 3.0 | (1.0–7.0) | |
| Cardiovascular diseases | Myocardial infarction | 13,272 | (13,270) | 3.8 | (0.7–8.7) |
| Unstable anginaa | 4330 | (4324) | 2.6 | (0.5–6.4) | |
| Stable anginaa | 14,807 | (14,800) | 3.6 | (1.0–7.9) | |
| Arrhythmiaa | 15,912 | (15,905) | 3.7 | (1.1–8.3) | |
| Heart failurea | 7610 | (7601) | 1.9 | (0.4–5.4) | |
| Peripheral arterial diseases | 2683 | (2679) | N/A | ||
| Liver diseases | Chronic hepatitis B | 1346 | (1345) | N/A | |
| Chronic hepatitis C | 5819 | (5817) | N/A | ||
| Liver cirrhosis | 2519 | (2509) | N/A | ||
| Urologic diseases | Nephrotic syndrome | 1056 | (1007) | N/A | |
| Urolithiasis | 6307 | (6306) | N/A | ||
| Metabolic diseases | Osteoporosis | 6743 | (6739) | N/A | |
| Diabetes mellitusb | 39,697 | (39,686) | 6.0 | (2.0–12.0) | |
| Dyslipidemiab | 43,812 | (43,787) | 3.0 | (1.0–7.0) | |
| Endocrine diseases | Graves' disease | 2323 | (2322) | N/A | |
| Connective tissue diseases | Rheumatoid arthritis | 4139 | (4139) | 7.7 | (2.6–15.3) |
| Allergic diseases | Hay fever | 5658 | (5645) | N/A | |
| Dermatologic diseases | Drug eruption | 585 | (586) | N/A | |
| Atopic dermatitisa | 2938 | (2896) | 16.0 | (5.0–25.0) | |
| Keloid | 809 | (808) | N/A | ||
| Gynecologic diseases | Uterine fibroid | 5904 | (5902) | N/A | |
| Endometriosis | 1843 | (1843) | N/A | ||
| Pediatric diseases | Febrile seizure | 333 | (275) | N/A | |
| Ophthalmologic diseases | Glaucoma | 4755 | (4753) | 3.6 | (1.1–8.2) |
| Cataract | 20,002 | (19,988) | 1.5 | (0.2–5.4) | |
| Dental diseases | Periodontitis | 3898 | (3896) | 0.1 | (0.0–1.7) |
| Other | Amyotrophic lateral sclerosisa | 782 | (771) | 4.0 | (2.1–6.9) |
Abbreviation: N/A, not available.
The duration of disease was calculated based on the time of disease onset to the time of entry into the registry.
Because we surveyed calendar year of diagnosis for these diseases, the periods from diagnosis were not measured in units of days and months.
Biobanking of biological samples and clinical information
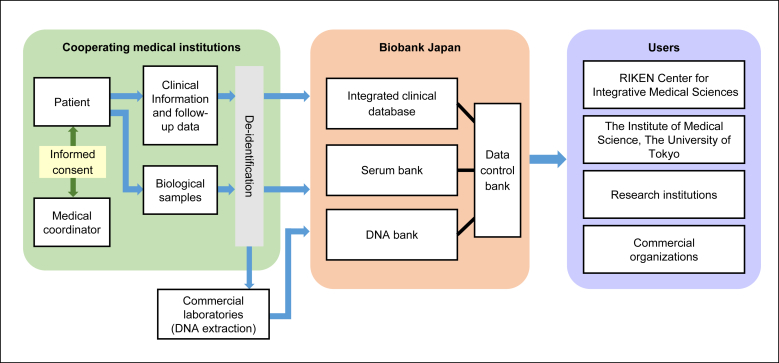
The study flow related to the collection of biological samples and clinical information is shown in Fig. 2. For biological samples, we collected DNA and serum samples from participants at the cooperating hospitals and stored them at the BBJ DNA bank and serum bank, respectively. Details of the blood sampling and storage processes are described in the Blood Sampling and Storage section. Clinical information was initially stored in the local server at the cooperating hospitals and was subsequently sent to the BBJ at the end of every year after anonymization.
Fig. 2.
The flow of the collection of the sample.
Samples stored in the BBJ were provided to research institutions and companies beginning in 2005 after receiving approval from the Sample Providing Committee of this project. Detailed information about the distribution of the samples is available at the project website (https://biobankjp.org/index.html; Japanese only).8
Baseline survey
We collected baseline clinical information through interviews and reviews of medical records using a standardized questionnaire (Table 1); full details of the questionnaire are available at the project website (https://biobankjp.org/sample/pdf/list7.1.4.pdf; Japanese only). The common items addressed during each interview included smoking and alcohol habits, height, weight, systolic and diastolic blood pressure, past history and family history of various diseases, frequency of food intake and physical exercise, and reproductive history (for female participants). The common items collected from medical record reviews included information about birth year, sex, drug use within one month before study entry, history of adverse drug events, and routine laboratory examination data (including complete blood count, urinalysis, and biochemical tests such as measurements of liver enzymes, kidney function, lipids, and blood sugar).
Table 1.
Clinical Information collected during the annual BioBank Japan survey.
| Categories | Classifications |
|---|---|
| Common items | Basic clinical Information |
| Current drug use | |
| History of adverse drug events | |
| Laboratory examination items | Malignant tumors |
| Cerebral diseases | |
| Respiratory diseases | |
| Cardiovascular diseases | |
| Liver diseases | |
| Urologic diseases | |
| Metabolic diseases | |
| Endocrine diseases | |
| Connective tissue diseases | |
| Allergic diseases | |
| Dermatological diseases | |
| Gynecological diseases | |
| Pediatric diseases | |
| Ophthalmologic diseases | |
| Dental diseases | |
| Disease-specific items | – |
Disease-specific laboratory tests and imaging data were collected for 15 disease groups. For example, we collected information related to surgical history, chemotherapy, hormonal therapy, radiotherapy, and tumor markers for 13 types of cancers. Echocardiogram and coronary angiography information were collected for cardiovascular diseases. Detailed clinical information for each case, such as disease onset, symptoms, subtypes, severity, and complications, was collected separately for disease-specific items. Because the BBJ included both incident and prevalent cases, the duration of disease at registration was calculated based on the date of onset or diagnosis of disease and the date of enrollment. All surveys were performed by medical coordinators at cooperating hospitals.
In this analysis, body mass index (kg/m2) was calculated on the basis of the self-reported height and body weight. Hypertension was defined as a systolic blood pressure ≥140 mmHg, a diastolic blood pressure ≥90 mmHg, or a prescription of antihypertensive drugs.
Data cleansing
To improve the quality of the baseline survey data, we reviewed all data items and performed data cleansing for 4627 out of 17,850 items that were available for >50% of participants or that were determined to be clinically important variables (e.g., TNM classification for cancer). We checked the distribution of all numeric data and excluded outliers for each variable. For categorical data, we checked the concordance of related variables and excluded discordant data in order to achieve consistency among the values.
Blood sampling and storage
A 14 mL sample of whole blood was obtained from each participant at baseline using two 7 mL EDTA-containing tubes. One 7 mL sample of whole blood was sent to one of three commercial laboratories (SRL, BML, and MBC, Japan) for DNA extraction. After DNA was extracted according to the standard procedures in each laboratory, DNA concentration was adjusted to 100 ng/μL and dispensed into three 1 mL tubes with 2D barcodes at the bottom. All DNA tubes were stored at 4–10 °C and delivered to the BBJ. After checking the concordance of the 2D barcodes on the DNA tubes with participant anonymized numbers, all DNA samples were stored in the DNA bank at 4 °C (Fig. 3). For children or participants with whom there were difficulties in drawing blood, we collected buccal swabs, nail trimmings, or hair trimmings to extract DNA.
Fig. 3.
Picture of DNA bank (automated DNA storage system).
Another 7 mL sample of whole blood was centrifuged according to standard procedures at each cooperating hospital, and the resultant serum was dispensed into three 1 mL tubes and labeled with 2D barcodes. All serum samples were initially stored at −80 °C at cooperating hospitals. After several samples were stored, these samples were then delivered to the BBJ. After checking the concordance of the 2D barcodes on the serum tubes with participant anonymized numbers, all serum samples were stored in the serum bank at −150 °C (Fig. 4). Participants were requested to provide serum samples once a year starting the year following enrollment until March 2013.
Fig. 4.
Picture of serum bank.
Follow-up surveys
After the baseline survey, we continuously collected clinical information from participants through reviews of medical records once a year until March 2013. If a participant developed a new disease from among the 47 target diseases, medical coordinators registered this new disease and collected relevant clinical information from medical records.
In addition, we collected survival data for participants with any of 32 of the 47 target diseases based on the participants' registered disease in the 2010 medical record survey. Medical coordinators initially checked the date of last visit and identified participants who had not visited the hospital in more than one year or who had died during the follow-up period. Hospitals requested a copy of the resident card for these participants from local government offices. Medical coordinators then recorded whether the participants were alive, had moved, were unidentified, or had died. For participants who had moved, new addresses were recorded for the next survival survey. For deceased participants, medical coordinators recorded the date of death. We obtained vital statistics from the Statistics and Information Department of the Ministry of Health, Labour and Welfare in Japan and identified the cause of death according to ICD-10 code by matching birth date, date of death, sex, and local government code.
Ethical review
The study protocol for the BBJ Project was approved by the research ethics committees at the Institute of Medical Science, the University of Tokyo, the RIKEN Yokohama Institute, and the 12 cooperating hospitals. All participants gave written consent to participate in the study.
Results
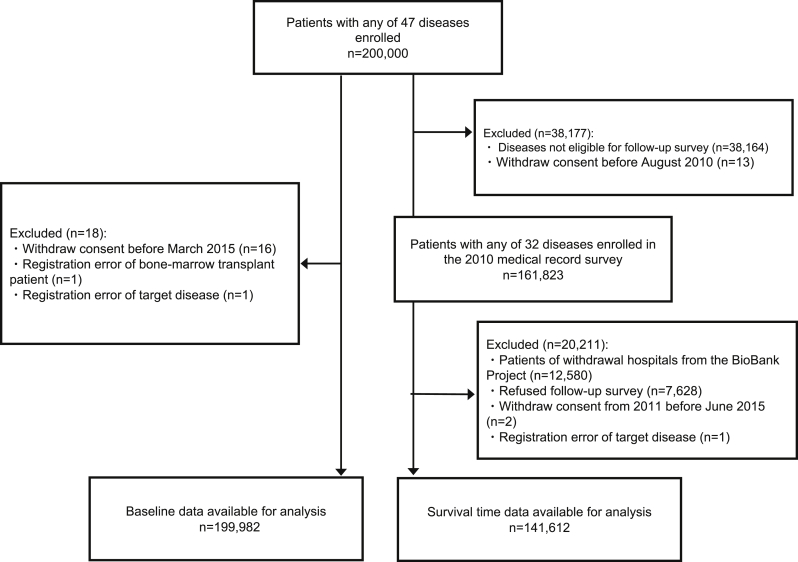
A flow diagram describing the process for the baseline survey and survival survey is shown in Fig. 5. There were 200,000 patients who were enrolled in the study. Since the clinical information of participants who withdrew informed consent or who were mistakenly enrolled was already removed from the clinical database, baseline data for 199,982 participants were available in this analysis.
Fig. 5.
Flow diagram for baseline and follow-up survey data of study participants.
Of the 200,000 participants, 161,823 participants were registered with any of 32 diseases during the 2010 medical record survey. Among them, we excluded 20,211 participants who were enrolled at hospitals that withdrew from the BioBank Project, those who refused to complete the follow-up survey, those who withdrew informed consent, or those who were mistakenly enrolled. Finally, survival data for 141,612 participants were available for analysis. The follow-up rate was 97.0%, and the mean follow-up period was 7.7 years. The details of the results of the follow-up survey are described in another report.9
The total number of cases was 291,274 at baseline (Table 2). The largest number of cases was related to dyslipidemia followed by diabetes mellitus. Information on disease onset or diagnosis was collected for 31 diseases. When we evaluated disease duration at registration, the median disease duration was short for most cancer cases, but was long for cases of atopic dermatitis, bronchial asthma, and rheumatoid arthritis, indicating that the proportion of prevalent cases was high for these diseases. Among the participants, 53.1% were male (Table 3). The average age was 62.7 years for male patients and 61.5 years for female patients. Among participants aged 20 years or older, 27.5% of male and 23.7% of female patients were overweight or obese. The proportion of current smokers was 27.3% for male patients and 10.3% for female patients; the proportion of current drinkers was 54.9% for male patients and 23.6% for female patients. About 51% of male patients and 43% of female patients were defined as having hypertension. Full details regarding baseline characteristics are described in another report.10
Table 3.
Characteristics of study participants in the BioBank Japan Study.
| Male |
Female |
|||
|---|---|---|---|---|
| N | % | N | % | |
| Sex | 106,093 | 53.1 | 93,696 | 46.9 |
| Age group at entry (years) | ||||
| <20 | 1413 | 1.3 | 982 | 1.0 |
| 20–29 | 2062 | 1.9 | 2842 | 3.0 |
| 30–39 | 4815 | 4.5 | 6534 | 7.0 |
| 40–49 | 8302 | 7.8 | 9483 | 10.1 |
| 50–59 | 19,515 | 18.4 | 16,410 | 17.5 |
| 60–69 | 31,637 | 29.8 | 23,796 | 25.4 |
| 70–79 | 30,065 | 28.3 | 24,106 | 25.7 |
| ≥80 | 8272 | 7.8 | 9522 | 10.2 |
| Unknown | 12 | 0.0 | 21 | 0.0 |
| BMIa (kg/m2) | ||||
| Lean (BMI < 18.5) | 5653 | 5.4 | 8525 | 9.2 |
| Normal (18.5 ≤ BMI < 25) | 63,137 | 60.3 | 55,330 | 59.7 |
| Overweight (25 ≤ BMI < 30) | 24,865 | 23.8 | 17,826 | 19.2 |
| Obese (30 ≤ BMI) | 3898 | 3.7 | 4202 | 4.5 |
| Unknown | 7115 | 6.8 | 6810 | 7.3 |
| Smoking statusa | ||||
| Never smoker | 25,690 | 24.5 | 71,637 | 77.3 |
| Ex-smoker | 44,919 | 42.9 | 8521 | 9.2 |
| Current smoker | 28,524 | 27.3 | 9505 | 10.3 |
| Smoker, unknown current status | 3547 | 3.4 | 1293 | 1.4 |
| Unknown if ever smoked | 1988 | 1.9 | 1737 | 1.9 |
| Alcohol intakea | ||||
| Never drinker | 30,933 | 29.6 | 64,987 | 70.1 |
| Ex-drinker | 13,618 | 13.0 | 3614 | 3.9 |
| Current drinker (0–10 g/day) | 14,610 | 14.0 | 11,818 | 12.7 |
| Current drinker (10–20 g/day) | 10,296 | 9.8 | 3801 | 4.1 |
| Current drinker (20–60 g/day) | 20,884 | 20.0 | 3251 | 3.5 |
| Current drinker (60 + g/day) | 7496 | 7.2 | 765 | 0.8 |
| Drinker, unknown current status | 4175 | 4.0 | 2277 | 2.5 |
| Unknown | 2656 | 2.5 | 2180 | 2.4 |
| Hypertension | ||||
| Normotension | 37,898 | 35.7 | 40,692 | 43.4 |
| Hypertension | 53,962 | 50.9 | 38,127 | 40.7 |
| Unknown | 14,233 | 13.4 | 14,877 | 15.9 |
| Physical exercise | ||||
| Three or more times a week | 21,558 | 20.3 | 20,159 | 21.5 |
| Once or twice a week | 3692 | 3.5 | 6132 | 6.5 |
| No habit | 68,367 | 64.4 | 56,796 | 60.6 |
| Unknown | 12,476 | 11.8 | 10,609 | 11.3 |
Because the sex was unknown, 193 participants were excluded from this analysis.
Abbreviation: BMI, body mass index.
People of unknown age or under the age of 20 were excluded.
Discussion
The BBJ Project enrolled approximately 200,000 participants during the first 5-year period of the study. DNA and serum samples stored in the BBJ have enabled investigation of novel genetic factors and new biomarkers related to common diseases. The collected clinical information and follow-up survey data have provided knowledge about the general clinical features of many common diseases in Japan, including both therapeutic conditions and the course of the diseases. Further analyses of the genomic data, clinical information, and follow-up surveys will provide important information regarding the interaction of genetic and clinical factors, and ideally, will eventually contribute to optimized medical treatment that takes into account the individual genetic makeup of the patient.
In recent years, many large-scale biobanks have been established in Europe11 and the United States,12 including the UK Biobank,13 the Auria Biobank,14 the Estonian Biobank,15 the Kaiser Biobank,12 the Million Veteran Program,16 and the Precision Medicine Initiative Cohort.17 The UK Biobank has already collected samples from 500,000 participants in the United Kingdom.13 The Precision Medicine Initiative Cohort plans to collect samples from 1 million participants in the United States.17 In Japan, the Tohoku Medical Megabank is collecting biological specimens from 150,000 residents.18 However, most of these biobanks are population-based. Large, patient-based biobanks, such as the BioVU19 and Mayo Clinic Biobank,20 are limited. To our knowledge, the BBJ is the first patient-based biobank in the world. This is an advantage in analyzing risk factors of disease prognosis because the BBJ enables examination of data with the longest follow-up period available.
Patient-based biobanks allow for the identification of susceptibility genes for common diseases because large numbers of cases are registered. In contrast, population-based biobanks allow for estimation of environmental exposures and gene–environment interactions, but long-term follow-up is needed because a large number of disease onset cases is needed to achieve sufficient statistical power.21 Therefore, patient-based biobanks and population-based biobanks should work in cooperation with the goal of implementing personalized, precision medicine in the future.
Conflicts of interest
The authors declare they have no conflict of interest with respect to this research study and paper.
Acknowledgements
We express our gratitude to all the participants in the BioBank Japan Project. We thank all the medical coordinators of the cooperating hospitals for collecting samples and clinical information, as well as Yasushi Yamashita and staff members of the BioBank Japan Project for administrative support. We also thank Dr. Kumao Toyoshima for his overall supervision of the BioBank Japan Project. This study was supported by funding from the Tailor-Made Medical Treatment with the BBJ Project from Japan Agency for Medical Research and development, AMED (since April 2015), and the Ministry of Education, Culture, Sports, Science and Technology (from April 2003 to March 2015).
Footnotes
Peer review under the responsibility of The Japan Epidemiological Association.
Contributor Information
Michiaki Kubo, Email: michiaki.kubo@riken.jp.
BioBank Japan Cooperative Hospital Group:
Masaki Shiono, Kazuo Misumi, Reiji Kaieda, Hiromasa Harada, Shiro Minami, Mitsuru Emi, Naoya Emoto, Hiroyuki Daida, Katsumi Miyauchi, Akira Murakami, Satoshi Asai, Mitsuhiko Moriyama, Yasuo Takahashi, Tomoaki Fujioka, Wataru Obara, Seijiro Mori, Hideki Ito, Satoshi Nagayama, Yoshio Miki, Akihide Masumoto, Akira Yamada, Yasuko Nishizawa, Ken Kodama, Hiromu Kutsumi, Yoshihisa Sugimoto, Yukihiro Koretsune, Hideo Kusuoka, and Hideki Yanai
Appendix A.
Author list for the BioBank Japan Cooperative Hospital Group.
Members of medical institutions cooperating on the BioBank Japan Project who coauthored this paper include Masaki Shiono, Kazuo Misumi, Reiji Kaieda and Hiromasa Harada (Tokushukai Hospitals); Shiro Minami, Mitsuru Emi and Naoya Emoto (Nippon Medical School); Hiroyuki Daida, Katsumi Miyauchi and Akira Murakami (Juntendo University); Satoshi Asai, Mitsuhiko Moriyama and Yasuo Takahashi (Nihon University); Tomoaki Fujioka and Wataru Obara (Iwate Medical University); Seijiro Mori and Hideki Ito (Tokyo Metropolitan Institute of Gerontology); Satoshi Nagayama and Yoshio Miki (The Cancer Institute Hospital of JFCR); Akihide Masumoto and Akira Yamada (Aso Iizuka Hospital); Yasuko Nishizawa and Ken Kodama (Osaka Medical Center for Cancer and Cardiovascular Diseases); Hiromu Kutsumi and Yoshihisa Sugimoto (Shiga University of Medical Science); Yukihiro Koretsune and Hideo Kusuoka (National Hospital Organization, Osaka National Hospital); and Hideki Yanai (Fukujuji Hospital).
References
- 1.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welter D., MacArthur J., Morales J. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins F.S., Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginsburg G.S., Willard H.F. Genomic and personalized medicine: foundations and applications. Transl Res. 2009;154:277–287. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Hamburg M.A., Collins F.S. The path to personalized medicine. N Engl J Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 6.BioBank Japan. Tailor-made Medical Treatment Program. https://biobankjp.org/work/public.html. Accessed 1 July 2016.
- 7.Nakamura Y. The BioBank Japan Project. Clin Adv Hematol Oncol. 2007;5:696–697. [PubMed] [Google Scholar]
- 8.BioBank Japan. A Brochure about the Distribution of Samples for Researchers [in Japanese]. Tailor-made Medical Treatment Program. https://biobankjp.org/sample/pdf/panf_v2.0.pdf. Updated 7 March 2015. Accessed 1 July 2016.
- 9.Hirata M., Nagai A., Kamatani Y. Overview of BioBank Japan follow-up data in 32 diseases. J Epidemiol. 2017;27:S22–S28. doi: 10.1016/j.je.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirata M., Kamatani Y., Nagai A. Cross-sectional analysis of BioBank Japan clinical data: a large cohort of 200,000 patients with 47 common diseases. J Epidemiol. 2017;27:S9–S21. doi: 10.1016/j.je.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaskell G., Gottweis H. Biobanks need publicity. Nature. 2011;471:159–160. doi: 10.1038/471159a. [DOI] [PubMed] [Google Scholar]
- 12.Scott C.T., Caulfield T., Borgelt E. Personal medicine – the new banking crisis. Nat Biotechnol. 2012;30:141–147. doi: 10.1038/nbt.2116. [DOI] [PubMed] [Google Scholar]
- 13.Collins R. What makes UK biobank special? Lancet. 2012;379:1173–1174. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 14.AURIA BIOPANKKI. What Is Auria Biobank? https://www.auriabiopankki.fi/auria-biobank/?lang=en. Accessed 5 July 2016.
- 15.Leitsalu L., Haller T., Esko T. Cohort profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol. 2015;44:1137–1147. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 16.Gaziano J.M., Concato J., Brophy M. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 17.National Institutes of Health. About the Precision Medicine Initiative Cohort Program. https://www.nih.gov/precision-medicine-initiative-cohort-program. Accessed 5 July 2016.
- 18.Kuriyama S., Yaegashi N., Nagami F. The Tohoku Medical Megabank Project: design and mission. J Epidemiol. 2016;26:493–511. doi: 10.2188/jea.JE20150268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roden D.M., Pulley J.M., Basford M.A. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharm Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson J.E., Ryu E., Johnson K.J. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clin Proc. 2013;88:952–962. doi: 10.1016/j.mayocp.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manolio T.A., Bailey-Wilson J.E., Collins F.S. Genes, environment and the value of prospective cohort studies. Nat Rev Genet. 2006;7:812–820. doi: 10.1038/nrg1919. [DOI] [PubMed] [Google Scholar]