Abstract
The role of human NK cells in viral infections is poorly understood. We used a cytokine flow-cytometry assay to simultaneously investigate the IFN-γ response of NK and T lymphocytes to influenza A virus (fluA). When PBMCs from fluA-immune adult donors were incubated with fluA, IFN-γ was produced by both CD56dim and CD56bright subsets of NK cells, as well as by fluA-specific T cells. Purified NK cells did not produce IFN-γ in response to fluA, while depletion of T lymphocytes reduced to background levels the fluA-induced IFN-γ production by NK cells, which indicates that T cells are required for the IFN-γ response of NK cells. The fluA-induced IFN-γ production of NK cells was suppressed by anti–IL-2 Ab, while recombinant IL-2 replaced the helper function of T cells for IFN-γ production by NK cells. This indicates that IL-2 produced by fluA-specific T cells is involved in the T cell–dependent IFN-γ response of NK cells to fluA. Taken together, these results suggest that at an early stage of recurrent viral infection, NK-mediated innate immunity to the virus is enhanced by preexisting virus-specific T cells.
Introduction
Influenza A virus (fluA) is the major pathogen of humans and several animal species causing annual winter epidemics in the United States and has the potential to cause worldwide pandemics (1). Studies in humans and mice have implicated adaptive immune responses including Ab responses and T cell responses in protective immunity against fluA infection (2–6). Previous studies in the mouse model suggested that NK cells were also involved in the control of fluA infection (7, 8). The role of the innate immune response in protective immunity against fluA is poorly understood, however, especially in humans.
NK cells are important effector cells in the innate immune response against infections and tumors. Two mechanisms are involved in the protective effects of NK cells against viral infections: cytokine production and cytotoxic activity (9–12). Human NK cells are characterized phenotypically by the presence of CD56 and the lack of CD3 expression (10). Two subsets of human peripheral blood NK cells have been identified and characterized. The majority subset (approximately 90%) expresses low levels of CD56 (CD56dim), whereas the minority subset (approximately 10%) expresses high levels of CD56 (CD56bright) (13). These 2 NK subsets are thought to have unique functional attributes and, therefore, distinct roles in the human immune response (13, 14). The CD56dim NK subset is more naturally cytotoxic and may serve as the major cytotoxic effectors. By contrast, the CD56bright subset has the capacity to produce abundant cytokines and may serve as immunoregulators (15, 16). One of the cytokines produced by CD56bright NK cells is IFN-γ, which has immune regulatory activity (17–21) as well as direct antiviral activity (22–25). The role of these 2 subsets of human NK cells in the context of a viral infection has not been extensively investigated.
In this study, we analyzed the production of cytokines by human NK cells and T cells during ex vivo incubation of PBMCs with fluA and explored the relationship between the cytokine response of NK cells and T cells to the virus. We demonstrate that both CD56bright and CD56dim NK cells produce IFN-γ in response to fluA and that IL-2 produced by virus-specific T cells influences the IFN-γ production of NK cells. These results indicate a role of adaptive immune lymphocytes in regulating the function of innate immune cells.
Results
The fluA virus induces production of IFN-γin the CD56bright and CD56dim subsets of NK cells as well as in T cells.
In our previous work, we developed a cytokine flow-cytometry assay for the detection and characterization of fluA-specific memory CD8+ T cells. PBMCs were incubated with fluA ex vivo, followed by intracellular staining for IFN-γ (26). In the current study, we modified the assay to simultaneously investigate the IFN-γ response of NK cells and T cells to fluA. When PBMCs from adult donors were incubated with purified fluA for 17 hours, IFN-γ–producing (IFN-γ+) cells were detected in CD3+ and CD3– lymphocyte subsets (Figure 1A). The majority of IFN-γ+ cells in the CD3+ T cell population did not express CD56 (Figure 1B). Most of the CD3– IFN-γ+ cells expressed CD56 (Figure 1C), indicating that they were NK cells. Incubating PBMCs with heat-inactivated (56°C/35 min) purified fluA induced similar IFN-γ response in CD3+ T cells and CD3–CD56+ NK cells (data not shown).
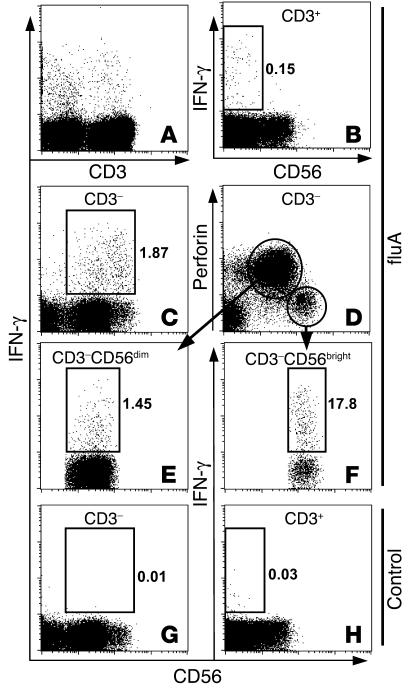
Figure 1.
IFN-γ production by T cells and CD56dim and CD56bright NK cell subsets in response to fluA. PBMCs from an adult donor were incubated with fluA (A–F) or SPG (negative control, G and H) for 17 hours, with brefeldin A added during the last 5 hours. The cells were stained for CD56, fixed and permeabilized, and then stained for CD3, IFN-γ, and perforin. See Methods for details. Displayed in the dot plots A–H are cells gated on different lymphocyte populations: (A) IFN-γ production of CD3– and CD3+ lymphocytes (gated by forward scattering and side scattering) in response to fluA; (B) IFN-γ production of CD3+ lymphocytes in response to fluA; (C) IFN-γ production of CD3– lymphocytes in response to fluA; (D) expression of CD56 and perforin by CD3– lymphocytes; (E and F) IFN-γ production of CD56dim perforin+ NK and CD56bright perforin– NK subsets, respectively, in response to fluA; (G and H) negative controls showing baseline levels of IFN-γ production by CD3– and CD3+ lymphocytes in the absence of fluA. Numbers in the dot plots refer to percentage of IFN-γ+ cells in the gated population.
To examine the IFN-γ production by the CD56bright and CD56dim subsets of NK cells, the fluA-stimulated PBMCs were costained for cell surface CD56 and intracellular perforin in addition to CD3 and IFN-γ. Perforin is a main effector molecule in rapid natural cytotoxicity. As previously reported (27), only the CD56dim subset of NK cells expressed perforin (Figure 1D). Thus, the CD3–CD56+ NK cell population could be further resolved into 2 subsets: CD56bright perforin– and CD56dim perforin+. Both subsets produced IFN-γ in response to fluA, with a higher percentage of IFN-γ+ cells in the CD56bright population than in the CD56dim population (Figure 1, E and F). When PBMCs were incubated in the absence of fluA, IFN-γ was produced neither in NK cells (Figure 1G) nor in CD3+ T cells (Figure 1H), indicating that IFN-γ production in both NK cells and T cells was induced by fluA.
The same experiments as those shown in Figure 1 were conducted with PBMCs from 25 adult donors. Perforin staining was incorporated in all experiments to improve the resolution of CD56bright and CD56dim NK populations, which, for some donors, overlapped significantly with each other when defined by CD56 signal intensity alone (data not shown). For all donors, IFN-γ production was detected for both NK subsets in PBMC samples incubated with fluA. Both the percentage of NK cells producing IFN-γ (Figure 2A) and the IFN-γ staining intensity (Figure 2B) varied among donors. The percentage of IFN-γ+ cells in the CD56bright perforin– and CD56dim perforin+ subsets were positively correlated (rS = 0.878, P < 0.001), with average percentage of IFN-γ+ cells in the CD56bright subset approximately 7-fold higher than that in the CD56dim subset (P < 0.001) (Figure 2A). The amount of IFN-γ per cell as indicated by the mean fluorescence intensity (MFI) of IFN-γ+ cells in the CD56bright and CD56dim NK subsets of each donor were also positively correlated (rS = 0.716, P < 0.001), with average MFI of the CD56bright subset approximately 2-fold higher than in the CD56dim subset (P < 0.001) (Figure 2B). Since the number of NK cells in the CD56dim subset was approximately 10-fold higher than that of the CD56bright subset (data not shown), the absolute number of IFN-γ+ cells in the 2 NK cell subsets from the same aliquot of PBMCs was approximately the same (Figure 2C).
Figure 2.
FluA-induced IFN-γ production by the CD3–CD56bright perforin– NK cell subset (labeled as CD56bright) and CD3–CD56dim perforin+ NK cell subset (labeled as CD56dim) in 25 donors. PBMCs were incubated with fluA for 17 hours, followed by cytokine flow-cytometric analysis (see Methods). Lines connect pairs of observations from the same donor. Black bars mark the positions of groups’ means, which were compared using paired Student’s t tests. The attained significance levels (P values) are reported. For A and C, the t tests were performed on logarithmic-transformed data. (A) The percentage of IFN-γ+ cells in the 2 NK cell subsets. (B) The IFN-γ MFI of IFN-γ+ cells in the 2 NK cell subsets. (C) The total numbers of IFN-γ+ cells detected simultaneously in the 2 NK cell subsets from the same PBMC aliquot of each donor.
T cells are required for the IFN-γresponse of NK cells to fluA.
To assess if the IFN-γ response of NK cells to fluA involves other cell types, we first determined if fluA could directly induce IFN-γ production in virus-exposed NK cells. We purified NK cells from PBMCs by depleting T cells, B cells, and monocytes prior to incubating them with fluA. Only background levels of IFN-γ were observed for the CD56bright and the CD56dim NK cell subsets in all 4 donors tested (Figure 3, A and B). IFN-γ production was induced from the purified NK cells by incubation with recombinant cytokines IL-12 and IL-2 (Figure 3B), indicating that NK cells remained functional after the purification procedure. When purified NK cells were incubated with recombinant IFN-γ in the presence of fluA, no IFN-γ+ NK cells were detected (Figure 3B), indicating that the detection of IFN-γ+ NK cells was not due to the uptake of extracellular IFN-γ by NK cells. These results demonstrate that the fluA-induced IFN-γ production of NK cells requires other leukocyte subset(s) present in PBMCs.
Figure 3.
Purified NK cells do not produce IFN-γ in response to fluA. NK cells were isolated from PBMCs by negative selection and incubated under different conditions, followed by intracellular staining for IFN-γ. Displayed in the bar graphs are frequencies of IFN-γ+ cells in the CD56bright perforin– (left panels) and CD56dim perforin+ (right panels) NK cell subsets. (A) PBMCs or purified NK cells from 3 donors (nos. 1–3) were incubated with fluA for 17 hours. (B) PBMCs or purified NK cells from a fourth donor were incubated with fluA for 17 hours. Purified NK cells were also incubated with recombinant IFN-γ (100 ng/ml) in the presence of fluA, or with recombinant IL-12 (100 ng/ml) and IL-2 (250 U/ml), for 17 hours.
To assess what other cells are necessary for the IFN-γ response of NK cells to fluA, we first investigated the contribution of T cells. PBMCs were depleted of T cells by negative selection with anti-CD3 Ab. The remaining CD3-depleted PBMCs were incubated with fluA and examined for IFN-γ production by NK cells. In the absence of CD3+ cells, IFN-γ production was reduced to background levels in both NK cell subsets, a result obtained for all donors tested (Figure 4). This result indicates that CD3+ cells are required for the IFN-γ response of NK cells to fluA.
Figure 4.
Depletion of CD3+ cells from PBMCs reduces to background levels the IFN-γ response of the NK cells to fluA. CD3+ cells were depleted from the PBMCs of 8 donors (PBMC-CD3). PBMCs and CD3-deleted PBMCs were incubated with fluA for 17 hours, followed by intracellular staining for IFN-γ. Displayed in the bar graphs are frequencies of IFN-γ+ cells in the CD56bright perforin– (left) and CD56dimperforin+ (right) NK cell subsets. Results of 3 donors (nos. 4–6) are presented in this figure, while those of other donors are presented in Figures 5 and 9 as part of other experiments.
A small subset of the CD3+ population expressed the NK cell marker CD56. Some of these cells have been defined as NK T cells. Of note, NK T cells have been suggested to provide helper functions for cytokine production by NK cells (28). To determine if the CD3+CD56+ subset was required for the production of IFN-γ by NK cells in response to fluA, we depleted all CD3+ cells or just CD3+CD56+ cells from PBMCs by a strategy shown in Figure 5, A and B. When CD3-depleted PBMCs, CD3+CD56+ cell-depleted PBMCs, and unfractionated PBMCs were incubated with fluA, IFN-γ production in NK cells was observed in the unfractionated PBMCs and the CD3+CD56+ cell-depleted PBMCs but not in the CD3-depleted PBMCs (Figure 5C). In all 3 donors tested, the levels of IFN-γ production in NK cells were reduced to less than 10% of the original level when all CD3+ cells were depleted, while remaining at greater than 80% of the original level when only CD3+CD56+ cells were depleted (Figure 5C and data not shown). This result indicates that only CD56– T cells are required for the IFN-γ production of NK cells in response to fluA.
Figure 5.
IFN-γ response of NK cells to fluA required CD3+CD56– cells but not CD3+CD56+ cells. (A) A blood sample was split into 2 fractions. CD3+ cells were depleted from fraction I. CD56+, CD36+, CD19+, and CD16+ cells were depleted from fraction II to yield enriched CD56– T cells. Combination of fractions I and II resulted in a population depleted of CD3+CD56+ cells. (B) Flow-cytometric analysis of the unfractionated PBMCs, CD3-depleted PBMCs, CD56– T cells, as well as CD3+CD56+ cell–depleted PBMCs. Displayed in the dot plots are cells gated on lymphocyte population by forward scattering and side scattering. (C) FluA-induced production of IFN-γ by NK cells in unfractionated PBMCs, CD3-depleted PBMCs, and CD3+CD56+ cell-depleted PBMCs. Cells were incubated with fluA for 17 hours. Displayed are cells gated on CD3– lymphocyte population. The numbers in the dot plots are percentages of IFN-γ+ cells among CD3–CD56+ NK cells. Similar results were obtained in experiments using blood samples from 2 other donors.
We also sought to estimate the correlation between the levels of fluA-induced IFN-γ production in the NK and T cell populations, taking advantage of the fact that both responses were measured simultaneously in the same PBMC aliquot from each donor. For the 25 donors tested, the percentage of IFN-γ+ cells in CD56bright and CD56dim NK subsets were each positively correlated with the percentage of IFN-γ+ T cells or fluA-specific T cells (P ≤ 0.002; Figure 6). Taken together, these results suggest that the IFN-γ response of NK cells to fluA is associated with the fluA-specific T cell subset.
Figure 6.
Level of IFN-γ response of the NK cell subsets correlates with frequency of fluA-specific T cells in PBMCs of 25 donors. PBMCs were incubated with fluA for 17 hours, followed by cytokine flow-cytometric analysis to determine the percentage of IFN-γ+ cells in the CD56bright perforin+ NK subset (labeled as CD56bright), CD56dim perforin– NK subset (labeled as CD56dim), and CD3+ T cell subset of each donor. The percentage of IFN-γ+ cells in each NK subset was plotted against that in the T cell population from the same donor, respectively. The estimated Spearman correlation coefficient rS and P value is reported in each plot.
IL-2 produced by fluA-specific T cells is involved in the IFN-γresponse of NK cells to fluA.
It has been reported that recombinant IL-2 induced IFN-γ production by mouse and human NK cells (9, 29) and enhanced IL-12–induced IFN-γ production by human CD56bright NK cells (16). To test the hypothesis that IL-2 is involved in the T cell–dependent IFN-γ production of NK cells in response to fluA, we first examined whether IL-2 was produced in PBMCs cultured with fluA. As shown in Figure 7, IL-2 was indeed produced by T cells after exposure to fluA, which preceded the onset of IFN-γ production by NK cells (Figure 7C). The majority of IL-2+ cells also produced IFN-γ, while the majority of IFN-γ+ cells did not produce IL-2 (Figure 7, A and B), indicating that only a subset of fluA-specific T cells was capable of producing IL-2.
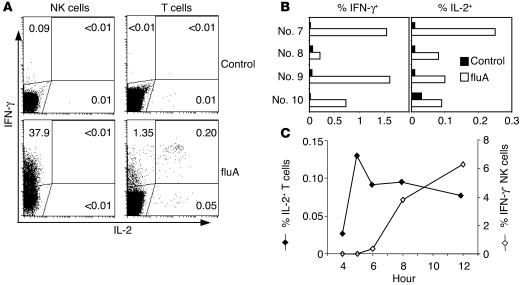
Figure 7.
IFN-γ and IL-2 production by NK cells and T cells in response to fluA. PBMCs from adult donors were incubated with fluA or SPG (negative control) for 12 hours, with brefeldin A added during the last 5 hours (A and B) or were incubated with fluA for 4–12 hours, with brefeldin A added for the last 4 hours (C). The cells were stained for CD56, fixed and permeabilized, and then stained intracellularly for CD3, IFN-γ, and IL-2. (A) Dot plots for a representative donor (no. 7) displaying cells gated on CD3–CD56+ NK cell population (left panels) or CD3+ T cell population (right panels). Numbers in the dot plots refer to the percentage of cytokine-producing cells in each quadrant. (B) Summary of the levels of IFN-γ and IL-2 production by T cells from 4 donors (nos. 7–10). IL-2 was not detected in NK cells from any of these 4 donors. (C) Kinetics of IL-2 production by T cells and IFN-γ production by NK cells (donor no. 20).
Next, we examined the effect of IL-2–neutralizing Ab on IFN-γ production by NK cells. In 5 of 6 donors tested, fluA-induced IFN-γ production by NK cells was suppressed by the IL-2–neutralizing Ab 12% to 67% (Figure 8). This result suggests that IL-2 produced by fluA-specific T cells is involved in the IFN-γ production of NK cells in the majority but not all of the donors.
Figure 8.
Suppression of IFN-γ production of NK cells in response to fluA by IL-2–neutralizing Ab. PBMCs from 6 donors (nos. 11–16) were incubated with fluA for 17 hours in the presence of anti–IL-2 Ab or its isotype control (4 μg/ml), respectively, followed by intracellular staining for IFN-γ to determine frequencies of IFN-γ+ CD3–CD56+ NK cells. Displayed in the bar graph is the percentage suppression for each donor, which is defined as [1 – (frequency of IFN-γ+ NK cells in the presence of anti–IL-2)/(frequency of IFN-γ+ NK cells in the presence of isotype control)] ×100.
Finally, we assessed if recombinant IL-2 could replace T cells in the induction of IFN-γ production by NK cells in response to fluA. As shown in Figure 9, while IFN-γ production of NK cells was reduced to background levels by depletion of CD3+ T cells, addition of recombinant IL-2 to the CD3-depleted PBMCs cultured with fluA restored IFN-γ production of NK cells. Taken together, these results indicate that IL-2 produced by fluA-specific T cells is one of the regulatory factors for the IFN-γ response of NK cells to fluA.
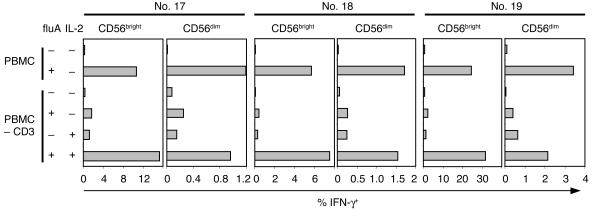
Figure 9.
The helper function of T cells for fluA-induced IFN-γ production in NK cells can be replaced by exogenous IL-2. PBMCs or CD3-depleted PBMCs from 3 donors (nos. 17–19) were incubated with fluA or control for 17 hours, with or without addition of recombinant IL-2 (250 U/ml). Displayed in the graphs are frequencies of IFN-γ+ cells in the CD56bright or CD56dim NK cell subsets under each condition.
Discussion
The host immune response to an infection involves orchestrated activities of different components of the immune system. Therefore, a comprehensive approach is necessary for understanding protective immunity to a virus, which is likely to encompass innate as well as adaptive immunity. In this set of experiments, we used a single cell-based flow-cytometry assay to detect and quantify IFN-γ+ cells in the 2 NK subsets and the T cell population from PBMCs exposed to fluA. We observed that in addition to fluA-specific T cells, CD56bright and CD56dim NK cells produced IFN-γ after a 17-hour ex vivo incubation of PBMCs with fluA. This IFN-γ response of NK cells depends on the T cell population in the PBMCs and was correlated with the level of the T cell response to fluA. IFN-γ production by NK cells responding to fluA could be suppressed by neutralizing Ab against IL-2, while addition of recombinant IL-2 could replace the effect of T cells on the IFN-γ production by NK cells, indicating that IL-2 produced by fluA-specific T cells is involved in the T cell–dependent IFN-γ response of NK cells to fluA.
CD56bright and CD56dim NK cells are thought to represent functionally distinct subsets of mature human NK cells in terms of cytotoxicity and cytokine production. CD56dim NK cells are more granular and have greater natural cytotoxic potential than CD56bright NK cells (14). Regarding cytokine production, previous studies showed that freshly isolated CD56bright human NK cells were the primary source of NK cell–derived IFN-γ and other cytokines in response to exogenous monokines including IL-12, IL-15, IL-18, and IL-1b alone or in combination, whereas the CD56dim NK cell subset produced negligible amounts of cytokines under the same culture condition for 72 hours (15, 30). It has also been observed that the IL-12–induced IFN-γ production of CD56bright NK cells depends on the costimulation by IL-2, either in the form of recombinant molecules or product of a CD4+ T cell clone specific for the tetanus toxoid antigen (16), suggesting a link between innate immunity and adaptive immunity. Of note, IL-2 is a cytokine primarily produced by activated antigen-specific T cells (31).
In the current study we demonstrate that after 17 hours of ex vivo incubation of PBMCs with fluA, both CD56bright and CD56dim NK cells produced IFN-γ. In agreement with previously published results, the CD56bright NK subset was the more potent IFN-γ producer as indicated by greater percentage of IFN-γ+ cells and amount of IFN-γ per cell in this subset (Figure 2, A and B). The number of IFN-γ+ cells contributed by the 2 NK subsets in PBMCs was similar (Figure 2C), however, and on average the difference between these 2 subsets in the amount of IFN-γ per cell was only approximately 2-fold (Figure 2B). There are 2 possible explanations for the apparently greater capability of IFN-γ production by the CD56dim NK subset observed in our study than that reported previously (15, 16). This subset of NK cells may require different regulatory factors for IFN-γ production, which were provided by fluA but not by the monokines used in the previous studies; alternatively, the IFN-γ production of CD56dim subset may be less sustained and may only be detected at the earlier stage of culturing.
IFN-γ is known for its immune regulatory activity as well as direct antiviral activity (17–25). Rapid production of IFN-γ and other inflammatory cytokines by NK cells is an important component of the innate immune response against viral infections (12), which has been shown to be mediated by IL-12 in a murine CMV-infected mouse model (32, 33). Our results suggest that both CD56bright and CD56dim NK subsets participate in the innate immune response against fluA by producing IFN-γ during the early stage of infection.
Resting CD56bright and CD56dim NK cells are known to express distinct panels of lymphocyte homing receptors, suggesting their different homing potential (34). CD56bright NK cells express the chemokine receptor CCR7 (35) and high levels of the adhesion molecule CD62L (L-selectin) (36); both are receptors mediating the homing of lymphocytes to secondary lymphoid organs. Consistent with their expression of lymph node homing receptors, CD56bright NK cells have been shown to constitute the major NK population in lymph nodes (16, 37). In contrast, NK cells in the peripheral blood and the spleen are overwhelmingly CD56dim (13, 37). Resting CD56dim NK cells lack the expression of CCR7 and CD62L but express high levels of the chemokine receptors CXCR1, CXCR2, CXCR3, CXCR4, and CX3CR1 (34). Although the homing potential of CD56dim NK cells is not clear, they have not been found in the lymph nodes and are likely to migrate to other sites in the body and exert their antiviral activity by killing infected cells and producing antiviral cytokines, including IFN-γ, during the early phase of fluA infection.
The infection of host cells by fluA is mediated by binding of viral HA molecules to the sialic acid residues present on cell surface receptors (38). Previous studies have shown that fluA infects different subsets of leukocyte and induces production of innate cytokines, including various IFNs and interleukins (39–41). Of particular interest, DCs are the major producer of type I IFN and IL-12, which have profound effects on other immune cell subsets (42). These innate cytokines can activate NK cells and induce production of IFN-γ (12, 15, 16). Therefore, they are likely to play a critical role in the fluA-induced IFN-γ production of NK cells observed in our current study.
The major finding of this study, however, is that the IFN-γ response of NK cells to fluA also depends on T cells. It has been reported that IL-2, a cytokine produced by activated T cells, enhances IL-12–induced IFN-γ production by CD56bright NK cells (16). In the experiments reported here, we observed that depletion of T cells always reduced to background the fluA-induced IFN-γ production by NK cells (Figure 4), which upon exposure to the virus, fluA-specific T cells produced IL-2 prior to production of IFN-γ by NK cells (Figure 7), and that the T cell–dependent IFN-γ production of NK cells can be suppressed by IL-2–neutralizing Ab for the majority of donors (Figure 8). We have also observed exogenous IL-2 to replace T cells in facilitating IFN-γ production of NK cells exposed to fluA (Figure 9). In addition, the level of IFN-γ response of NK cells appears to correlate positively with the level of fluA-specific T cells (Figure 6).
Based on these results, as well as previously reported effects of DC-derived innate cytokines on the activation and IFN-γ production of NK cells (15, 16), we propose the following model for the IFN-γ response of NK cells to fluA. The production of IFN-γ by NK cells requires regulatory signals from both DC and T cells. Incubation of PBMCs with fluA results in the infection of DCs (43, 44), which produces innate cytokines including IFN-α, IFN-β, IL-12, and other monokines with the potential to activate NK cells (12, 15, 16, 45–47). On the other hand, fluA-infected DCs process and present fluA antigens to fluA-specific T cells, which produce IL-2 and other cytokines. Under the collective actions of DC-derived monokines and T cell–derived cytokines, the NK cells respond by producing IFN-γ (16). Of note, while important proof of concept for this model was provided by the experiments of purified NK cells incubated with recombinant IL-12 plus IL-2 (ref. 16 and Figure 3B) and T cell–depleted PBMCs incubated with fluA plus IL-2 (Figure 9), other cytokines derived from DCs and T cells could be involved in the underlying mechanism for this model as well. In particular, the fact that IL-2–neutralizing Ab only partially blocked the IFN-γ production by NK cells (Figure 8) suggests that IL-2 is not the only T cell–derived regulatory factor for the IFN-γ response of NK cells.
Taken together, our results suggest a dependence of one of the innate immune functions, that is, IFN-γ production by NK cells, on the fluA-specific T cell recall reaction, which is a part of adaptive immunity. FluA infection does not persist, but occurs at multiple times throughout the life of an individual. Since only donors without recent flulike disease were used in this study, the IFN-γ+ T cells detected in PBMCs stimulated ex vivo for 17 hours or fewer with our cytokine-flow cytometric assay are likely to represent preexisting fluA-specific memory T cells.
The innate immune response, which is rapid but not thought to be antigen specific, provides a first line of defense against viral infection and influences the subsequent adaptive T cell response (48). Recent studies have revealed a complex interaction between NK cells and DCs that may lead to NK cell activation, DC activation, or NK cell–mediated killing of DCs under different circumstances (49–53), indicating an important role of NK cells in the regulation of adaptive immunity to infections. It has been shown in a mouse model that NK cells are necessary for optimal priming of adenovirus-specific T cells (54). Of particular interest, depletion of NK cells abrogated fluA-specific CD8+ T cell responses both in vitro and in vivo (55). Conversely, the experiments reported here suggest that at the very early stage of infection, preexisting memory T cells specific for the infecting virus may also play a critical role in regulating the antiviral functions of NK cells. In addition, the correlation we observed between the levels of IFN-γ responses in NK cells and T cells to fluA suggests that at a later stage of infection, when the virus-specific T cell population has expanded, the NK response to fluA will be enhanced further. Thus the strength of innate immunity to a viral infection may be modulated by the quantitative and qualitative nature of adaptive immunity specific for the infecting virus, especially in situations such as influenza where multiple reinfections are the rule. Taken together with previous work, our results support the notion that extensive reciprocal interactions exist between the components of innate immunity and adaptive immunity, which collectively constitute a successful immune response to clear an infection.
Methods
Human subjects and blood samples.
Twenty-three adult donors (ages 25–65) without recent flulike symptoms were enrolled with informed consent. The study protocol was approved by the institutional review board at Stanford University. Venous blood samples were collected using Vacutainer tubes with sodium heparin (Vacutainer Systems; BD). In addition, buffy coats obtained from 3 healthy blood donors at a blood bank were also included in the study, yielding a total of 26 subjects.
Preparation and fractionation of PBMCs.
PBMCs were prepared with standard Ficoll-Paque (Pharmacia Biotech Inc.) gradient centrifugation from the whole blood or buffy coats. Depletion of CD3+ cells or enrichment of NK cells and T cells were conducted using respective RosetteSep reagents (StemCell Technologies Inc.) or MACS MicroBeads (Miltenyi Biotec) following the manufacturer’s instructions.
Preparation of influenza virus.
Purified fluA Panama/2007/99 strain (H3N2) was prepared as previously described (26). In brief, virus was grown in 11-day-old embryonated specific pathogen-free hen eggs (Charles River Laboratories Inc.). Allantoic fluid was harvested 48 hours after infection and assayed for virus by measuring the concentration of influenza HA. Virus-containing allantoic fluid was pooled and centrifuged to pellet fluA particles. The virus pellet was resuspended in PBS and further purified by a continuous 15–60% sucrose gradient centrifugation. The purified virus was reconstituted in PBS, stabilized with sucrose-phosphate-glutamate (SPG) (BioWhittaker Inc.), dispensed into single-use aliquots, and stored at –70°C. The virus titer was determined with Madin-Darby canine kidney cells by standard procedures (56).
Cytokine flow cytometry.
Unfractionated or fractionated PBMCs or lymphocyte subsets were incubated with fluA as previously described (26). In brief, 2 × 106 cells were resuspended in 0.1 ml of RPMI-1640 medium without serum. Purified fluA virus was added to the cells at a MOI of 3 and incubated at 37°C with 5% CO2 for 1 hour. The same volume of SPG was used as the negative control. RPMI-1640 medium supplemented with 10% FCS and antibiotics was then added to a final volume of 0.7 ml, with or without the addition of 1 of the following reagents: rat anti-human IL-2–neutralizing Ab or its isotype control (BD Biosciences — Pharmingen), recombinant human IL-2 (Chiron Inc.), recombinant human IL-12 (Sigma-Aldrich), or recombinant human IFN-γ (a gift from L. Blatt, Intermune Inc.). The cells were incubated for another 11 hours (for detection of IL-2 and IFN-γ) or 16 hours (for detection of IFN-γ). Brefeldin A (Sigma-Aldrich) was added to a final concentration of 10 μg/ml for the last 5 hours of incubation. Staining and flow-cytometric analysis were done as described previously (26). In brief, the cells were first stained with allophycocyanin-labeled anti-CD56 (BD Biosciences — Pharmingen) and then treated with FACS Lysing Solution and FACS Permeabilizing Solution (BD Biosciences). The permeabilized cells were subsequently stained with different combinations of the following Abs: phycoerythrin-labeled or FITC-labeled anti–IFN-γ, phycoerythrin-labeled anti–IL-2 (BD Biosciences), FITC-labeled anti-perforin, and PerCP-labeled anti-CD3 (BD Biosciences — Pharmingen). Surface and intracellular staining were each carried out by incubating at room temperature for 30 minutes. Stained cells were then washed, fixed with 1% paraformaldehyde in PBS, and analyzed using a FACSCalibur flow cytometer, with Cellquest software (BD Biosciences).
Statistical analyses.
Subsets’ means were compared using paired Student’s t tests, in some cases after logarithmic transformation of the data. Correlation testing was based on Spearman’s rank-correlation statistic rS, a robust, distribution-free measure of correlation (57). Individual test’s P-value thresholds for declaring statistical significance were set at or below 0.05 to control the total type I error rate collectively across all statistical tests at 0.05 (58).
Acknowledgments
We thank C. Zhang and L. Rott for technical assistance, L. Blatt for providing recombinant IFN-γ, and E.D. Mellins and D.B. Lewis for useful discussions. This work was supported by the NIH grants AI-057229 and DK-56339.
Footnotes
Nonstandard abbreviations used: fluA, influenza A virus; IFN-γ+, IFN-γ–producing; MFI, mean fluorescence intensity; SPG, sucrose-phosphate-glutamate.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Lamb, R.A., and Krug, R.M. 2001. Orthomyxoviridae: the viruses and their replication. In Fields virology. D.M. Knipe and P.M. Howley, editors. Lippincott Williams & Wilkins. Philadelphia, Pennsylvania, USA. 1533–1579.
- 2.Yap KL, Ada GL, McKenzie IF. Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature. 1978;273:238–239. doi: 10.1038/273238a0. [DOI] [PubMed] [Google Scholar]
- 3.McMichael AJ, Gotch FM, Noble GR, Beare PA. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 1983;309:13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 4.Doherty PC, et al. Effector CD4+ and CD8+ T-cell mechanisms in the control of respiratory virus infections. Immunol. Rev. 1997;159:105–117. doi: 10.1111/j.1600-065x.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 5.Graham MB, Braciale TJ. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. J. Exp. Med. 1997;186:2063–2068. doi: 10.1084/jem.186.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benton KA, et al. Heterosubtypic immunity to influenza A virus in mice lacking IgA, all Ig, NKT cells, or gamma delta T cells. J. Immunol. 2001;166:7437–7445. doi: 10.4049/jimmunol.166.12.7437. [DOI] [PubMed] [Google Scholar]
- 7.Dong L, Mori I, Hossain MJ, Kimura Y. The senescence-accelerated mouse shows aging-related defects in cellular but not humoral immunity against influenza virus infection. J. Infect. Dis. 2000;182:391–396. doi: 10.1086/315727. [DOI] [PubMed] [Google Scholar]
- 8.Liu B, et al. Interleukin-18 improves the early defence system against influenza virus infection by augmenting natural killer cell-mediated cytotoxicity. J. Gen. Virol. 2004;85:423–428. doi: 10.1099/vir.0.19596-0. [DOI] [PubMed] [Google Scholar]
- 9.Trinchieri G. Biology of natural killer cells. Adv. Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson MJ, Ritz J. Biology and clinical relevance of human natural killer cells. Blood. 1990;76:2421–2438. [PubMed] [Google Scholar]
- 11.Tay CH, Szomolanyi-Tsuda E, Welsh RM. Control of infections by NK cells. Curr. Top. Microbiol. Immunol. 1998;230:193–220. doi: 10.1007/978-3-642-46859-9_12. [DOI] [PubMed] [Google Scholar]
- 12.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 13.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 14.Nagler A, Lanier LL, Cwirla S, Phillips JH. Comparative studies of human FcRIII-positive and negative natural killer cells. J. Immunol. 1989;143:3183–3191. [PubMed] [Google Scholar]
- 15.Cooper MA, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001;97:3146–3151. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 16.Fehniger TA, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood. 2003;101:3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Botran R, Sanders VM, Mosmann TR, Vitetta ES. Lymphokine-mediated regulation of the proliferative response of clones of T helper 1 and T helper 2 cells. J. Exp. Med. 1988;168:543–558. doi: 10.1084/jem.168.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gajewski TF, Goldwasser E, Fitch FW. Anti-proliferative effect of IFN-gamma in immune regulation. II. IFN-gamma inhibits the proliferation of murine bone marrow cells stimulated with IL-3, IL-4, or granulocyte-macrophage colony-stimulating factor. J. Immunol. 1988;141:2635–2642. [PubMed] [Google Scholar]
- 19.Parronchi P, et al. IL-4 and IFN (alpha and gamma) exert opposite regulatory effects on the development of cytolytic potential by Th1 or Th2 human T cell clones. J. Immunol. 1992;149:2977–2983. [PubMed] [Google Scholar]
- 20.Maggi E, et al. Reciprocal regulatory effects of IFN-gamma and IL-4 on the in vitro development of human Th1 and Th2 clones. J. Immunol. 1992;148:2142–2147. [PubMed] [Google Scholar]
- 21.Bradley LM, Dalton DK, Croft M. A direct role for IFN-gamma in regulation of Th1 cell development. J. Immunol. 1996;157:1350–1358. [PubMed] [Google Scholar]
- 22.Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. [DOI] [PubMed] [Google Scholar]
- 23.Frese M, et al. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology. 2002;35:694–703. doi: 10.1053/jhep.2002.31770. [DOI] [PubMed] [Google Scholar]
- 24.Cheney IW, et al. Comparative analysis of anti-hepatitis C virus activity and gene expression mediated by alpha, beta, and gamma interferons. J. Virol. 2002;76:11148–11154. doi: 10.1128/JVI.76.21.11148-11154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanford RE, et al. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 2003;77:1092–1104. doi: 10.1128/JVI.77.2.1092-1104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He XS, et al. Analysis of the frequencies and of the memory T cell phenotypes of human CD8+ T cells specific for influenza A viruses. J. Infect. Dis. 2003;187:1075–1084. doi: 10.1086/368218. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs R, et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur. J. Immunol. 2001;31:3121–3127. doi: 10.1002/1521-4141(2001010)31:10<3121::aid-immu3121>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Carnaud C, et al. Cutting edge: cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J. Immunol. 1999;163:4647–4650. [PubMed] [Google Scholar]
- 29.Biron CA, Young HA, Kasaian MT. Interleukin 2-induced proliferation of murine natural killer cells in vivo. J. Exp. Med. 1990;171:173–188. doi: 10.1084/jem.171.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper MA, et al. Interleukin-1beta costimulates interferon-gamma production by human natural killer cells. Eur. J. Immunol. 2001;31:792–801. doi: 10.1002/1521-4141(200103)31:3<792::aid-immu792>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 31.Smith KA. Interleukin-2: inception, impact, and implications. Science. 1988;240:1169–1176. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- 32.Orange JS, Biron CA. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J. Immunol. 1996;156:4746–4756. [PubMed] [Google Scholar]
- 33.Orange JS, Biron CA. An absolute and restricted requirement for IL-12 in natural killer cell IFN-gamma production and antiviral defense. Studies of natural killer and T cell responses in contrasting viral infections. J. Immunol. 1996;156:1138–1142. [PubMed] [Google Scholar]
- 34.Robertson MJ. Role of chemokines in the biology of natural killer cells. J. Leukoc. Biol. 2002;71:173–183. [PubMed] [Google Scholar]
- 35.Campbell JJ, et al. Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. J. Immunol. 2001;166:6477–6482. doi: 10.4049/jimmunol.166.11.6477. [DOI] [PubMed] [Google Scholar]
- 36.Frey M, et al. Differential expression and function of L-selectin on CD56bright and CD56dim natural killer cell subsets. J. Immunol. 1998;161:400–408. [PubMed] [Google Scholar]
- 37.Ferlazzo G, et al. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J. Immunol. 2004;172:1455–1462. doi: 10.4049/jimmunol.172.3.1455. [DOI] [PubMed] [Google Scholar]
- 38.Weis W, et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 39.Ronni T, Sareneva T, Pirhonen J, Julkunen I. Activation of IFN-alpha, IFN-gamma, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J. Immunol. 1995;154:2764–2774. [PubMed] [Google Scholar]
- 40.Bender A, et al. The distinctive features of influenza virus infection of dendritic cells. Immunobiology. 1998;198:552–567. doi: 10.1016/S0171-2985(98)80078-8. [DOI] [PubMed] [Google Scholar]
- 41.Coccia EM, et al. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- 42.Decker T, Stockinger S, Karaghiosoff M, Müller M, Kovarik P. IFNs and STATs in innate immunity to microorganisms. J. Clin. Invest. 2002;109:1271–1277. doi:10.1172/JCI200215770. doi: 10.1172/JCI15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsson M, et al. Requirement of mature dendritic cells for efficient activation of influenza A-specific memory CD8+ T cells. J. Immunol. 2000;165:1182–1190. doi: 10.4049/jimmunol.165.3.1182. [DOI] [PubMed] [Google Scholar]
- 44.Fonteneau JF, et al. Activation of influenza virus-specific CD4+ and CD8+ T cells: a new role for plasmacytoid dendritic cells in adaptive immunity. Blood. 2003;101:3520–3526. doi: 10.1182/blood-2002-10-3063. [DOI] [PubMed] [Google Scholar]
- 45.Scharton-Kersten T, Afonso LC, Wysocka M, Trinchieri G, Scott P. IL-12 is required for natural killer cell activation and subsequent T helper 1 cell development in experimental leishmaniasis. J. Immunol. 1995;154:5320–5330. [PubMed] [Google Scholar]
- 46.Zhang T, et al. Interleukin-12 (IL-12) and IL-18 synergistically induce the fungicidal activity of murine peritoneal exudate cells against Cryptococcus neoformans through production of gamma interferon by natural killer cells. Infect. Immun. 1997;65:3594–3599. doi: 10.1128/iai.65.9.3594-3599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mailliard RB, et al. Complementary dendritic cell-activating function of CD8+ and CD4+ T cells: helper role of CD8+ T cells in the development of T helper type 1 responses. J. Exp. Med. 2002;195:473–483. doi: 10.1084/jem.20011662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medzhitov R, Janeway CA., Jr Innate immunity: impact on the adaptive immune response. Curr. Opin. Immunol. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- 49.Ferlazzo G, et al. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J. Exp. Med. 2002;195:343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerosa F, et al. Reciprocal activating interaction between natural killer cells and dendritic cells. J. Exp. Med. 2002;195:327–333. doi: 10.1084/jem.20010938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piccioli D, Sbrana S, Melandri E, Valiante NM. Contact-dependent stimulation and inhibition of dendritic cells by natural killer cells. J. Exp. Med. 2002;195:335–341. doi: 10.1084/jem.20010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zitvogel L. Dendritic and natural killer cells cooperate in the control/switch of innate immunity. J. Exp. Med. 2002;195:F9–F14. doi: 10.1084/jem.20012040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moretta A. Natural killer cells and dendritic cells: rendezvous in abused tissues. Nat. Rev. Immunol. 2002;2:957–964. doi: 10.1038/nri956. [DOI] [PubMed] [Google Scholar]
- 54.Liu ZX, Govindarajan S, Okamoto S, Dennert G. NK cells cause liver injury and facilitate the induction of T cell-mediated immunity to a viral liver infection. J. Immunol. 2000;164:6480–6486. doi: 10.4049/jimmunol.164.12.6480. [DOI] [PubMed] [Google Scholar]
- 55.Kos FJ, Engleman EG. Role of natural killer cells in the generation of influenza virus-specific cytotoxic T cells. Cell. Immunol. 1996;173:1–6. doi: 10.1006/cimm.1996.0245. [DOI] [PubMed] [Google Scholar]
- 56.Lennette, D.A. 1995. General principles for laboratory diagnosis of viral, rickettsial, and chlamydial infections. In Diagnostic procedures for viral, rickettsial, and chlamydial infections. E.H. Lennette, D.A. Lennette, and E.T. Lennette, editors. American Public Health Association. Washington, D.C., USA. 3–25.
- 57.Daniel, W.W. 1990. Applied nonparametric statistics. PWS-Kent. Boston, Massachusetts, USA. 358–363.
- 58.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]