Abstract
The lack of standardized methods for human phenotyping is a major obstacle in translational science. We have developed a bleeding history phenotyping system comprising an ontology, a questionnaire, a Web‐based phenotype recording instrument (PRI), and a database. The ontology facilitates transparency, collaboration, aggregation of data, and data analysis. The integrated system allows investigators worldwide to use the PRI, add their de‐identified data to the database, and query the aggregated data. Thus, this system can increase the power to detect genotype–phenotype–environment relationships and help new investigators begin their studies. We anticipate that this approach may be applicable to other disorders.
Keywords: bleeding disorders, bleeding symptoms, hematology, hemostasis, phenotyping, bioinformatics
Introduction
The lack of standardized methods for human phenotyping is a major obstacle to translational science. The benefits of carefully collecting and organizing phenotypic information for research purposes are exemplified by the Framingham study, which has directly influenced medical practice and led to improvements in human health. 1
Advances in genomics and proteomics offer extraordinary opportunities for translational research. However, their successful application requires correlation with high‐quality phenotypic information, especially with regard to subtle and/or complex gene–gene and gene–environment interactions. 2 Yet phenotyping as a scientific discipline has not kept pace with advances in genetics, prompting Freimer and Sabatti to call for a “human phenome project.” 3
Rockefeller University investigators have therefore undertaken an initiative to enhance human phenotyping under the auspices of a Clinical and Translational Science Award (CTSA). To address the deficiencies in current practices—including the lack of standardized, rigorous, and comprehensive data recording instruments, the common practice of discarding case report forms after study completion, and the use of different instruments by different investigators—we developed an electronic phenotyping system that could be used by investigators worldwide. This prototype system uses the bleeding history as a paradigm.
In order to promote standardization, collaboration, transparency, and aggregation of data from multiple sources, the bleeding history phenotyping system was grounded in the creation of a domain ontology. Ontologies help to achieve these goals by explicitly defining the existing knowledge about the disorder and formally encoding that information. In this way, the ontology helps a community of investigators develop a common understanding of the disorder, and in the process makes assumptions about the disorder explicit. The ontology's structure facilitates the organization, retrieval, and analysis of the encoded knowledge, including database design and merging of databases. Examples of valuable ontologies include the gene ontology (GO), 4 which organizes rapidly expanding genetic data, and the Internet search engine Yahoo.
In addition, we set the following goals for the system: (1) ensure the quality of the instrument by expert review (2) maximize the use of standardized vocabulary for medical terms (3) ensure the security of the system (4) ensure transparency by making the instrument publicly available (5) facilitate adoption of the recording instrument by investigators at other sites by making it Web‐accessible, and (6) connect the instrument to a scalable database. To achieve these goals, in addition to the bleeding history ontology (BHO), we developed a comprehensive bleeding history questionnaire (BHQ), an electronic phenotype recording instrument (PRI), and a database. Together, these comprise the bleeding history phenotyping system (BHPS) ( Figure 1 ).
Figure 1.
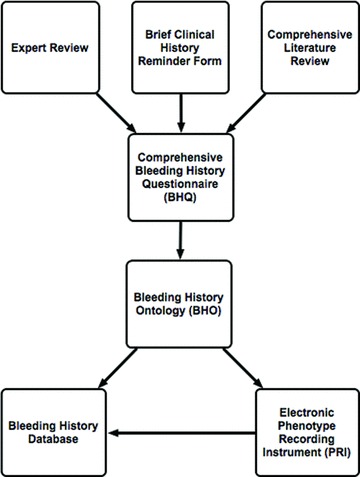
Creation of the bleeding history phenotyping system. After an extensive literature search and review by experts, a paper‐based bleeding history clinical reminder form was expanded into a comprehensive bleeding history questionnaire (BHQ). The questionnaire was used to derive a bleeding history ontology (BHO) as well as a bleeding history database and a graphical user interface and electronic recording instrument, the phenotype recording instrument (PRI).
Methods
Bleeding history questionnaire
The BHPS was based on a comprehensive questionnaire (the BHQ), which in turn was derived from a paper‐based reminder form developed by one of the authors for clinical use. 5 To ensure that it was comprehensive, this original document was dramatically expanded based on an extensive literature review 5 and comments from expert hematologists. The questionnaire was formatted to include both yes/no and multiple choice questions, and to allow for the entry of data on multiple episodes of bleeding in the same category (e.g., surgery). A comprehensive list of treatments was also developed for select questions. The question format in the BHQ was reviewed by an expert in questionnaire development. To standardize the language used in the questionnaire (and subsequently, the ontology), 168 terms were mapped to Unified Medical Language System (UMLS) codes. 6
Bleeding history ontology
Since none of the ontologies in the BioPortal 7 (http://bioportal.bioontology.org/) or Open Biomedical Ontologies (OBO) Foundry 8 (http://www.obofoundry.org/) repositories included a representation of bleeding symptoms, a new ontology was constructed from the BHQ. The ontology construction process adhered to the principles proposed by Uschold. 9 In particular, we identified the purpose of the ontology as supporting the discovery of new research and clinical knowledge about bleeding disorders and the user group as investigators in this discipline.
We chose an intermediate level of formality to strike a balance between intuitiveness and facilitation of computer processing. The BHQ defined the scope of the ontology and it was constructed using Protégé, 10 an open‐source ontology editor supported by the National Center for Biomedical Ontology (NCBO, http://www.bioontology.org/). After completion, the BHO was made publicly available on BioPortal (http://bioportal.bioontology.org/visualize/38563) to allow others to comment on it and offer suggestions for improvement and updating.
The ontology was initially developed in Protégé Frames, then later converted to the Web Ontology Language (OWL 11 ) format in order to meet Semantic Web standards. It consists of Classes (i.e., concepts), Individuals within Classes (i.e., concrete entities of the concepts), and Properties (definitions of the interrelationship between and among Classes and, by definition, the interrelationship of their Individuals).
The ontology comprises two “subontologies:” a bleeding history application ontology (BHAO) and a bleeding history domain ontology (BHDO) ( Figure 2 ). An application ontology represents a specific application, for example the questions used in the BHQ. In contrast, a domain ontology organizes information on a subject independent of how one might acquire or use the information. 9 Thus, the BHAO captures the exact phrasing and format of the BHQ, whereas the BHDO is a generic ontology that defines an idealized conceptualization of the bleeding history, independent of the specific questions asked to elicit the information.
Figure 2.
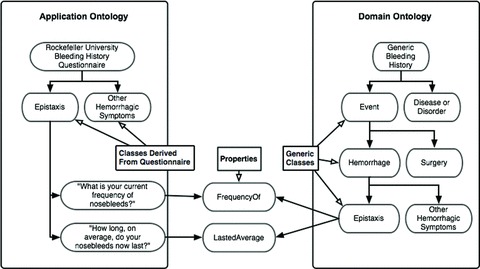
Conceptual schematic of the bleeding history application ontology and bleeding history domain ontology. The examples of epistaxis and two associated questions are depicted. By deriving its structure directly from the bleeding history questionnaire, the bleeding history application ontology captures exact question syntax. In contrast, the bleeding history domain ontology is an idealized conceptualization of bleeding events, divorced from the exact syntax of any particular question. The bleeding history application ontology is linked to the bleeding history domain ontology via shared Properties. These Properties, in combination with Classes from either ontology, defi ne Individuals, which is a technical term that should not be confused with a specifi c person.
To create the BHAO, question categories were used to define Classes, and the wording of individual questions was entered into Protégé under those Classes. To create the BHDO, medical terms used in the questionnaire were organized into idealized, hierarchically structured Classes. These generic Classes were designed to be similar in content to the questionnaire but independent of specific question syntax. To link the BHAO to the BHDO, shared Properties were defined. These Properties connect Classes from the two ontologies to one another.
Combinations of Classes and Properties define Individuals. As used in the context of an ontology, the term Individual does not refer to a unique person, but rather a specific question response or bleeding episode. For instance, in the example depicted in Figure 2 , information about an episode of epistaxis is collected via questions from the BHAO Class “Epistaxis.” One question within this Class is described by the Property “FrequencyOf.” The combination of the BHAO Class “Epistaxis” and BHAO Property “FrequencyOf” defines an Individual, i.e., a specific episode of epistaxis. The same Property links that BHAO Individual to the BHDO Class “Epistaxis,” defining the same episode in generic terms. This organization separates the specific questionnaire syntax from generic knowledge, a useful property that can be leveraged to support a variety of applications, including database mergers. The latter may be vital in aggregating legacy data with data obtained prospectively with newer instruments.
Database
A MySQL database was generated from the finished ontology. 12 First, the ontology structure was exported to an Extensive Markup Language (XML) file. A parsing program was used to convert this XML file to a Structured Query Language (SQL) statement containing the structures of questions and answers represented in the ontology. These SQL statements were entered into MySQL to populate table structures and define tabular relationships. Because this process produced a database congruent with the ontology, the database can be easily updated as knowledge about bleeding phenotypes (and therefore the bleeding history ontology) evolves. The database was made Web‐accessible and housed on a Rockefeller University server behind an institutional firewall to ensure security. This organization permits investigators worldwide to contribute and query their de‐identified data.
Phenotype recording instrument
To facilitate the entry and retrieval of data by investigators, we created a separate Web‐based PRI using Python and the Django Web Application Framework 13 ( Figure 3 ). Access to the PRI requires a password and identification of the site and protocol. Data are entered via a system‐generated unique personal identification number (UPIN) rather than by name or other personal identifier. The individual's age, rather than date of birth, is obtained to further minimize the risk of identification. Genealogic information, which poses a theoretical risk of personal identification, is obtained because it is vital to the scientific and clinical goals of the project. A timestamp is incorporated into the PRI to permit analysis of the time required to complete the study. Subjects can log in and out at their convenience without data loss. Visual aids such as high‐quality photographs are included to help individuals understand the questions and ensure standardization. Within the PRI, each group of phenotypic symptoms is independently accessible so as to create convenient, modular questionnaire sections. Within sections, logical axioms are implemented to speed questionnaire completion. For instance, a research subject answering “Yes” to the question “Have you ever had or do you currently have spontaneous nosebleeds?” would be directed to appropriate follow‐up questions ( Figure 2 ). In contrast, a subject answering “No” would not be asked any of the 14 additional questions about nosebleeds and instead would be directed to the next question module. These axioms allowed the BHQ to be completed in a median time of 30 minutes in the first study conducted with 500 healthy individuals. 14
Figure 3.
Phenotype recording instrument.
Data representation and query utilities are included to help investigators retrieve and review their data. These permit investigators to query the database for questionnaire responses and generate printable pdf files containing individual responses or graphical representations of data collated from the responses of multiple individuals. One goal of this project is to encourage individual investigators to add the de‐identified data from their studies into a common data repository that will benefit from the increase in sample size. Investigators who elect to participate in creating the aggregated database will be granted authority to query the entire database, filter the query by subject characteristics (such as bleeding disorder diagnoses), and generate a downloadable spreadsheet that is suitable for statistical analyses.
Discussion
The BHPS offers investigators with Internet access a comprehensive and standardized format for collecting, storing, and retrieving bleeding phenotypes. Use of this system will permit investigators to collect and compare data across different institutions and studies. In addition, it will facilitate data aggregation from a large number of subjects, which is necessary for detailed genotype–phenotype–environment correlations. The ontology on which the system is built not only provides an effective tool to organize and analyze data, but also provides a transparent mechanism for the hemostasis community to continually update its understanding of the relationships among and between bleeding signs and symptoms, bleeding disorders, laboratory data, and therapies.
The many potential applications for this system include: defining the range of bleeding symptoms in normal populations; defining patterns of bleeding symptoms in different disorders; optimizing diagnostic criteria for differentiating normals from individuals with bleeding disorders developing streamlined sets of questions to assess the likelihood of bleeding from invasive procedures or treatment with anticoagulants or antiplatelet agents; assessing whether to initiate laboratory evaluations; correlating bleeding symptoms with biochemical, genetic, proteomic, and other data; and identifying patients with discordances between bleeding symptoms and biochemical data for further analysis of genetic and environmental influences. We have begun our own studies by analyzing the bleeding symptoms of 500 normal individuals of different ages, sexes, races, and ethnic groups. We plan to also publicly share these data when completed to help other investigators establish the normal ranges in their populations. 14 We especially hope that the ready availability of the BHPS and the normal control data will encourage and help junior investigators to begin careers in studying bleeding disorders. We anticipate that the approach we have taken may be applicable to a number of other disorders and we have prepared materials to help others create their own phenotyping systems for these disorders.
Acknowledgments
This study was supported by grants KL2RR024142 and 1U54RR023419‐01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. Contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. This project was also aided by the tools made available by the National Center for Biomedical Ontology, one of seven National Centers for Biomedical Computing funded by the NIH Roadmap.
We would like to thank Dr. Daniel Rubin and Dr. Mark Musen of Stanford University for their valuable conceptual and practical contributions to the initial design of the Bleeding History Phenotyping System.
References
- 1. Shindler E. Framingham heart study. Available at: http://www.framinghamheartstudy.org/about/milestones.html. Accessed September 14, 2009.
- 2. Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008; 322(5903): 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Freimer N, Sabatti C. The human phenome project. Nat Genet. 2003; 34(1): 15–21. [DOI] [PubMed] [Google Scholar]
- 4. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel‐Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000; 25(1): 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coller BS, Schneiderman PI. Clinical evaluation of hemorrhagic disorders: The bleeding history and differential diagnosis of purpura In: Hoffman R, Benz EJ, Shattil SJ, Furie B, Silberstein LE, McGlave P, eds. Hematology: Basic Principles and Practice. New York : Churchill Livingstone, 2009, 1851–1876. [Google Scholar]
- 6. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004; 32(Database issue): D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Musen M, Shah N, Noy N, Dai B, Dorf M, Griffith N, Buntrock JD, Junquet C, Montegut MJ, Rubin DL. BioPortal: ontologies and data resources with the click of a mouse. AMIA Annu Symp Proc. 2008; 1223–1224. [PubMed] [Google Scholar]
- 8. Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, The OBI Consortium , Leontis N, Rocca‐Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007; 25(11): 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uschold, M . Building ontologies: towards a unified methodology. The 16th Annual Conference of the British Computer Society Specialist Group of Expert Systems; Cambridge , UK , Dec 16, 1996.
- 10. Noy NF, Crubezy M, Fergerson RW, Knublauch H, Tu SW, Vendetti J, Musen MA. Protege‐2000: an open‐source ontology‐development and knowledge‐acquisition environment. AMIA Annu Symp Proc. 2003; 953. [PMC free article] [PubMed] [Google Scholar]
- 11. Dean M, Schreiber G, Bechhofer S, Van Harmelen F, Hendler J, Horrocks I, McGuinness DL, Patel‐Schneider PF, Stein LA. OWL web ontology language reference, W3C Recommendation. Available at: http://www.w3.org/TR/owl‐ref/. 2004. Accessed January 5, 2009.
- 12. MySQL . Available at: http://www.mysql.com. Accessed September 14, 2009.
- 13. Django . Available at: http://www.djangoproject.com/. Accessed September 14, 2009.
- 14. Mauer AC, Barbour EM, Khazanov NA, Levenkova N, Mollah SA, Coller BS. Initial deployment of a comprehensive, ontology‐backed, Web‐based bleeding history phenotyping instrument in normal individuals. J Thromb Haemost 2009; 7(Suppl 2): 14. 19090814 [Google Scholar]


