Abstract
Polyamines are essential organic cations with multiple cellular functions. Their synthesis is controlled by a feedback regulation whose main target is ornithine decarboxylase (ODC), the rate-limiting enzyme in polyamine biosynthesis. In mammals, ODC has been shown to be inhibited and targeted for ubiquitin-independent degradation by ODC antizyme (AZ). The synthesis of mammalian AZ was reported to involve a polyamine-induced ribosomal frameshifting mechanism. High levels of polyamine therefore inhibit new synthesis of polyamines by inducing ODC degradation. We identified a previously unrecognized sequence in the genome of Saccharomyces cerevisiae encoding an orthologue of mammalian AZ. We show that synthesis of yeast AZ (Oaz1) involves polyamine-regulated frameshifting as well. Degradation of yeast ODC by the proteasome depends on Oaz1. Using this novel model system for polyamine regulation, we discovered another level of its control. Oaz1 itself is subject to ubiquitin-mediated proteolysis by the proteasome. Degradation of Oaz1, however, is inhibited by polyamines. We propose a model, in which polyamines inhibit their ODC-mediated biosynthesis by two mechanisms, the control of Oaz1 synthesis and inhibition of its degradation.
Keywords: antizyme, ODC, polyamines, proteasome, ubiquitin
Introduction
Proteolysis by the 26S proteasome is the main pathway for ATP-dependent nonlysosomal degradation of intracellular proteins in eukaryotes (Hershko and Ciechanover, 1998). Usually, the substrates destined for degradation by this multi-subunit protease are marked by the attachment of polyubiquitin chains, which are recognized by binding sites in the 19S caps (Verma et al, 2004). Several exceptions to this principle of ubiquitin-dependent degradation by the proteasome have been reported. The first described and best-studied substrate of ubiquitin-independent degradation by the proteasome is ornithine decarboxylase (ODC) (Coffino, 2001b). Other examples are c-jun in mammals and Rpn4 in Saccharomyces cerevisiae (Jariel-Encontre et al, 1995; Xie and Varshavsky, 2001). In the latter two cases, however, ubiquitylation has been shown to be relevant to proteolysis in vivo (Treier et al, 1994; Ju and Xie, 2004). ODC thus remains the only characterized bona fide substrate of ubiquitin-independent degradation by the proteasome.
ODC is the rate-limiting enzyme in the biosynthesis of the polyamines spermine and spermidine (Coffino, 2001b; Wallace et al, 2003). It catalyses the conversion of ornithine derived from arginine into the diamine putrescine, which is converted to spermidine by spermidine synthase. Spermine is derived from spermidine by the action of spermine synthase (Wallace et al, 2003). These polyamines are essential organic polycations that have been implicated in stabilization of chromatin and the cytoskeleton, as well as in processes ranging from DNA replication, transcription and translation: ion transport, to the regulation of cell growth and apoptosis (Coffino, 2001b; Childs et al, 2003; Wallace et al, 2003).
The effects of lowering the intracellular production of polyamines have been extensively studied in transgenic models (Janne et al, 2004). Inactivation of the ODC gene in mice, for example, led to embryonic lethality (Pendeville et al, 2001). S. cerevisiae mutants lacking the SPE1 gene encoding ODC are viable but cease to grow and become morphologically abnormal upon transfer to polyamine-free media (Schwartz et al, 1995). Studies that employed overexpression of genes involved in polyamine synthesis in mice on the other hand demonstrated that too high levels of polyamines are detrimental as well and result in a variety of defects including sterility and the promotion of malignant transformation (Janne et al, 2004). Consistent with the latter notion, elevated levels of ODC have been associated with cancer, and polyamine analogues as well as drugs that influence intracellular polyamine levels have been considered as anticancer agents (Childs et al, 2003; Wallace et al, 2003).
The intracellular concentration of polyamines is controlled at several steps, including their uptake and their biosynthesis. The latter is mainly achieved by controlling the cellular ODC activity via an unusual mechanism involving ODC antizyme (AZ) (Hayashi et al, 1996; Coffino, 2001b). This protein was first identified in mammals, where it is now known to exist in several isoforms (AZ1, AZ2, AZ3, AZ4) (Heller et al, 1976; Ivanov et al, 1998a, 2000). AZ1 disrupts enzymatically active ODC homodimers by forming ODC/AZ heterodimers (Mitchell and Chen, 1990; Li and Coffino, 1992). AZ1 binding moreover mediates ODC degradation by the 26S proteasome (Li and Coffino, 1992; Murakami et al, 1992; Elias et al, 1995). In contrast, binding of AZ2 does not result in degradation of ODC (Zhu et al, 1999). The modes of action of AZ3, whose expression appears to be limited to the testis, and of AZ4 have not been analysed in detail (Ivanov et al, 2000; Coffino, 2001a). AZ1-dependent degradation of ODC was shown both in vitro and in vivo not to require its ubiquitylation (Rosenberg-Hasson et al, 1989; Murakami et al, 1992). It was reported that, instead, a C-terminal degradation signal in ODC is exposed upon AZ1 binding that mediates binding to a ubiquitin recognition site in the 19S cap of the proteasome (Zhang et al, 2003).
AZ levels increase with rising intracellular polyamine concentrations. Polyamine induction of AZ thus constitutes a feedback control in polyamine homeostasis. For mammals, it was shown that polyamines mediate increased AZ expression by promoting +1 ribosomal frameshifting during decoding of the mRNA (Matsufuji et al, 1995). ODC AZ is widespread among eukaryotes ranging from fungi to mammals. Despite the availability of the complete genome sequence of S. cerevisiae since 1996, previous attempts to identify a sequence encoding an ODC AZ orthologue in this model eukaryote have been unsuccessful (Zhu et al, 2000). Experimental data, however, indicated that regulation of ODC in S. cerevisiae appears to be similar to that in mammals in that treatment with polyamines induces its rapid degradation. These data suggested the presence of a functional analogue of AZ in S. cerevisiae (Toth and Coffino, 1999; Gupta et al, 2001).
In this study, we used a profile-based sequence analysis method to identify an S. cerevisiae ODC AZ orthologue, termed Oaz1. Oaz1 resembles its mammalian counterparts in that it mediates degradation of ODC and in that its synthesis involves polyamine-controlled ribosomal frameshifting. We show that polyamines in addition control the level of AZ by inhibiting its ubiquitin-dependent degradation, thereby providing a mechanism that allows cells to rapidly reinitiate polyamine synthesis after its transient inhibition by Oaz1.
Results
Identification of an ODC AZ homologue (Oaz1) in S. cerevisiae
The family of obvious AZ orthologues detectable in the databases currently comprises more than 40 members. An inspection of their alignment revealed a conserved architecture with two homologous regions that are separated from each other by divergent linker sequences of variable length (shown in Figure 1C for selected AZ orthologues). A conserved N-terminal sequence (D1) is centred at the +1 frameshifting site present in all AZs (Figure 1C). The larger C-terminal region D2 has been linked to ODC binding and degradation (Ichiba et al, 1994; Chen et al, 2002).
Figure 1.
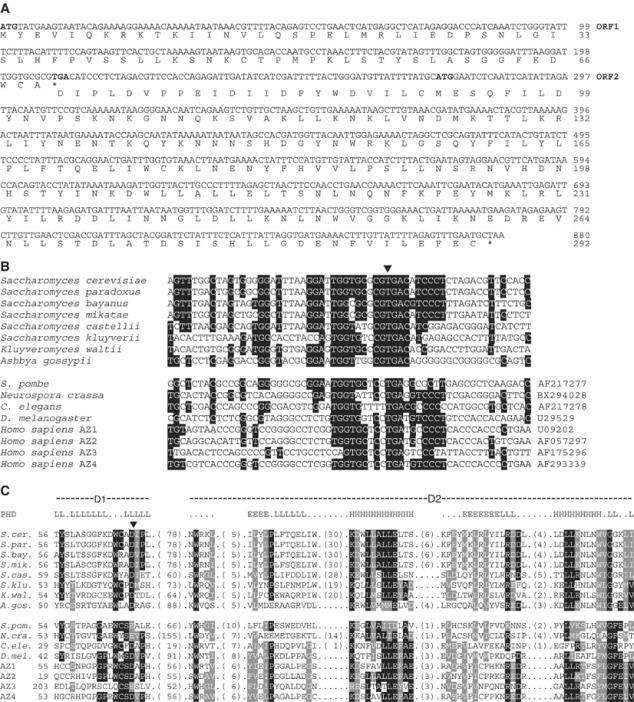
Sequence analysis of S. cerevisiae Oaz1. (A) Coding sequence of the OAZ1 genomic locus together with the encoded polypeptides. ORF1 including a TGA stop codon (shown in bold) encompasses the sequence until position 210. Upon a predicted ribosomal frameshifting resulting from skipping the first nucleotide in this stop codon (marked by an asterisk), translation continues in the +1 frame (ORF2). The ATG codon of the annotated ORF ‘YPL052w' is shown in bold. (B) Alignment of genomic or cDNA sequences encompassing the frameshifting sites in established AZ genes (lower part, accession numbers are given on the right side), as well as of previously unrecognized orthologues in Hemiascomycetes (upper part, sequences found as homologues of YPL052w in the Saccharomyces genome database, www.yeastgenome.org). A triangle marks the base that is presumed to be skipped during +1 ribosomal frameshifting. Conserved positions are printed on black background. (C) Alignment of AZs orthologues from the same species as in (B). A triangle indicates the position of the frameshifting site. Conserved residues are printed on black background, and positions assigned to amino acids with similar physicochemical properties are shaded in grey if supported by at least 50% of a total of 40 AZ family members that were compared in this analysis (only eight are shown). The sequences of a region termed D1 were aligned such that the frameshifting sites occupy equivalent positions. In the top row, the secondary structure as calculated via PHD (Rost, 1996) is presented. Only positions with an expected average accuracy >82% were considered. The abbreviations denote the following secondary structure types: E, extended (beta-sheet), A, alpha-helix and L, loop.
In order to identify a sequence encoding a presumptive ODC AZ in S. cerevisiae, we searched its genome with generalized profiles that were based upon a multiple alignment of AZ sequences derived from various fungal species. The generalized profile method constitutes a sensitive means to identify distant homologues based on family-wide conserved sequence features (Bucher et al, 1996; see Supplementary data). After a few iterations of profile construction, we found the previously uncharacterized S. cerevisiae open reading frame (ORF) ‘YPL052w' to encode a polypeptide related to C-terminal parts of AZs in other species. Obvious orthologues of ‘YPL052w' are also present in the genome of other closely related Hemiascomycetes (Figure 1 and Supplementary Figure S1). Upon close inspection, the genomic locus of ‘YPL052w' and its upstream sequences exhibit several features that are strikingly similar to those found in known AZ genes (Figure 1A). A putative ATG start codon is located 274 base pairs upstream of the annotated start codon of ORF ‘YPL052w'. The upstream ATG represents the translational start site of a short ORF that ends with a TGA stop codon after 207 bases. This ORF, which we refer to as ‘ORF1', is too short to be annotated in the Saccharomyces genome database. The stop codon is embedded in a sequence stretch of ∼20 nucleotides, which is well conserved around the established ribosomal frameshifting site in mammalian AZ mRNAs (Figure 1B). If an analogous +1 ribosomal frameshifting that results in the omission of the U in the UGA stop codon in the mRNA is assumed in S. cerevisiae, translation would shift to a second reading frame (ORF2) to produce a putative 292 residue polypeptide. The C-terminal 202 amino acids of this protein are encoded by the annotated ORF ‘YPL052w'. In support of the inferred frameshifting site, the resulting protein sequence that is encoded by the sequence encompassing this site exhibits a striking similarity to the D1 motif in known AZs (Figure 1C). The presumed AZs of S. cerevisiae and its relatives moreover display homology to the D2 domain of established AZ orthologues from other eukaryotes, which is likely to have an αβα fold (Figure 1C). Taken together, these data led us to hypothesize that S. cerevisiae ODC AZ is encoded by a locus, termed OAZ1 for ODC AZ, which extends from the beginning of ORF1 to the end of ‘YPL052w' on chromosome XVI. According to this hypothesis, and similar to its counterparts in other eukaryotes, synthesis of Oaz1 in S. cerevisiae would also involve a ribosomal frameshifting event.
Oaz1 mediates degradation of ODC by the proteasome
To test our hypothesis that S. cerevisiae OAZ1 encodes a putative ODC AZ experimentally, we first asked whether this gene, as some of its counterparts in other eukaryotes including mammals (see Introduction), is indeed required for the regulated turnover of ODC. To address this question, we generated genomic tags, leading to an expression of ODC marked with three copies of the ha epitope at the C-terminus. These tags were introduced into a wild-type strain and a congenic strain deleted for sequences encompassing the annotated ORF ‘YPL052w', which encodes about two-thirds of the presumptive Oaz1. The steady-state levels of ODC in these two strains grown with and without treatment with 100 μM of the polyamine spermidine for 3 h were determined by anti-ha Western blot analysis (Figure 2A). Quantification of the signals revealed that the wild type (wt) contained only ∼50% of the ODC as compared to the oaz1-Δ strain (Figure 2B). The presence of 100 μM spermidine in the culture medium resulted in a further reduction of ODC levels in wt to ∼12%, whereas no changes were observed in oaz1-Δ. Next we studied the kinetics of ODC disappearance after addition of spermidine to a culture of wild-type cells. ODC levels dropped rapidly to an extent that they were below detection already 60 min after spermidine addition (Figure 2C). In the oaz1-Δ mutant, in contrast, ODC levels were not affected by spermidine. We conclude that the presence of Oaz1 is required for controlling the levels of ODC in response to changes in polyamine concentrations, consistent with its presumed function of an ODC AZ and a role in ODC turnover. To verify that the observed effect reflects differences of ODC half-life, we performed pulse-chase analyses to determine ODC turnover rates in the same strains. As shown in Figure 2D and quantified in Figure 2E, addition of spermidine to the culture of the wild-type strain resulted in the induction of a rapid turnover of ODC (t1/2∼9 min). Deletion of OAZ1 resulted in a drastic stabilization of ODC, with spermidine having no effect on its stability. The observation that spermidine-induced degradation of ODC required the presence of the OAZ1 gene suggested that levels of Oaz1 are controlled by polyamines. To test for spermidine induction of Oaz1, we fused a sequence encoding three copies of the myc epitope in frame to the 3′ end of ORF2 on chromosome XVI. When extracts of the so-marked strain were assayed with anti-myc antibodies, a protein with an apparent molecular weight of ∼40 kDa was detected (Figure 2F). The size is consistent with the calculated molecular weight of the predicted Oaz1-myc3 polypeptide encoded from the inferred ATG start codon of ORF1 to the end of ORF2 (34 kDa) plus the C-terminal myc tag (∼6 kDa). The level of Oaz1-myc3 increased when spermidine was added to the culture media. The effect was even more striking when the spe1-Δ mutant lacking ODC was used in the experiment. In the absence of supplemented spermidine, this strain, which is unable to synthesize spermidine, did not contain detectable amounts of Oaz1 (Figure 2F). Addition of high amounts of spermidine, however, also resulted in a strong induction of Oaz1 in this mutant. Together, these results demonstrated that a regulation of Oaz1 levels is underlying the observed effects of spermidine on ODC stability.
Figure 2.
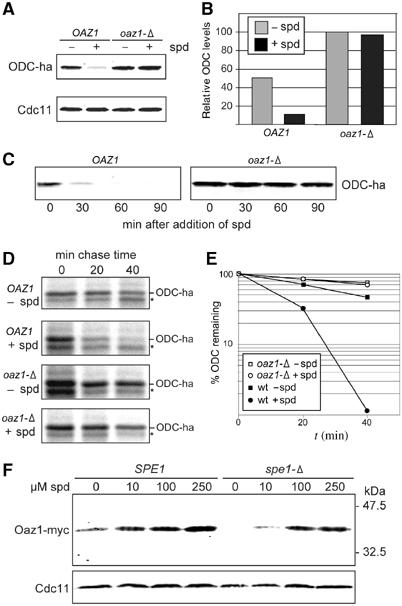
Oaz1 mediates degradation of ODC. (A) Steady-state levels of ODC-ha in strains PMY1 (wt) and PMY2 (oaz1-Δ) grown for 3 h in the absence (−spd) or presence of 100 μM spermidine (+spd) were analysed by anti-ha Western blotting. The blot was simultaneously probed with anti-Cdc11 antibodies to control for differences in protein loading. (B) Quantitation of fluorescence signals shown in (A). Values were normalized using the data obtained for Cdc11 and are given in % of the signal detected for oaz1-Δ grown in the absence of spermidine, which was set to 100%. (C) ‘Spermidine chase' of ODC-ha. ODC-ha in cell extracts was detected as in (A), but at the indicated time points after adding spermidine to a concentration of 100 μM to the media. (D) Pulse-chase analysis of ODC turnover. The same strains as in (A) were grown for 3 h in the absence or presence of 100 μM spermidine before labelling. An asterisk marks the position of a nonspecific band. (E) Quantitation of radioactive ODC-ha signals shown in (D), which were normalized using data for nonspecific background bands. (F) Spermidine induction of Oaz1-myc. Extracts from chromosomally tagged wt or spe1-Δ (=odc-Δ) strains (Table I) were analysed by anti-myc Western blotting. Both strains were incubated in the presence of the indicated concentrations of spermidine in the media.
Consistent with previous reports (Elias et al, 1995; Gandre and Kahana, 2002; Hoyt et al, 2003), we observed that an intact proteasome is required for spermidine-induced degradation of ODC. In strains carrying either the cim3-1/rpt6 mutation affecting an essential ATPase subunit (Rpt6) of the 19S activator complex of the 26S proteasome, or in pre1-1 and pre2-1 mutants, in which the β4 or β5 subunits of the 20S proteasome core are affected, spermidine-induced degradation of ODC was severely impaired (Figure 3A). We conclude that Oaz1 is required for spermidine-induced degradation of ODC by the proteasome establishing its role as an ODC AZ in S. cerevisiae. Degradation of ODC in mammals has been the paradigm of ubiquitin-independent degradation by the proteasome. It has been reported recently that degradation of ODC by the proteasome in S. cerevisiae does not require ubiquitin either (Gandre and Kahana, 2002; Hoyt et al, 2003). Together, these data indicated that regulated AZ-mediated and ubiquitin-independent degradation of ODC is conserved from S. cerevisiae to humans.
Figure 3.
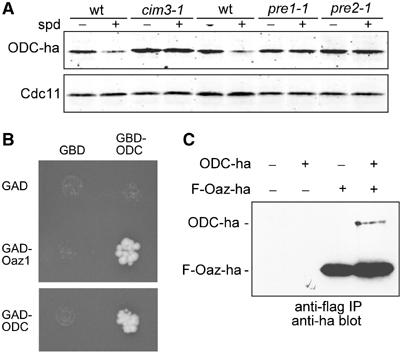
ODC binding to Oaz1 underlies its degradation by the proteasome. (A) Steady-state levels of ODC-ha expressed from PODC in the centromeric plasmid pPM67 in wt or proteasome mutants grown for 3 h in the absence (−spd) or presence of 100 μM spermidine (+spd) were analysed by anti-ha Western blotting. Lower part, anti-Cdc11 loading control. (B) Two-hybrid interaction of Oaz1 and ODC. Strain PJ64-4A was transformed with plasmids expressing Oaz1 or ODC as fusions to the Gal4-activating domain (GAD) or the Gal4 DNA-binding domain (GBD). Plasmids expressing just GAD or GBD were used as controls. Interaction was assayed on SD media lacking histidine to monitor the expression of a HIS3 reporter gene that is under the control of PGAL1. Colony growth indicates interaction. (C) ODC coimmunoprecipitates with Oaz1. Extracts from strain YHI29/1 (pre1-1) expressing either ODC-ha, flag(F)-Oaz1-ha or both were subjected to immunoprecipitation with anti-flag resin. Immunoprecipitated proteins were analysed by SDS–PAGE and anti-ha Western blotting.
Oaz1 physically interacts with ODC
Having established that Oaz1 is required for regulated proteolysis of ODC in S. cerevisiae, we next asked whether its mode of action is similar to that of its counterpart AZ1 in mammals (see Introduction). We therefore tested whether S. cerevisiae ODC and Oaz1 interact in vivo. Using the two-hybrid assay, we detected a strong interaction of ODC with Oaz1, as well as of ODC with itself (Figure 3B). The latter result demonstrated that dimerization of yeast ODC could be detected with this procedure. The former result was consistent with a model, in which, in analogy to mammalian systems, heterodimer formation underlies the targeted degradation of ODC in S. cerevisiae. To confirm this result biochemically, we co-expressed epitope-tagged versions of ODC and Oaz1 in yeast cells. Both proteins carried ha epitopes at their C-termini. Oaz1 was in addition tagged with a flag epitope at the N-terminus. Immunoprecipitations were carried out with anti-flag antibodies. ODC-ha was detected in precipitates from extracts of flag-Oaz1-ha co-expressing cells, but was absent from cell extracts lacking it (Figure 3C). Together, these data showed that Oaz1 forms a complex with ODC in S. cerevisiae, consistent with its function as an ODC AZ.
Polyamines induce ribosomal frameshifting during translation of OAZ1 mRNA
As outlined above, the complex genomic structure of the OAZ1 gene suggested that synthesis of Oaz1 protein involves a ribosomal frameshifting event. For ODC AZ in mammals, it has been demonstrated that translational frameshifting is induced by polyamines (see Introduction). The observed spermidine inducibility of Oaz1 suggested that a similar mechanism might operate in S. cerevisiae as well. To investigate the mechanism underlying the inducible synthesis of Oaz1, we generated fusions of a sequence encoding two copies of the myc epitope with the 5′ end of ORF1 and of a sequence encoding two copies of the ha epitope with the 3′ end of ORF2. Two otherwise identical constructs were generated that carried both tags but were distinguished either by the presence (OAZ1) or the absence of the frameshifting site (OAZ1-if; ‘if' denotes an in-frame fusion of ORF1 and ORF2). To generate the latter construct, the T nucleotide within the TGA stop codon of ORF1 was deleted. Both constructs were expressed from the unrelated PCUP1 promoter to exclude any putative effects of polyamine on transcriptional regulation. In addition, we chose the proteasome-deficient pre1-1 mutant (Heinemeyer et al, 1991) as a host strain because it turned out that Oaz1 is degraded by the proteasome and that this process is influenced by polyamines (described in a later section). The effects of spermidine on ribosomal frameshifting were therefore studied in pre1-1 cells, which were transformed with centromeric plasmids carrying the constructs described above. The presence of tagged polypeptides in cell extracts was analysed by Western blotting (Figure 4A and B). The cells expressing the construct containing the frameshift site showed increasing myc-Oaz1-ha signals with rising spermidine concentration in the growth media. The strain expressing the in-frame fusion, in contrast, yielded myc-Oaz1-ha signals that did not respond to changes of the spermidine concentration in the media (Figure 4B). Note that the levels of tagged Oaz1 in the latter strain were ∼10-fold higher than those obtained with the strain requiring induced frameshifting to synthesize Oaz1. In both strains, these signals were detectable with anti-myc as well as with anti-ha antibodies, supporting the notion that spermidine-induced ribosomal frameshifting had taken place during expression from the OAZ1 construct. This conclusion was supported further by the detection of the faster migrating myc-Orf1 polypeptide, in this case only with the anti-myc antibody, in the extracts of the strain expressing OAZ1 bearing the frameshifting site. The intracellular concentration of this polypeptide declined with increasing spermidine concentration in the growth media (Figure 4A and C). Together, these data proved our prediction that Oaz1 is expressed from ORF1 and ORF2 as a result of a spermidine-induced ribosomal frameshifting. This notion is supported by the ability of an OAZ1-if construct expressed from PCUP1 to complement the defect of the oaz1-Δ mutant in the degradation of ODC (Figure 4E). An otherwise identical construct bearing the authentic frameshifting site resulted in efficient complementation of oaz1-Δ even without supplemented spermidine. This result indicated that PCUP1 provided an expression level of OAZ1 sufficient for efficient targeting of ODC without additional polyamines in the media for the induction of ribosomal frameshifting. A construct expressing only the annotated ORF ‘YPL052w', in contrast, showed no complementation of oaz1-Δ, supporting the notion that this ORF does not encode a functional AZ. The experiment shown in Figure 4E also indicated that relatively low amounts of Oaz1 are sufficient to mediate degradation of the bulk of ODC. The residual ODC appears to be fairly resistant to Oaz1-induced degradation. The constructs used in Figure 4E carried N-terminal flag-His6 tags. Similar results were obtained with N-terminal myc2, although this tag appeared to reduce the efficiency of Oaz1 in ODC targeting (data not shown).
Figure 4.
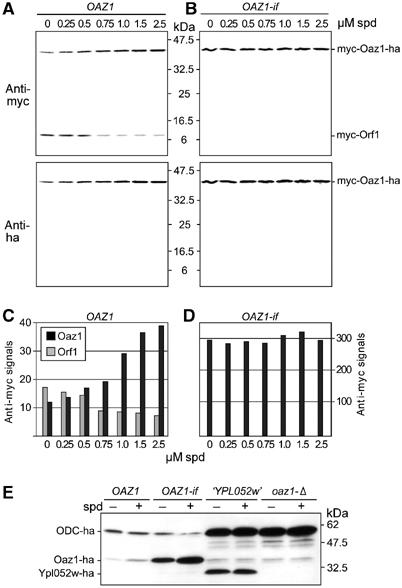
Synthesis of Oaz1 involves spermidine-inducible frameshifting. (A, B) Western blot analysis of pre1-1 mutant strain YHI29/1 transformed with centromeric plasmids expressing, from PCUP1, either (A) OAZ1 including the frameshift site or (B) OAZ1-if, an in-frame fusion of ORF1 and ORF2 that was generated by deleting the T nucleotide marked with an asterisk in Figure 1A. Both constructs were fused at their 5′ ends to a sequence encoding two copies of the myc tag and at their 3′ ends to a sequence encoding two copies of the ha tag. Cells were grown in the presence of the indicated concentrations of spermidine and 100 μM CuSO4 to induce expression from PCUP1. Extracts were analysed simultaneously for myc of ha tagged proteins by immunoblotting. (C, D) Quantitation of fluorescence signals shown in (A, B), respectively. (E) An OAZ1 gene lacking the frameshift mediates ODC degradation. Strain PMY2 (oaz1-Δ ODC-ha) was transformed with an empty vector or with plasmids expressing, from PCUP1, either OAZ1, OAZ1-if or ORF ‘YPL052w'. All three constructs were fused at their 5′ ends to a sequence encoding a flag-6His tag and at their 3′ ends to a sequence encoding two copies of the ha tag. Transformants were grown in the presence of 100 μM CuSO4 and either in the absence or presence of 100 μM spermidine as indicated. Yeast cell extracts were analysed by anti-ha immunoblotting.
Control of Oaz1 levels involves its ubiquitin-mediated degradation by the proteasome
While we were studying the effects of mutations affecting proteasome function on ODC levels, we observed that Oaz1 levels were also increased. As shown in Figure 5A for the pre1-1 mutant, Oaz1 accumulated to much higher levels in such strains. We next asked whether degradation of Oaz1 by the proteasome was ubiquitin-dependent. To test this, we expressed Oaz1-ha from an in-frame fusion of ORF1 and ORF2 (Oaz1-if, see above) in a mutant with a temperature-sensitive ubiquitin-activating enzyme (uba1) and in mutants lacking various ubiquitin-conjugating enzymes (ubc) (Jentsch, 1992). Increased levels of Oaz1 were detected in the uba1 mutant (grown at the semi-permissive temperature of 30°C) and in a strain lacking UBC4 and UBC5 (Seufert and Jentsch, 1990; McGrath et al, 1991) (Figure 5A). Mutants lacking other Ubc-encoding genes such as ubc1, ubc2, ubc6 ubc7, ubc8, ubc10, and ubc13 had Oaz1 levels comparable to the wild type (data not shown). Pulse-chase experiments showed that the elevated steady-state levels in pre1-1, uba1, and ubc4 ubc5 mutants were due to a drastic stabilization of the Oaz1 protein (Figure 5B and C). These data suggested that Oaz1 levels are directly controlled by Ubc4/Ubc5-mediated ubiquitin-dependent proteolysis by the proteasome. A prediction of this interpretation is that Oaz1 should be ubiquitylated prior to degradation. To verify this, we co-expressed Oaz1-ha with myc-tagged ubiquitin (myc-Ub) and performed anti-ha immunoprecipitations. Myc-Ub conjugates were precipitated from cells co-expressing these two tagged proteins, but were absent from control strains expressing either of the two proteins alone (Figure 5D). The more abundant of these conjugates was also detectable with anti-Ha antibody, but only after overexposing the Western blot (data not shown). Since the extracts were boiled in the presence of 2% SDS prior to immunoprecipitation, we conclude that myc-Ub was covalently attached to Oaz1.
Figure 5.
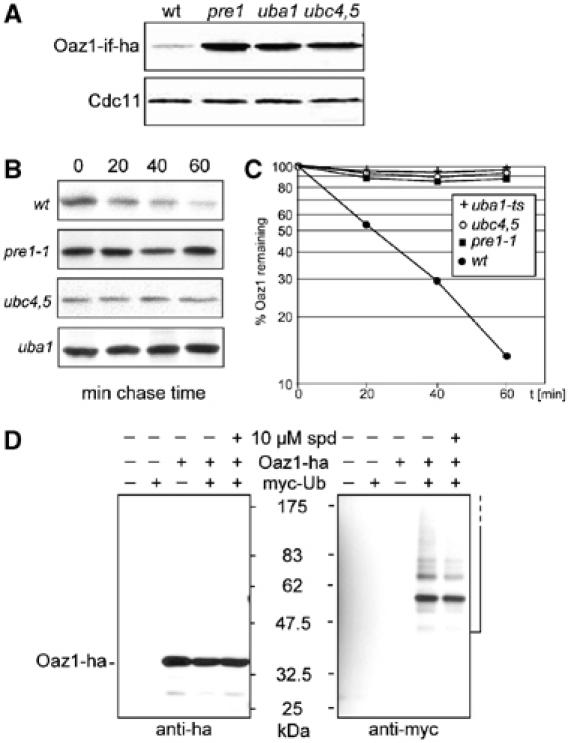
Ubiquitin-mediated degradation of Oaz1. (A–C) Degradation of Oaz1 requires ubiquitylation enzymes and the proteasome. (A) Oaz1-ha steady-state levels detected by anti-ha Western blotting in the strains indicated. OAZ1-if-ha was expressed from PCUP1 in plasmid pPM58. Lower part, anti-Cdc11-loading control. (B) Pulse-chase analysis of Oaz1 turnover in the same strains as in (A). (C) Quantitation of data shown in (B). (D) Oaz1 is ubiquitylated in vivo. JD47-13C transformants expressing either OAZ1-if-ha, myc-Ub, or both were grown in the presence of 100 μM CuSO4. Where indicated, spermidine (10 μM) was added to the media 1 h before extraction. After immunoprecipitation with anti-ha antibodies, precipitates were analysed by anti-myc and anti-ha Western blotting. Ubiquitylated forms of Oaz1-ha are indicated by an open-ended bracket.
Polyamines block degradation of Oaz1
While we were studying ribosomal frameshifting during the synthesis of Oaz1, we observed that spermidine addition to the growth media increased Oaz1 levels even when it was expressed from the control construct (OAZ1-if) that lacked the frameshift (Figure 4E). This effect of spermidine on Oaz1 levels, however, was not detected in the proteasome-deficient pre1-1 mutant (Figure 4A). Since expression of OAZ1-if was driven from PCUP1, an effect of spermidine on the transcriptional regulation of this construct appeared unlikely. These observations suggested that ubiquitin-mediated degradation of Oaz1 by the proteasome is influenced by polyamine levels. To follow up on this initial finding, we applied spermidine in concentrations ranging from 0 to 10 μM to a culture expressing OAZ1-if from PCUP1. As a result, we observed a dose-dependent increase in Oaz1 signal (Figure 6A and B). We therefore asked whether spermidine addition affected the half-life of Oaz1. Pulse-chase analyses demonstrated that spermidine inhibited Oaz1 degradation in a dose-dependent manner (Figure 6C and D). In order to test whether the observed inhibitory effect of spermidine was specific for Oaz1 degradation or whether this polyamine acted as a general inhibitor of proteolysis, we used established test substrates of ubiquitin-mediated proteolysis. R-β-galactosidase (R-βgal) is degraded by the N-end rule pathway, whereas Ub-P-βgal is degraded by the ubiquitin-fusion degradation (UFD) pathway (Johnson et al, 1995; Varshavsky, 1996). M-βgal served as a stable control protein. In marked contrast to the effect on Oaz1, spermidine addition to the growth media of cells expressing the β-galactosidase (βgal) test proteins had no effects on their steady-state levels (Figure 7A and B) or their turnover rates (Figure 7C and D). Since, as described above, Oaz1 degradation is ubiquitin-dependent, we asked whether spermidine inhibited ubiquitylation of Oaz1 or whether the turnover of ubiquitylated Oaz1 is blocked. To address this question, we again co-expressed myc-Ub with Oaz1-ha, this time in the presence of spermidine in the medium. Addition of the polyamine resulted in a reduction of detectable ubiquitylated forms of Oaz1-ha (Figure 5D). These data indicated that spermidine interferes with degradation of Oaz1 at least in part by inhibiting its ubiquitylation.
Figure 6.
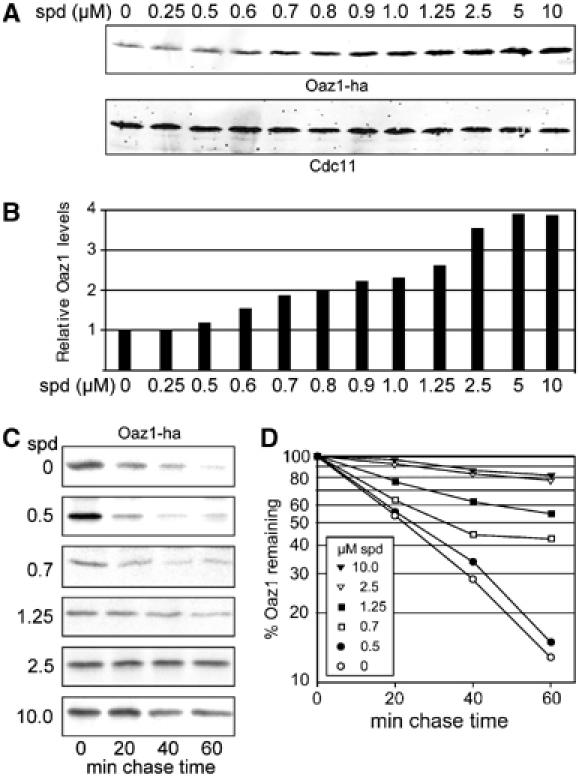
Spermidine inhibits degradation of Oaz1. (A) Analysis of the effects of spermidine on Oaz1 levels in the absence of ribosomal frameshifting. Wild-type strain JD47-13C transformed with pPM58 expressing OAZ1-if-ha was grown for 1 h in the presence of spermidine at the indicated concentrations. Extracts were analysed by anti-ha Western blotting. Cdc11 was detected simultaneously as a loading control. (B) Quantitation of data shown in (A). (C) Pulse-chase detection of concentration-dependent inhibition of Oaz1 degradation by polyamines in the same strain as in (A). Spermidine at the indicated concentrations was added 1 h before pulse labelling. (D) Quantitation of data shown in (C).
Figure 7.
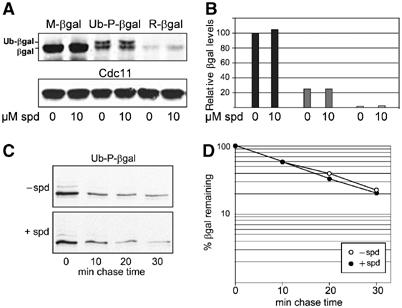
Spermidine has no general effect on turnover rates of proteolytic substrates. (A) Steady-state levels of proteolytic test substrate detected by anti-ha Western blotting (upper part). Anti-Cdc11 loading control (lower part). βgal variants were expressed as ubiquitin fusions (Ub-X-βgal). (B) Quantitation of data in (A). (C) Pulse-chase detection of Ub-P-βgal turnover rates. (D) Quantitation of data in (C).
Discussion
We report the discovery of OAZ1, an S. cerevisiae gene encoding an orthologue of mammalian ODC AZs. In addition, we detected closely related genes in the genomes of other Hemiascomycetes as well, thereby extending the AZ family by a set of fungal sequences, whose existence has been postulated (see Introduction). The sequence similarity, the conservation of the domain structure and the presence of a putative frameshifting site, all point to a common evolutionary origin of S. cerevisiae OAZ1 and ODC AZ genes in other eukaryotes.
Conservation of regulation and function of ODC AZ
Similar to its extensively studied orthologues in mammals, S. cerevisiae Oaz1 mediates ubiquitin-independent degradation of ODC by the proteasome (Figures 2 and 3). Polyamine induces Oaz1 levels without affecting transcription of the OAZ1 gene (data not shown). Synthesis of Oaz1 from two ORFs, instead, is controlled by polyamine-regulated programmed ribosomal frameshifting (Figure 4). Disruption of ORF2 resulted in abrogation of ODC degradation. Ectopic expression of the annotated ORF ‘YPL052w' did not restore degradation. An in-frame fusion of ORF1 and ORF2, in contrast, resulted in a complementation of this defect in ODC degradation. These and related results established that ribosomal frameshifting is essential for synthesis of a functional Oaz1 in S. cerevisiae.
Whereas many viruses utilize programmed frameshifting (commonly −1) to decode their RNAs, there are only few examples for an employment of such mechanisms in the synthesis of cellular proteins. In S. cerevisiae, aside of the retrotransposon Ty, two genes have been proposed to utilize a programmed +1 ribosomal frameshifting in their decoding. EST3 encodes a protein required for telomer replication (Morris and Lundblad, 1997). ABP140 encodes an actin filament-binding protein (Asakura et al, 1998). In all these cases, a tRNA slippage at a CUU Leu codon in a sequence (CUU AGG/A), in which it is followed by a slowly recognized codon, appears to underlie the frameshifting event (Sundararajan et al, 1999). A similar sequence element is absent from the frameshifting sites in AZ genes from yeast and other species (Figure 1B). Here an occlusion model has been proposed, in which sequences upstream of the frameshifting site and a downstream pseudoknot modify the structure in the A site of the ribosome occupied by the UGA termination codon, such that an Asp or a Glu codon in the +1 frame is recognized by the respective tRNAs (Namy et al, 2004). Despite the differences in the sequences of the frameshifting sites, +1 ribosomal frameshifting in decoding of both Ty1 and Oaz1 is modulated by polyamines (Balasundaram et al, 1994a, 1994b) (Figure 4). Whether polyamines more generally stimulate +1 frameshifting in S. cerevisiae, however, is unclear as no data are available on the effects of polyamines on the rates of frameshifting in decoding of EST3 and ABP140. It also remains to be investigated whether ribosomal frameshifting signals and their recognition are conserved among mRNAs of AZ orthologues. It was reported that decoding of a test construct containing the rat AZ cDNA in S. cerevisiae involves a −2 ribosomal frameshifting. Whether this event is stimulated by polyamines was not tested (Ivanov et al, 1998b). The discovery of the OAZ1 gene in S. cerevisiae provides an easy to manipulate in vivo system that will help to understand underlying mechanisms and the sequence requirements for polyamine-induced ribosomal frameshifting.
Ubiquitin-mediated degradation of Oaz1 is inhibited by spermidine
We noticed that polyamine addition to the media resulted in increased Oaz1 protein levels even when an in-frame construct that did not require frameshifting was used. Our analysis revealed that Oaz1 is subject to ubiquitin-mediated proteolysis by the proteasome. Degradation of Oaz1 is inhibited by mutations affecting the ubiquitin-activating enzyme Uba1 (E1), the ubiquitin-conjugating enzymes Ubc4 and Ubc5, or the proteasome. Similarly, an E1 requirement was previously reported for degradation of AZ1 in mammalian cells, but ubiquitylated forms of it were not detected (Gandre et al, 2002). In yeast, we detected such forms for Oaz1, indicating that ubiquitylation is essential for its proteasomal degradation (Figure 5D). Upon addition of spermidine to the growth media, we observed a dose-dependent inhibition of Oaz1 degradation. How might polyamines interfere with Oaz1 degradation? There are examples in the literature both for the inhibition of proteasomal degradation of a ubiquitylated substrate and for the inhibition of the ubiquitylation of a protein by small organic compounds. Degradation of a ubiquitylated dihydrofolate reductase was shown to be inhibited by the folic acid analogue methotrexate (Johnston et al, 1995). Our results, in which we detected a reduction rather than a stabilization of ubiquitylated forms of Oaz1 upon spermidine addition, suggested that polyamines, in contrast, stabilize Oaz1 at least in part by interfering with its ubiquitylation (Figure 5D). Similar observations were made for mammalian spermidine/spermine N-acetyltransferase (SSAT), a key enzyme in polyamine catabolism. Ubiquitylation of SSAT was inhibited in vitro by polyamine analogues (Coleman and Pegg, 2001). It was proposed that binding of the analogue brings about a conformational change of SSAT that inhibits its ubiquitylation. A similar mechanism may underlie or contribute to the inhibition of Oaz1 degradation by spermidine.
Alternatively, the enzymes that mediate ubiquitylation of Oaz1 may be inhibited by spermidine. One example of an E3 ubiquitin ligase whose activity is regulated by a small organic compound is Ubr1 in S. cerevisiae. This ligase indirectly regulates the uptake of dipeptides by controlling the stability of the transcriptional repressor Cup9 that blocks expression of the PTR1 dipeptide transporter gene. Dipeptides at low concentrations act as allosteric activators of the Ubr1 ligase activity towards Cup9 and thereby induce their uptake (Turner et al, 2000). In a diversion of such a mechanism, an as yet to be identified E3 enzyme responsible for Oaz1 ubiquitylation might be inhibited by polyamines.
We favor a model in which spermidine binding to Oaz1 inhibits its ubiquitylation, based on our observation that SSATs were detected in searches that used AZ-based profiles. In a reverse approach, profiles constructed from acetyl transferases alignments moreover retrieved several AZs. In both cases, the sequence similarity, however, was below the threshold that would establish a clear evolutionary relationship. Secondary structure prediction for the putative homologous region in acetyl transferases and AZs, however, indicated that they share a common αβα motif (data not shown). In the crystal structure of yeast N-acetyltransferase Gna1, this αβα motif overlaps with the binding site for Acetyl-CoA and the substrate (Peneff et al, 2001). By analogy, this domain is likely to be involved in spermidine binding within SSAT enzymes. We hypothesize that the spermidine-dependent stabilization of AZ may be due to a direct binding of spermidine to an αβα motif in the C-terminal region of AZ.
Based on our data, we propose that polyamines regulate their synthesis by controlling Oaz1 levels via two separate mechanisms. The first mechanism is the induction of ribosomal frameshifting in decoding of OAZ1 mRNA by a mechanism that is poorly understood. The second mechanism is the inhibition of Oaz1 proteolysis. We propose that the latter mechanism ensures a rapid recovery from a state in which ODC is downregulated by AZ. In this model, a drop of polyamine levels below a critical threshold does not only prevent de novo synthesis of AZ but also results in a turnover of its existing pools, thereby allowing for a rapid recovery of ODC activity. Polyamines thus appear to mediate an efficient feedback control of their formation by affecting both the synthesis and turnover rates of ODC AZ.
Materials and methods
Yeast media
Yeast-rich (YPD) and synthetic (S) minimal media with 2% dextrose (SD) were prepared as described (Ramos et al, 1998). Spermidine (Sigma) was added to SD media at various concentrations as indicated.
Yeast strains and plasmids
Table I lists the strains used in this study. Strains expressing C-terminally ha3- or myc3-tagged variants of ODC or Oaz1 were constructed by a PCR-based method using short-flanking oligos and plasmids pYM2 and pYM5, respectively, as templates (Knop et al, 1999). The following plasmids were constructed in the background of the CEN/URA3-based plasmid YCplac33 (Gietz and Sugino, 1988). pPM52 expressed flag-His6-OAZ1-ha2 from PCUP1. Plasmid pPM53 was identical to pPM52, except that it expressed Oaz1 as an in-frame fusion of ORF1 and ORF2 (Oaz1-if). Plasmid pPM54 instead expressed flag-His6-YPL052w-ha2; pPM58 expressed OAZ1-if-ha2. Plasmids pPM64 and pPM67 were based on the CEN/LEU plasmid pRS315 (Sikorski and Hieter, 1989) and expressed ODC-ha3 from PCUP1 or PODC, respectively. Plasmids pPM84 and pPM85 were based on the CEN/URA3 plasmid pRS316 (Sikorski and Hieter, 1989). pPM84 expressed, from PCUP1, myc2-OAZ1-ha2 with a frameshift in the OAZ1 ORF, whereas pPM85 expressed an otherwise identical in-frame fusion lacking the frameshift site.
Table 1.
Yeast strains
| Strain | Relevant genotype | Source/comment |
|---|---|---|
| BY4741 | MATa his3-Δ1 leu2Δ0 met15-Δ0 ura3-Δ0 | EUROSCARF (‘wt') |
| BY4742 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 | EUROSCARF (‘wt') |
| Y02776 | oaz1-Δ∷KanMX4 (=ypl052w-Δ) | Derivative of BY4741, EUROSCARF |
| Y15034 | spe1-Δ∷KanMX4 (=‘odc-Δ') | Derivative of BY4742, EUROSCARF |
| PMY1 | ODC-ha3∷HISMX6 | Derivative of BY4741 |
| PMY2 | ODC-ha3∷HISMX6 oaz1-Δ∷KanMX4 | Derivative of BY4741 |
| PMY17 | OAZ1-myc3∷HISMX6 | Derivative of BY4741 |
| PMY15 | OAZ1-myc3∷HISMX6 spe1-Δ∷KanMX4 | Derivative of Y15034 |
| WGC4a | MATα ura3 his3-11 leu2-3,112 | (Heinemeyer et al, 1991) |
| YHI29/1 | pre1-1 | Derivative of WGC4a (Heinemeyer et al, 1991) |
| YPH500 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | (Sikorski and Hieter, 1989) |
| CMY762 | cim3-1 | Derivative of YPH500 (Ghislain et al, 1993) |
| ubc4 ubc5 | ubc4-Δ∷HIS3 ubc5-Δ∷LEU2 | (Seufert and Jentsch, 1990) |
| JD47-13C | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 | (Ramos et al, 1998) |
| JD77-1-1 | uba1-Δ∷HIS3 pRSts64-1(uba1-ts) | Derivative of JD47-13C (McGrath et al, 1991) |
| PJ64-4A | MATa his3-Δ200 leu2-3,112 trp1-901 ura3-52 gal4-Δ gal80-Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ | (James et al, 1996) |
Immunoblot analysis and determination of protein stability by pulse-chase analysis
For detection of steady-state levels, S. cerevisiae cells were grown at 30°C in SD media to an optical density measured at 600 nm (OD600) of 1.0±0.2. Proteins were extracted from yeast cells by boiling in loading buffer (0.0625 M Tris–HCl (pH 6.8), 2% SDS, 1% β-mercaptoethanol, 10% glycerol, 0.002% bromophenol blue) for 5 min. Extracts from cells corresponding to O.5 OD600 (≈15 μg) were loaded per lane. SDS–PAGE and immunoblots were performed as described (Ramos et al, 1998). Proteins were either detected using secondary anti-mouse or anti-rabbit IgG coupled to peroxidase (Boehringer Mannheim), Lumi-LightPLUS chemiluminescent substrate (Roche), and X-ray films (Figures 3C, 4E and 5D), or with anti-mouse, anti-rat, or anti-rabbit IgG coupled to near-infrared fluorophores (Rockland), and the Odyssey Infrared Imaging System. The latter system was also used for signal quantification. For detection of epitope tags, we used the following monoclonals as primary antibodies. The ha epitope was detected with 16B12 from mouse (Covance) in all experiments except those shown in Figure 4A and B, in which 3F10 from rat (Boehringer Mannheim) was chosen instead. Mouse monoclonal 9B11 (Cell Signaling Technology) was used for myc epitope detection. Cdc11 was detected with polyclonal rabbit antibodies (Santa Cruz Biotechnology), and βgal with a mouse monoclonal antibody (Promega). Pulse-chase analyses were carried out as described (Ramos et al, 1998).
Assay of ODC Oaz1 complex formation
Yeast two-hybrid assays were carried out as described (James et al, 1996). For coimmunoprecipitation, proteins were extracted from yeast cells by glass bead lysis in ice-cold lysis buffer (50 mM Na-HEPES (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% Triton X-100) containing the ‘Complete' Protease-Inhibitor cocktail (Roche). In all, 300 μg of protein was subjected to immunoprecipitation using anti-flag resin (Sigma). Precipitated proteins were washed six times with lysis buffer and eluted with 300 μg/ml flag peptide (Sigma).
Detection of ubiquitin conjugates
Yeast cells co-expressing OAZ1-if-ha (pPM58) and myc-Ub (plasmid YEp105) (Ellison and Hochstrasser, 1991), both from PCUP1, were lysed by boiling for 5 min in lysis buffer (see above) with 2% SDS and 1% β-mercaptoethanol. Empty vector transformants were used as controls. Samples were diluted 1:20 with lysis buffer and subjected to immunoprecipitation with an anti-ha high-affinity matrix (Roche). Precipitated proteins were washed six times with a lysis buffer containing 0.1% SDS, eluted by boiling in loading buffer and analysed by SDS–PAGE and anti-myc and, after stripping, with anti-ha Western blotting.
Supplementary Material
Supplementary material
Supplementary Figure
Acknowledgments
We thank Michael Ellison, Stefan Jentsch, Mark Hochstrasser, Carl Mann, Alexander Varshavsky, and Dieter H Wolf for yeast strains and plasmids. RP is supported by a predoctoral fellowship from the NRW graduate school in Genetics and Functional Genomics. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Do 649) to RJD.
References
- Asakura T, Sasaki T, Nagano F, Satoh A, Obaishi H, Nishioka H, Imamura H, Hotta K, Tanaka K, Nakanishi H, Takai Y (1998) Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene 16: 121–130 [DOI] [PubMed] [Google Scholar]
- Balasundaram D, Dinman JD, Tabor CW, Tabor H (1994b) SPE1 and SPE2: two essential genes in the biosynthesis of polyamines that modulate +1 ribosomal frameshifting in Saccharomyces cerevisiae. J Bacteriol 176: 7126–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaram D, Dinman JD, Wickner RB, Tabor CW, Tabor H (1994a) Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 91: 172–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P, Karplus K, Moeri N, Hofmann K (1996) A flexible motif search technique based on generalized profiles. Comput Chem 20: 3–23 [DOI] [PubMed] [Google Scholar]
- Chen H, MacDonald A, Coffino P (2002) Structural elements of antizymes 1 and 2 are required for proteasomal degradation of ornithine decarboxylase. J Biol Chem 277: 45957–45961 [DOI] [PubMed] [Google Scholar]
- Childs AC, Mehta DJ, Gerner EW (2003) Polyamine-dependent gene expression. Cell Mol Life Sci 60: 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffino P (2001a) Antizyme, a mediator of ubiquitin-independent proteasomal degradation. Biochimie 83: 319–323 [DOI] [PubMed] [Google Scholar]
- Coffino P (2001b) Regulation of cellular polyamines by antizyme. Nat Rev Mol Cell Biol 2: 188–194 [DOI] [PubMed] [Google Scholar]
- Coleman CS, Pegg AE (2001) Polyamine analogues inhibit the ubiquitination of spermidine/spermine N1-acetyltransferase and prevent its targeting to the proteasome for degradation. Biochem J 358: 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias S, Bercovich B, Kahana C, Coffino P, Fischer M, Hilt W, Wolf DH, Ciechanover A (1995) Degradation of ornithine decarboxylase by the mammalian and yeast 26S proteasome complexes requires all the components of the protease. Eur J Biochem 229: 276–283 [PubMed] [Google Scholar]
- Ellison MJ, Hochstrasser M (1991) Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem 266: 21150–21157 [PubMed] [Google Scholar]
- Gandre S, Bercovich Z, Kahana C (2002) Ornithine decarboxylase-antizyme is rapidly degraded through a mechanism that requires functional ubiquitin-dependent proteolytic activity. Eur J Biochem 269: 1316–1322 [DOI] [PubMed] [Google Scholar]
- Gandre S, Kahana C (2002) Degradation of ornithine decarboxylase in Saccharomyces cerevisiae is ubiquitin independent. Biochem Biophys Res Commun 293: 139–144 [DOI] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C (1993) S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366: 358–362 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Gupta R, Hamasaki-Katagiri N, White Tabor C, Tabor H (2001) Effect of spermidine on the in vivo degradation of ornithine decarboxylase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 98: 10620–10623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Murakami Y, Matsufuji S (1996) Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends Biochem Sci 21: 27–30 [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH (1991) Proteinase yscE, the yeast proteasome/multicatalytic–multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J 10: 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller JS, Fong WF, Canellakis ES (1976) Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci USA 73: 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Zhang M, Coffino P (2003) Ubiquitin-independent mechanisms of mouse ornithine decarboxylase degradation are conserved between mammalian and fungal cells. J Biol Chem 278: 12135–12143 [DOI] [PubMed] [Google Scholar]
- Ichiba T, Matsufuji S, Miyazaki Y, Murakami Y, Tanaka K, Ichihara A, Hayashi S (1994) Functional regions of ornithine decarboxylase antizyme. Biochem Biophys Res Commun 200: 1721–1727 [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Gesteland RF, Atkins JF (1998a) A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics 52: 119–129 [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Gesteland RF, Matsufuji S, Atkins JF (1998b) Programmed frameshifting in the synthesis of mammalian antizyme is +1 in mammals, predominantly +1 in fission yeast, but −2 in budding yeast. RNA 4: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Rohrwasser A, Terreros DA, Gesteland RF, Atkins JF (2000) Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: antizyme 3. Proc Natl Acad Sci USA 97: 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janne J, Alhonen L, Pietila M, Keinanen TA (2004) Genetic approaches to the cellular functions of polyamines in mammals. Eur J Biochem 271: 877–894 [DOI] [PubMed] [Google Scholar]
- Jariel-Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Piechaczyk M (1995) Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J Biol Chem 270: 11623–11627 [DOI] [PubMed] [Google Scholar]
- Jentsch S (1992) The ubiquitin-conjugation system. Annu Rev Genet 26: 179–207 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Johnston JA, Johnson ES, Waller PR, Varshavsky A (1995) Methotrexate inhibits proteolysis of dihydrofolate reductase by the N-end rule pathway. J Biol Chem 270: 8172–8178 [DOI] [PubMed] [Google Scholar]
- Ju D, Xie Y (2004) Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin-dependent and -independent. J Biol Chem 279: 23851–23854 [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Li X, Coffino P (1992) Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol Cell Biol 12: 3556–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S (1995) Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 80: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavsky A (1991) UBA 1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J 10: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JL, Chen HJ (1990) Conformational changes in ornithine decarboxylase enable recognition by antizyme. Biochim Biophys Acta 1037: 115–121 [DOI] [PubMed] [Google Scholar]
- Morris DK, Lundblad V (1997) Programmed translational frameshifting in a gene required for yeast telomere replication. Curr Biol 7: 969–976 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A (1992) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360: 597–599 [DOI] [PubMed] [Google Scholar]
- Namy O, Rousset JP, Napthine S, Brierley I (2004) Reprogrammed genetic decoding in cellular gene expression. Mol Cell 13: 157–168 [DOI] [PubMed] [Google Scholar]
- Pendeville H, Carpino N, Marine JC, Takahashi Y, Muller M, Martial JA, Cleveland JL (2001) The ornithine decarboxylase gene is essential for cell survival during early murine development. Mol Cell Biol 21: 6549–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peneff C, Mengin-Lecreulx D, Bourne Y (2001) The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J Biol Chem 276: 16328–16334 [DOI] [PubMed] [Google Scholar]
- Ramos PC, Höckendorff J, Johnson ES, Varshavsky A, Dohmen RJ (1998) Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 92: 489–499 [DOI] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y, Bercovich Z, Ciechanover A, Kahana C (1989) Degradation of ornithine decarboxylase in mammalian cells is ATP dependent but ubiquitin independent. Eur J Biochem 185: 469–474 [DOI] [PubMed] [Google Scholar]
- Rost B (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Meth Enzymol 266: 525–539 [DOI] [PubMed] [Google Scholar]
- Schwartz B, Hittelman A, Daneshvar L, Basu HS, Marton LJ, Feuerstein BG (1995) A new model for disruption of the ornithine decarboxylase gene, SPE1, in Saccharomyces cerevisiae exhibits growth arrest and genetic instability at the MAT locus. Biochem J 312: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W, Jentsch S (1990) Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 9: 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan A, Michaud WA, Qian Q, Stahl G, Farabaugh PJ (1999) Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol Cell 4: 1005–1015 [DOI] [PubMed] [Google Scholar]
- Toth C, Coffino P (1999) Regulated degradation of yeast ornithine decarboxylase. J Biol Chem 274: 25921–25926 [DOI] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78: 787–798 [DOI] [PubMed] [Google Scholar]
- Turner GC, Du F, Varshavsky A (2000) Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405: 579–583 [DOI] [PubMed] [Google Scholar]
- Varshavsky A (1996) The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA 93: 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin–proteasome system. Cell 118: 99–110 [DOI] [PubMed] [Google Scholar]
- Wallace HM, Fraser AV, Hughes A (2003) A perspective of polyamine metabolism. Biochem J 376: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci USA 98: 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Pickart CM, Coffino P (2003) Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J 22: 1488–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Karplus K, Grate L, Coffino P (2000) A homolog of mammalian antizyme is present in fission yeast Schizosaccharomyces pombe but not detected in budding yeast Saccharomyces cerevisiae. Bioinformatics 16: 478–481 [DOI] [PubMed] [Google Scholar]
- Zhu C, Lang DW, Coffino P (1999) Antizyme2 is a negative regulator of ornithine decarboxylase and polyamine transport. J Biol Chem 274: 26425–26430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary Figure


