Abstract
In this study, methylation‐specific polymerase chain reaction was used to investigate the potential prognostic significance of the methylation status of p15, p16, MGMT, and DAPK genes in 51 specimens of diffuse large B‐cell lymphoma (DLBCL). Hypermethylation of p15 gene was significantly more prevalent in patients without relapse (p = 0.001) and there was a trend toward more frequent presence of p15 methylation in patients without death outcome within 5‐year follow‐up period (p = 0.086) Also, there was a trend toward accumulation of p15 methylation with favorable clinicopathological parameters including: age ≤ 60 years (p = 0.091), normal levels of lactate dehydrogenase (p = 0.090), Eastern Cooperative Oncology Group performance status < 2 (p = 0.095), and low/intermediate low International Prognostic Index (p = 0.076). In the female group and group of the patients without bulky tumor mass, treated with chemotherapeutic regimens including rituximab, methylation of p15 was significantly related to longer overall survival (p = 0.036 and 0.027, respectively). Our results suggest that promoter methylation of p15 gene could have prognostic value in DLBCL patients treated with rituximab when used in combination with gender and tumor size.
Keywords: diffuse large B‐cell lymphoma, methylation, p15, prognosis, rituximab
Introduction
Diffuse large B‐cell lymphoma (DLBCL) is the aggressive subgroup of non‐Hodgkin's lymphoma (NHL) and comprises approximately 30–40% of all cases. It is clinically and biologically heterogeneous disease, characterized with highly variable response to the treatment and clinical outcome.1 The International Prognostic Index (IPI) defines risk groups based on the clinical parameters at presentation.2 However, patients with identical IPI still exhibit marked variability in survival, suggesting the presence of significant biological heterogeneity within the same IPI category.3 In the recent years, much effort has been made to establish new, molecular prognostic parameters in order to further stratify patients into different risk groups and choose appropriate treatment strategy.4, 5, 6 The addition of rituximab to the standard CHOP chemotherapy (R‐CHOP) has led to a marked improvement in clinical outcome of patients with DLBCL, but in the same time, it's introduction has altered the significance of previously recognized prognostic parameters.7 So, it is important to investigate additional molecular markers and reexamine the formerly established in aim to improve prognosis in the era of rituximab.
Aberrant DNA methylation of CpG islands in the promoter region of tumor suppressor genes represents an important mechanism of their inactivation. There are increasing evidences that hypermethylation of certain genes in cancer could serve as marker for monitoring the clinical behavior of the disease and prediction of patients outcome.8 Recent investigations showed that multiple genes could be affected through aberrant DNA methylation in DLBCL, leading to deregulation of multiple cell pathways, including cell cycle control,9, 10 DNA damage repair, apoptosis,11 and many others. Though there are strong evidences that such epigenetic changes could contribute to lymphomagenesis and the biological behavior of the disease,12, 13, 14 the impact of particular genes methylation on patients’ prognosis and outcome, especially in the era of new treatment strategies, remains unsolved. Hence, the aim of our study was to investigate the methylation status of four selected, cancer‐related genes in the group of 51 DLBCL patients, in order to determine the possible association of their methylation with clinicopathogical features and outcome. The selected genes included: p15 and p16 tumor suppressor genes, encoding for cyclin‐dependent kinase inhibitors important for G1 cell cycle arrest,10 gene for O6‐methylguanine‐DNA methyltransferase (MGMT), a DNA repair enzyme that removes mutagenic and cytotoxic adducts from the O6 position of guanine,15 and DAPK (death‐associated protein kinase) gene that encodes a serine‐threonine kinase involved in the extrinsic pathway of apoptosis, initiated by γ‐interferon, FAS ligand, and tumor necrosis factor‐α.16 The promoter hypermethylation of these four genes has frequently been observed in DLBCL, indicating the important role of such epigenetic changes in the pathogenesis of this tumor type.10, 11, 12, 13, 14 However, the data considering the relationship between methylation status of selected genes and patients’ prognosis are contradictory, which needs more comprehensive investigations.
Methods
Patients and samples
Fifty‐one patients (26 male, 25 female; median age 52.4 years, range 19–83 years) with DLBCL included in this study were diagnosed and treated in the Institute of Hematology, Clinical Center of Serbia, Belgrade, Clinic of Hematology MMA, Belgrade, Serbia and Oncology Institute of Vojvodina, Sremska Kamenica, Serbia, from 2001 to 2012. Biopsy samples of lymph node, bone marrow, or other involved organs from patients were collected at diagnosis. The data according to the all clinicopathological parameters, overall survival (OS), as well as the presence of Bcl2, Bcl6, CD10, and Ki67 expression were taken from medical documentation of listed institutions. Tumor samples were considered positive for Bcl‐2, Bcl‐6, or CD10 when at least 50%, 10%, or 20% of tumor cells expressed Bcl2, Bcl6, or CD10 protein, respectively. Expression of Ki67 protein in 0–30% of tumor cells was considered as weak, in 30–60% of tumor cells as moderate, and in more than 60% of the cells as strong. All procedures were carried out with the prior informed consent of the patients and with the approval of the local Ethic Committee. Treatment consisted of CHOP/R‐CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone with or without rituximab) regimen. Some patients had adjuvant radiotherapy and/or surgery, and some of them underwent autologous stem‐cell transplantation. Response criteria and survival outcomes were defined according to the recommendation of Cheson et al.17
Analysis of methylation status of the p15, p16, MGMT, and DAPK genes
DNA methylation patterns in the promoter CpG islands of the p15, p16, MGMT, and DAPK genes were determined in all of 51 samples by methylation‐specific polymerase chain reaction (MSP) following the bisulfite modification of isolated genomic DNA, as described earlier.18 Briefly, DNA was isolated from deparaffined tumor specimens using standard proteinase K, phenol/chloroform/isoamyl alcohol extraction, and ethanol precipitation.19 Two micrograms of isolated DNA were denatured by NaOH (final 0.3 mol/L) at 42°C for 30 minutes and modified by sodium bisulfite (5.20–5.69 mol/L, pH 5.0, Sigma, St Louis, MO, USA) for 18 hours at 50°C. After incubation, DNA was purified using the DNA extraction KIT (MBI Fermentas, Lithuania), again treated by NaOH (final 0.3 mol/L), at 37°C for 20 minutes, precipitated with ethanol/ammonium acetate and resuspended in 40 μL of 1 mmol/L Tris‐HCl, pH 8.0. Aliquots of 4 μL of bisulfite‐modified DNA were used for MSP reactions. The polymerase chain reaction (PCR) mixture contained 1 × PCR buffer (16 mmol/L ammonium sulfate, 67 mmol/L Tris‐HCL, pH 8.8, 10 mmol/L 2‐mercaptoethanol), 6.7 mmol/L MgCl2, dNTP (each at 1.25 mmol/L), and primers (300 ng each per reaction) in a final volume of 50 μL. Reactions were hot‐started at 95°C for 5 minutes before the addition of 1.25 units of Taq polymerase (MBI Fermentas). Amplification was carried out in an Applied Biosystems (Foster City, CA, USA) 2720 temperature cycler for 40 cycles (45 seconds at 95°C, 45 seconds at the annealing temperature being specific for each reaction, and 60 seconds at 72°, followed by final extension for 4 minutes at 72°C). Primers sequences used for each reaction are listed in Table 1. DNA from peripheral blood lymphocytes from a healthy donor was used as negative control for methylated alleles. The same leukocyte DNA was methylated in vitro with excess SssI methyltransferase (New England Biolabs, Ipswich, MA, USA) to generate completely methylated DNA at all CpG sites and used as positive control for all genes. PCR products were separated by electrophoresis on 6% acrylamide gels, stained with silver nitrate and visualized by sodium carbonate.
Table 1.
Primer sets used for MSP
| Primer set | Sense primer | Antisense primer | Size (bp)/AT | References |
|---|---|---|---|---|
| p15‐M | 5’‐GCGTTCGTATTTTGCGGTT‐3’ | 5’‐CGTACAATAACCGAACGACCGA‐3’ | 148 bp/57°C | 20 |
| p15‐U | 5’‐TGTGATGTGTTTGTATTTTGTGGTT‐3’ | 5’‐CCATACAATAACCAAACAACCAA‐3’ | 154 bp/57°C | 20 |
| p16‐M | 5’‐TTATTAGAGGGTGGGGCGGATCGC‐3’ | 5’‐GACCCCGAACCGCGACCGTAA‐3’ | 150 bp/65°C | 20 |
| p16‐U | 5’‐TTATTAGAGGGTGGGGTGGATTGT‐3’ | 5’‐CAACCCCAAACCACAACCATAA‐3’ | 151 bp/60°C | 20 |
| MGMT‐M | 5’‐TTTCGACGTTCGTAGGTTTTCGC‐3’ | 5’‐GCACTCTTCCGAAAACGAAACG‐3’ | 81 bp/57°C | 21 |
| MGMT‐U | 5’‐TTTGTGTTTGATGTTTGTAGGTTTTTGT‐3’ | 5’‐AACTCCACACTCTTCCAAAAACAAAAA‐3’ | 93 bp/57°C | 21 |
| DAPK‐M | 5’‐GGATAGTCGGATCGAGTTAACGTC‐3’ | 5’‐CCCTCCCAAACGCCGA‐3’ | 98 bp/60°C | 22 |
| DAPK‐U | 5’‐GGAGGATAGTTGGATTGAGTTAATGTT‐3’ | 5’‐CAAATCCCTCCCAAACACCAA‐3’ | 106 bp/60°C | 22 |
M = primer set for methylated modified DNA sequence; U = primer set for unmethylated modified DNA sequence; AT = annealing temperature.
Statistical analysis
Contingency tables were analyzed using Pearson’ s χ2‐test or Fisher’ s exact two‐tailed test, when expected frequencies were lower than five. Continuous variables were compared with the use of Student’ s t‐test. OS distributions were estimated by the Kaplan‐Meier method and differences were evaluated by the Log‐rank test. In all tests, a p‐value less than 0.05 were considered as statistically significant. All statistical analyses were performed using the Sigma Plot 10.0 licensed statistical analysis software package.
Results
Correlation between promoter methylation status and clinicopathological features
Hypermethylation of p15, p16, MGMT, and DAPK genes was detected in 23% (12/51), 37%, (19/51), 39% (20/51), and 55% (28/51) of samples, respectively. Representative examples of the methylation analysis are shown in Figure 1. Overall, 74% (38/51) of cases showed at least one hypermethylated gene, and 25% (13/51) of cases show no methylation of any genes examined. We observed the significant correlations between methylation of p15 and p16 genes (p = 0.037), p16 and DAPK genes (p = 0.047), and MGMT and DAPK genes (p = 0.025), and observed comethylations were present in 16%, 27%, and 29% of cases, respectively.
Figure 1.
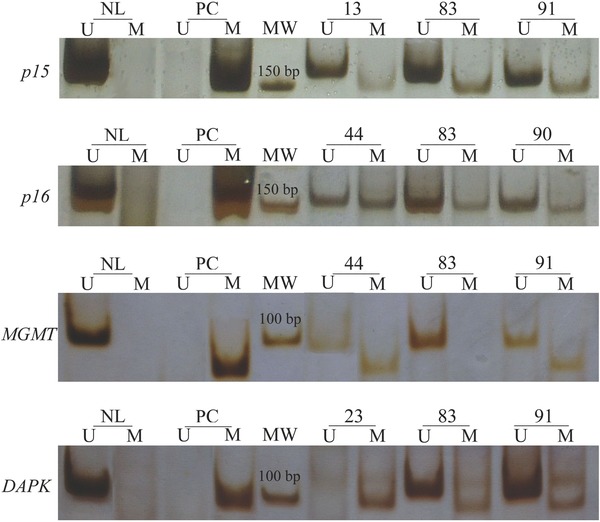
Analysis of p15, p16, MGMT, and DAPK genes methylation by MSP in representative cases of DLBCL. The presence of a visible PCR product in lanes U indicates the presence of unmethylated allels (154, 151, 93, and 106 bp, respectively), while the presence of product in lanes M indicates the presence of methylated allels (148, 150, 81, and 98 bp, respectively). NL = normal lymphocytes as a positive control for unmethylated allels; PC = in vitro methylated DNA from normal lymphocytes as a positive control for methylated allels; MW = molecular weight marker (50 bp).
Study of correlations between promoter methylation status of each gene and clinicopathological features are summarized in Table 2. Methylation of p16 gene was significantly more prevalent in patients with normal level of lactate dehydrogenase (LDH) (p = 0.035) and low/intermediate low IPI score (p = 0.034). We observed a trend toward more prevalent methylation of p15 gene in patients younger than 60 years (p = 0.091), normal level of LDH (p = 0.090), Eastern Cooperative Oncology Group (ECOG) performance status <2 (p = 0.095), and with low/intermediate low IPI score (p = 0.076). In the same time, we observed significant correlation between p15 methylation status and relapse of the disease; no one patient with relapsed DLBCL showed p15 methylation (p = 0.001, Table 2). Also, there was a trend toward more frequent presence of p15 methylation in patients without death outcome within 5‐year follow‐up period (p = 0.086). There was no correlation between methylation of MGMT or DAPK genes with any clinicopathological characteristic, Bcl2, Bcl6, CD10, and Ki67 expression, response to the therapy or patients’ outcome.
Table 2.
Correlation of p15, p16, MGMT, and DAPK methylation status with clinicopathological features in DLBCL
| Variable | p15m | p16m | MGMTm | DAPKm | p |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 4/26 (15%) | 9/26 (35%) | 11/26 (42%) | 14/26 (54%) | |
| Female | 8/25 (32%) | 10/25 (40%) | 9/25 (36%) | 14/25 (56%) | NS |
| Age (yr) | |||||
| ≤60 | 10/30 (33%) | 14/30 (47%) | 14/30 (47%) | 19/30 (63%) | |
| >60 | 2/21 (9%)* | 5/21 (24%) | 6/21 (29%) | 9/21 (43%) | 0.091* |
| Stage | |||||
| I, II | 2/8 (25%) | 4/8 (50%) | 2/8 (25%) | 5/8 (62%) | |
| III, IV | 10/43 (23%) | 15/43 (35%) | 18/43 (42%) | 23/43 (53%) | NS |
| LDH | |||||
| Normal | 7/20 (35%)** | 11/20 (55%)* | 7/20 (35%) | 12/20 (60%) | 0.035* |
| Elevated (>450 u/L) | 4/29 (14%) | 6/29 (21%) | 12/29 (41%) | 14/29 (48%) | 0.090** |
| Extranodal sites | |||||
| 0, 1 | 10/39 (26%) | 14/39 (36%) | 14/39 (36%) | 20/39 (51%) | |
| >1 | 2/12 (17%) | 5/12 (42%) | 6/12 (50%) | 8/12 (67%) | NS |
| ECOG performance status | |||||
| <2 | 10/35 (29%)* | 14/35 (40%) | 13/35 (37%) | 21/35 (60%) | |
| ≥2 | 2/16 (12%) | 5/16 (31%) | 7/16 (44%) | 7/16 (45%) | 0.095* |
| IPI score | |||||
| Low/intermediate low | 7/17 (41%)** | 10/17(59%)* | 7/17 (41%) | 12/17 (71%) | 0.034* |
| Intermediate high/high | 5/34 (15%) | 9/34 (26%) | 13/34 (38%) | 16/34 (47%) | 0.076** |
| B symptoms | |||||
| Absent | 4/16 (25%) | 7/16 (44%) | 4/16 (25%) | 8/16 (50%) | |
| Present | 7/33 (21%) | 12/33 (36%) | 15/33 (45%) | 18/33 (54%) | NS |
| BM involvement | |||||
| Absent | 9/32 (28%) | 13/32 (41%) | 13/32 (41%) | 20/32 (62%) | |
| Present | 3/19 (16%) | 6/19 (32%) | 7/19 (37%) | 8/19 (42%) | NS |
| Bulky tumor (≥10 cm) | |||||
| Absent | 9/38 (24%) | 15/38 (39%) | 15/38 (39%) | 21/38 (55%) | |
| Present | 3/11 (27%) | 4/11 (36%) | 4/11 (36%) | 5/11 (45%) | NS |
| Bcl‐2 expression | |||||
| Absent | 6/28 (21%) | 11/28 (39%) | 9/28 (32%) | 14/28 (50%) | |
| Present | 4/18 (22%) | 4/18 (22%) | 8/18 (44%) | 9/18 (50%) | NS |
| Bcl‐6 expression | |||||
| Absent | 1/12 (8%) | 5/12 (42%) | 5/12 (42%) | 7/12 (58%) | |
| Present | 5/23 (22%) | 8/23 (35%) | 6/23 (26%) | 11/23 (48%) | NS |
| CD10 expression | |||||
| Absent | 1/12 (8%) | 4/12 (33%) | 5/12 (42%) | 6/12 (50%) | |
| Present | 3/11 (27%) | 3/11 (27%) | 5/11 (45%) | 7/11 (64%) | NS |
| Ki67 expression | |||||
| Weak/moderate | 3/15 (20%) | 5/15 (33%) | 5/15 (33%) | 7/15 (47%) | |
| High | 6/21 (29%) | 7/21 (33%) | 8/21 (38%) | 12/21 (57%) | NS |
| Response to treatment | |||||
| CR/PR | 10/41 (24%) | 14/41 (34%) | 16/41 (39%) | 21/41 (51%) | |
| NR | 1/8 (12%) | 4/8 (50%) | 3/8 (37%) | 6/8 (75%) | NS |
| Relapse | |||||
| Absent | 11/31 (35%)* | 13/31 (42%) | 10/31 (32%) | 17/31 (55%) | |
| Present | 0/18 (0%) | 5/18 (28%) | 8/18 (44%) | 8/18 (44%) | 0.001* |
| Death outcome within 5 years | |||||
| Absent | 9/27 (33%)* | 11/27 (41%) | 10/27 (37%) | 16/27 (59%) | |
| Present | 2/20 (10%) | 6/20 (30%) | 8/20 (40%) | 9/20 (45%) | 0.086* |
LDH = lactate dehydrogenase; ECOG = Eastern Cooperative Oncology Group; BM = bone marrow; CR = complete remission; PR = partial remission; NR = no response; NS = not significant. * and ** relates significant p values with appropriate gene and examined clinicopathological characteristics. For the cases where methylation status of two genes is related to appropriate clinicopathological characteristic (LDH and IPI score), * relates appropriate p value with p16 gene, and ** with p15 gene.
Clinicopathological features of the group with concomitant p15 and p16 methylation (p15m/p16m)
We found that methylation of p15 gene occurs significantly more frequent with simultaneous methylation of p16 gene than as a single event (67% vs. 33%, p = 0.037). We have also observed a tendency toward accumulation of p15m/p16m with clinicopathological characteristics related to less aggressiveness of the disease, including: female gender (p = 0.140), normal LDH levels (p = 0.100), age less than 60 years (p = 0.119), ECOG performance status < 2 (p = 0.127), low/intermediate low IPI score (p = 0.099). In addition, none of the patients with p15m/p16m had relapsed disease during follow‐up period (p = 0.031).
Survival analysis in the whole group of DLBCL patients
Follow‐up data were available for 46/51 patients and the median follow‐up period was 30.5 months (range 1–111 months). Five‐year survival for the entire group was 61% (95% CI: 45.25–74.42). OS was significantly worse for patients who were in any of the following categories: intermediate high/high IPI risk group (p = 0.033), ECOG performance status ≥ 2 (p < 0.001) B symptoms present (p = 0.027), no Bcl6 expression (p = 0.050), no response to the initial therapy (p < 0.001), and not treated with rituximab (p = 0.031). Among all patients with DLBCL, there was no significant difference in the OS between those with hypermethylated and unmethylated of any examined genes (p > 0.05 in all cases, Figure 2). However, patients with methylated p15 tended to have longer OS in contrast to those with unmethylated p15 gene, though this difference was not statistically significant (p = 0.155, Figure 2A). There was no significant difference in OS between patients with hypermethylated and unmethylated of any examined genes in the different groups according to the IPI or any other group of the patients classified according to the parameters given in the Table 2 .
Figure 2.
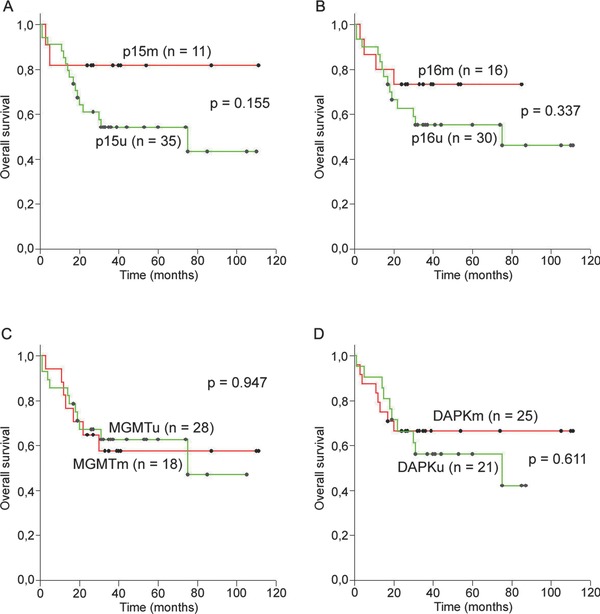
Overall survival (OS) in the entire group of DLBCL patients according to the methylation status of (A) p15, (B) p16, (C) MGMT, and (D) DAPK gene. No significant differences were observed in OS between patients with methylated and unmethylated any of examined genes. The greatest difference in OS is observed in the case of p15 gene, where patients with methylated p15 tended to have longer OS than the patients with unmethylated p15 gene.
Survival analysis in the group of the patients treated with multidrug regimens including rituximab
Considering our finding that DLBCL patients treated with rituximab in addition to the standard chemotherapy had significantly longer OS (p = 0.031, Figure 3A), we further evaluated the impact of the methylation status of four examined genes on survival in this group of the patients. Five‐year survival for the 38 patients who received rituximab was 65% (95% CI: 47.93–79.32). OS was significantly worse for patients with ECOG performance status ≥ 2 (p < 0.001), no Bcl6 expression (p = 0.043), no response to the initial therapy (p < 0.001), and with relapsed disease (p = 0.009). As in the whole group of the patients, we observed a trend toward longer OS for the patients with methylated p15 gene (p = 0.093, Figure 3B). We found no significant difference in OS between patients with hypermethylated and unmethylated p15 gene according to the IPI (results not shown). However, p15 methylation was significantly related to longer OS in the female group of the patients (p = 0.036, Figure 3C) and in those with no bulky tumor mass (p = 0.027, Figure 3D). Moreover, in both cases, all patients with p15 methylation were alive during follow‐up period. No significant impact on OS was seen for the other three genes in the any group of the patients treated with R‐CHOP (results not shown).
Figure 3.
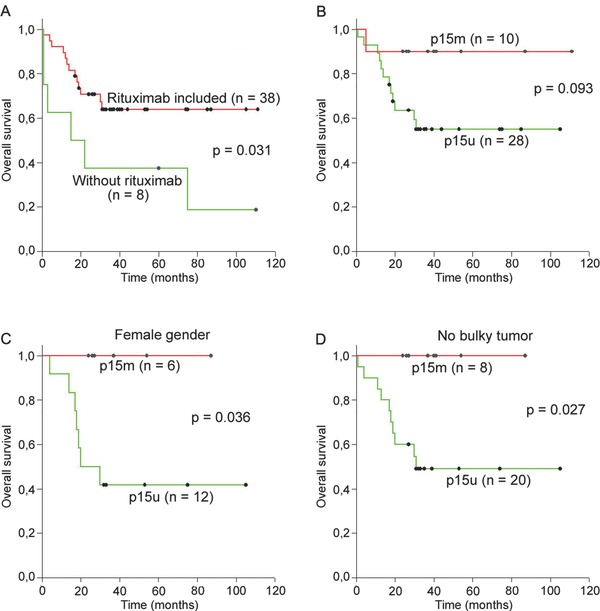
Overall survival (OS) in the DLBCL patients treated with rituximab. (A) OS is significantly longer in patients treated with rituximab in addition to the standard chemotherapy; (B) patients with methylated p15 tended to have longer OS than patients with unmethylated p15 gene; (C) female patients with methylated p15 had significantly longer OS than those with unmethylated p15 gene; (D) patients with no bulky tumor mass with methylated p15 had significantly longer OS than those with unmethylated p15 gene.
Discussion
Prognostic utility of epigenetic changes in malignant lymphoma, especially in the era of new therapy regimens, is still not clearly determined. In the present study, we investigated the biological significance and the prognostic relevance of the methylation status of four cancer‐related genes in a series of 51 DLBCL, most of them treated with rituximab in addition to standard chemotherapy. Aberrant methylation of p15, p16, MGMT, and DAPK genes was detected in 23%, 37%, 39%, and 55% of patients, respectively. Thirty‐eight of 51 patients (75%) had methylation of more than one examined genes. Our findings indicate that epigenetic alterations are common phenomenon in DLBCL and may be important for pathogenesis of this group of NHL.
Tumor suppressor genes p15 and p16 are the members of cyclin‐dependent kinase inhibitors and are frequently inactivated by aberrant hypermethylation of promoter CpG islands in various hematological tumors.23, 24 In DLBCL, methylation of p15 and p16 genes has been found in 32–77% and 27–54% of cases, respectively10, 13, 14, 24, 25, 26 and the results obtained in our study are similar to those in previous reports. However, prognostic significance of p15 and p16 methylation in DLBCL is unclear. While some studies have found p16 methylation as a marker of worse prognosis13, 14, 27 some others have not.26, 28 In our study, methylation of p16 gene was not related with patients outcome. However, we demonstrated a significant correlation between p16 hypermethylation and normal level of LDH and low/intermediate low IPI score. These two parameters have been related to better prognosis in many studies,2, 13, 14, 29 which is confirmed for IPI in ours. To our knowledge, we are the first to detect such associations, so the importance of our results should be confirmed in the further studies on the larger number of samples.
While there are reports about impact of p15 promoter methylation on patient's prognosis in various cancer types,30, 31, 32 such influence in DLBCL, to our knowledge, was not found till now.13, 14, 26 In the present study, we observed a significant correlation between p15 hypermethylation and longer OS in the female patients and those with no bulky tumor mass, treated with rituximab in addition to standard chemotherapy. In the same time, we observed a trend toward longer OS in the whole group of the patients under given treatment. The precise mechanism underlying such favorable impact is unknown. Of notice, the prevalence of p15 methylation was twice as higher in female than in male patients from our study, though this difference was not statistically significant (32% vs. 15%, Table 2). There are some reports about gender‐associated differences in DNA methylation at specific loci, but the results are controversial.33 In addition, male gender was recently reported as an adverse prognostic factor in DLBCL patients treated with R‐CHOP. The possible explanation for observed differences among genders in our study could be the difference in blood clearance of rituximab between males and females, which cause the better response of women to rituximab.34 Though not significant, a tendency toward accumulation of p15 and simultaneous p15/p16 methylation with clinicopathological characteristics related to better prognosis is observed in the whole group of the patients. So, we suggest that p16 and especially p15 methylation could be related to better prognosis in DLBCL, though the precise mechanism is unknown. Considering that methylation of both, p15 and p16, genes is more prevalent in lower IPI patients from our study, it is possible that their inactivation in higher IPI DLBCLs is achieved by some other mechanisms, including deletions. As described earlier, homozygous deletions of p15 and p16 are present in approximately one‐third of the DLBCL, predominantly in activated B‐cell subtype of DLBCL (ABC), where they are related to a poor prognosis.27, 35, 36, 37 On the contrary, methylation of these two genes is predominant mechanism of their inactivation in germinal center B‐cell subtype of DLBCL (GCB),37 which has better prognosis and distinct epigenetic and genetic signature than ABC‐DLBCL.4, 5 Moreover, Guney et al.37 have found that inactivation of p15 and p16 genes in GBC subtype is monoallelic, while in ABC subtype is biallelic. In addition, they demonstrated higher methylation levels of p15 and p16 promoters in the DLBCL cases with simultaneous heterozygous deletions of the second allel, compared to the methylation level of undeleted cases. Considering these results, we suppose that the expression of p15 and/or p16 in such types of DLBCL is not completely suppressed. It could be supported by finding of Cameron et al.,38 who showed that the extent of methylation is a critical determinant of the degree to which p15 is silenced in primary acute leukemia. It is possible that DLBCL patients with p15 and/or p16 methylation from our study belong to the GCB subtype, so it could be, at least, partially explanation for our results. Also, it is possible that in more aggressive DLBCL some other genetic and/or epigenetic alterations exist that are related to more aggressive behavior of the disease. Previous studies have shown that p16 methylation is related to more aggressive phenotype only when simultaneous inactivation of p53, p14, and p27 genes is present.25, 39 However, as we did not have enough data to distinguish our DLBCL samples on GBC and ABC subtypes, further investigations are needed to confirm our hypothesis and completely elucidate the role of p15 and p16 hypermethylation in DLBCL.
MGMT is a well‐known DNA repair gene, important for protecting cells from mutagenic and cytotoxic adducts originated from the environmental and therapeutic alkylating agents.15 Inactivation of MGMT due to promoter hypermethylation occurs at varying frequencies throughout the entire spectrum of B‐cell neoplasms.11, 40 We detected MGMT methylation in 39% of our cases, which is in line with observed frequencies in other studies on DLBCL.11, 21, 41 Previous studies on DLBCL demonstrated that MGMT methylation could be a useful marker for predicting survival in patients treated with multidrug regimens, including the alkylation agent cyclophosphamide.12 However, these results were obtained before the introduction of rituximab to the chemotherapy. Recent studies on DLBCL patients who received R‐CHOP showed that inactivation of MGMT gene does not play a role as a predictive marker of response to this treatment and in predicting survival. It is postulated that rituximab may play an important role in overcoming chemoresistance to cyclophosphamide, especially in the MGMT unmethylated group.42 Our results are in concordance with this finding, as we observed no difference in the response to the therapy or OS according to the methylation status of MGMT gene.
Inactivation of DAPK gene by aberrant promoter hypermethylation is common event in B‐cell malignancies, including DLBCL, where it is occurring in approximately 60% of cases.11, 14, 22, 41, 43 Our results are similar with those in previous reports. DAPK is a pro‐apoptotic serine/threonine kinase, involved in multiple apoptosis pathways, so its inactivation could be a key factor modulating the response to chemotherapy in human cancer.16, 44 While Amara et al.14 have found that DAPK hypermethylation is an independent prognostic factor in predicting shortened OS of DLBCL patients, Nakamichi et al.45 have found no impact of DAPK methylation on patients’ outcome. The latter finding is in concordance with our results, so further analyses are necessary to elucidate the prognostic utility of DAPK in DLBCL.
Conclusion
Although obtained results need to be confirmed in larger series, our study suggests that methylation of p15 gene could have prognostic value in DLBCL patients treated with rituximab when used in combination with gender and tumor size. It is possible that p15 hypermethylation is not a prognostic marker by itself, but identifies a specific pathogenetic subset of lymphomas with a more favorable outcome. However, further investigations are needed to confirm our hypothesis and clarify the role of p15 gene methylation in the pathogenesis of DLBCL.
Acknowledgments
This work was supported by the grant 173049 from the Ministry of Education, Science and Technological Development of the Republic of Serbia.
References
- 1. Gatter KC, Warnke RA. Diffuse large B‐cell lymphoma In: Jaffe E, Harris NL, Stein H, Vardiman JW, eds. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietics and Lymphoid Tissues IARC Press, Lyon, 2001, pp, 171–174. [Google Scholar]
- 2. The International Non‐Hodgkin's Lymphoma Prognostic Factors Project . A predictive model for aggressive non‐Hodgkin's lymphoma. N Engl J Med. 1993; 329: 987–994. [DOI] [PubMed] [Google Scholar]
- 3. Armitage JO, Weisenburger DD. New approach to classifying non‐Hodgkin's lymphomas: clinical features of the major histologic subtypes. Non‐Hodgkin's Lymphoma Classification Project. J Clin Oncol. 1998; 16: 2780–2795. [DOI] [PubMed] [Google Scholar]
- 4. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B‐cell lymphoma identified by gene expression profiling. Nature. 2000; 403: 503–511. [DOI] [PubMed] [Google Scholar]
- 5. Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller‐Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large B‐cell lymphoma. N Engl J Med. 2002; 346: 1937–1947. [DOI] [PubMed] [Google Scholar]
- 6. Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005; 87: 163–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sehn LH, Berry B, Chhanabhai M, Fitzgerald C, Gill K, Hoskins P, Klasa R, Savage KJ, Shenkier T, Sutherland J, et al. The revised International Prognostic Index (R‐IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B‐cell lymphomatreated with R‐CHOP. Blood. 2007; 109: 1857–1861. [DOI] [PubMed] [Google Scholar]
- 8. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003; 349: 2042–2054. [DOI] [PubMed] [Google Scholar]
- 9. Martinez‐Delgado B, Robledo M, Arranz E, Osorio A, Garcia MJ, Echezarreta G, Rivas C, Benitez J. Hypermethylation of p15/ink4b/MTS2 gene is differentially implicated among non‐Hodgkin's lymphoma. Leukemia. 1998; 12: 937–941. [DOI] [PubMed] [Google Scholar]
- 10. Baur AS, Shaw P, Burri N, Delacrétaz F, Bosman FT, Chaubert P. Frequent methylation silencing of p15 INK4b (MTS2) and p16 INK4a (MTS1) in B‐cell and T‐cell lymphomas. Blood. 1999; 94: 1773–1781. [PubMed] [Google Scholar]
- 11. Rossi D, Capello D, Gloghini A, Franceschetti S, Paulli M, Bhatia K, Saglio G, Vitolo U, Pileri SA, Esteller M, et al. Aberrant promoter methylation of multiple genes throughout the clinico‐pathologic spectrum of B‐cell neoplasia. Haematologica. 2004; 89: 154–164. [PubMed] [Google Scholar]
- 12. Esteller M, Gaidano G, Goodman SN, Zagonel V, Capello D, Botto B, Rossi D, Gloghini A, Vitolo U, Carbone A, et al. Hypermethylation of the DNA repair gene O6‐methylguanine DNA methyltransferase and survival of patients with diffuse large B‐cell lymphoma. J Natl Cancer Inst. 2002; 94: 26–32. [DOI] [PubMed] [Google Scholar]
- 13. Shiozawa E, Takimoto M, Makino R, Adachi D, Saito B, Yamochi‐Oniyuka T, Yamochi T, Shimozuma J, Maeda T, Kohno Y, et al. Hypermethylation of CpG islands in p16 as a prognostic factor for diffuse large B‐cell lymphoma in a high‐risk group. Leuk Res. 2006; 30: 859–867. [DOI] [PubMed] [Google Scholar]
- 14. Amara K, Trimeche M, Ziadi S, Laatiri A, Hachana M, Korbi S. Prognostic significance of aberrant promoter hypermethylation of CpG islands in patients with diffuse large B‐cell lymphomas. Ann Oncol. 2008; 19: 1774–1786. [DOI] [PubMed] [Google Scholar]
- 15. Pegg AE. Repair of O(6)‐alkylguanine by alkyltransferases. Mutat Res. 2000; 462: 83–100. [DOI] [PubMed] [Google Scholar]
- 16. Ng MH. Death associated protein kinase: from regulation of apoptosis to tumor suppresive function and B cell malignancies. Apoptosis. 2002; 7: 261–270. [DOI] [PubMed] [Google Scholar]
- 17. Cheson BD, Horning SJ, Coiffier B, Shipp MA, Fisher RI, Connors JM, Lister TA, Vose J, Grillo‐Lopez A, Hagenbeek A, et al. Report of an International Workshop to standardize response criteria for non‐Hodgkin's lymphomas. J Clin Oncol. 1999; 17: 1244–1253. [DOI] [PubMed] [Google Scholar]
- 18. Krtolica K, Krajnović M, Ušaj‐Knežević S, Babić D, Jovanović D, Dimitrijević B. Comethylation of p16 and MGMT genes in colorectal carcinoma: correlation with clinicopathological features and prognostic value. World J Gastroenterol. 2007; 13: 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sambrook J, Fritsch EF, Maniatis T. Analysis and cloning of eukariotic genomic DNA. In: Ford N, Nolan C, Ferguson M, eds. Molecular Cloning. A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989, pp, 16–19. [Google Scholar]
- 20. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation‐specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996; 93: 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6‐methylguanine‐DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999; 59: 793–797. [PubMed] [Google Scholar]
- 22. Katzenellenbogen RA, Baylin SB, Herman JG. Hypermethylation of the DAP‐kinase CpG island is a common alteration in B‐cell malignancies. Blood. 1999; 93: 4347–4353. [PubMed] [Google Scholar]
- 23. Drexler HG. Review of alterations of the cyclin‐dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia‐lymphoma cells. Leukemia. 1998; 12: 845–859. [DOI] [PubMed] [Google Scholar]
- 24. Garcia MJ, Martinez‐Delgado B, Cebrian A, Martinez A, Benitez J, Rivas C. Different incidence and pattern of p15INK4b and p16INK4a promoter region hypermethylation in Hodgkin's lymphoma and CD30‐positive non‐Hodgkin lymphomas. Am J Pathol. 2002; 161: 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanchez‐Beato M, Saez AI, Navas IC, Algara P, Sol Mateo M, Villuendas R, Camacho F, Sanchez‐Aguilera A, Sanchez E, Piris MA. Overall survival in aggressive B‐cell lymphomas is dependent on the accumulation of alterations in p53, p16, and p27. Am J Pathol. 2001; 159: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee SM, Lee EJ, Ko YH, Lee SH, Maeng L, Kim KM. Prognostic significance of O6‐methylguanine DNA methyltransferase and p57 methylation in patients with diffuse large B‐cell lymphoma. APMIS. 2009; 117: 87–94. [DOI] [PubMed] [Google Scholar]
- 27. Jardin F, Jais JP, Molina TJ, Parmentier F, Picquenot JM, Ruminy P, Tilly H, Bastard C, Salles GA, Feugier P, et al. Diffuse large B‐cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R‐CHOP treatment: a GELA study. Blood. 2010; 116: 1092–1104. [DOI] [PubMed] [Google Scholar]
- 28. Zainuddin N, Kanduri M, Berglund M, Lindell M, Amini RM, Roos G, Sundstrom C, Enblad G, Rosenquist R. Quantitative evaluation of p16INK4a promoter methylation using pyrosequencing in de novo diffuse large B‐cell lymphoma. Leuk Res. 2011; 35: 438–443. [DOI] [PubMed] [Google Scholar]
- 29. Perunicˇić Jovanović M, Jaković Lj, Bogdanović A, Marković O, Cˇemerikić V, Mihaljevićć B. Poor outcome in patients with diffuse large B‐cell lymphoma is associated with high percentage of bcl‐2 and Ki 67‐positive tumor cells. Vojnosanit Pregl. 2009; 66: 738–743. [DOI] [PubMed] [Google Scholar]
- 30. Wemmert S, Bettscheider M, Alt S, Ketter R, Kammers K, Feiden W, Steudel WI, Rahnenführer J, Urbschat S. p15 promoter methylation – a novel prognostic marker in glioblastoma patients. Int Journal Oncol. 2009; 34: 1743–1748. [DOI] [PubMed] [Google Scholar]
- 31. Deneberg S, Grovdal M, Karimi M, Jansson M, Nahi H, Corbaciogly A, Gaidzik V, Dohner K, Paul C, Ekstrom TJ, et al. Gene‐specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia. 2010; 24: 932–941. [DOI] [PubMed] [Google Scholar]
- 32. Kim M, Oh B, Kim SY, Park HK, Hwang SM, Kim TY, She CJ, Yang I, Yoon SS, Yoon JH, et al. p15INK4b methylation correlates with thrombocytopenia, blast percentage, and survival in myelodysplastic syndrome in a dose dependent manner: quantitation using pyrosequencing study. Leuk Res. 2010; 34: 718–722. [DOI] [PubMed] [Google Scholar]
- 33. El‐Maarri O, Becker T, Junen J, Manzoor SS, Diaz‐Lacava A, Schwaab R, Wienker T, Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007; 122: 505–514. [DOI] [PubMed] [Google Scholar]
- 34. Muller C, Murawski N, Wiesen MHJ, Held G, Poeschel V, Zeynalova S, Wenger M, Nickenig C, Peter N, Lengfelder E, et al. The role of gender and weight on rituximab clearance and serum elimination half life in elderly patients with DLBCL. Blood. 2012; 119: 3276–3284. [DOI] [PubMed] [Google Scholar]
- 35. Tagawa H, Suguro M, Tsuzuki S, Matsuo K, Karnan S, Ohshima K, Okamoto M, Morishima Y, Nakamura S, Seto M. Comparison of genome profiles for identification of distinct subgroups of diffuse large B‐cell lymphoma. Blood. 2005; 106: 1770–1777. [DOI] [PubMed] [Google Scholar]
- 36. Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. Molecular subtypes of diffuse large B‐cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA. 2008; 105: 13520–13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guney S, Jardin F, Bertrand P, Mareschal S, Parmentier F, Picquenot JM, Tilly H, Bastard C. Several mechanisms lead to the inactivation of the CDKN2A (p16), p14ARF, or CDKN2B (p15) genes in the GCB and ABC molecular DLBCL subtypes. Genes Chromosomes Cancer. 2012; 51: 858–867. [DOI] [PubMed] [Google Scholar]
- 38. Cameron EE, Baylin BS, Herman JG. p15 (INK4B) CpG island methylation in primary acute leukemia is heterogenous and suggests density as a critical factor for trancriptional silencing. Blood. 1999; 94: 2445–2451. [PubMed] [Google Scholar]
- 39. Gronbaek K, de Nully Brown P, Moller MB, Nedergaard T, Ralfkiaer E, Moller P, Zeuthen J, Guldberg P. Concurrent disruption of p16INK4a and the ARF‐p53 pathway predicts poor prognosis in aggressive non‐Hodgkin's lymphoma. Leukemia. 2000; 14: 1727–1735. [DOI] [PubMed] [Google Scholar]
- 40. Kraguljac N, Krajnović M, Dimitrijević B, Mihaljević B, Gotić M, Krtolica K. Frequency of aberrant promoter methylation of p15INK4B and O6‐methylguanine‐DNA methyltransferase genes in B‐cell non‐Hodgkin lymphoma: a pilot study. Arch Biol Sci. 2010; 62: 211–221. [Google Scholar]
- 41. Kim SS, Choi YH, Han CW, Choi YD, Park Y, Lee JJ, Kim HJ, Lee IK, Lee JS, Juhng SW, et al. DNA methylation profiles of MGMT, DAPK1, bhMLH1, CDH1, SHP1, and HIC1 in B‐cell lymphomas. Korean J Pathol. 2009; 43: 420–427. [Google Scholar]
- 42. Lee GW, Kang JH, Kim IS, Kim HG, Ko GH, Lee JH, Kim DC, Song DH, Yang JW, Lee JS. Is inactivation of O6‐methylguanine DNA methyltransferase still a favorable prognostic factor of patients with diffuse large B‐cell lymphoma in the era of R‐CHOP chemotherapy? Leuk Lymphoma. 2009; 50: 1992–1998. [DOI] [PubMed] [Google Scholar]
- 43. Voso MT, Gumiero D, D'Alo' F, Guidi F, Mansueto G, Di Febo AL, Massini G, Martini M, Larocca LM, Hohaus S, et al. DAP‐kinase hypermethylation in the bone marrow of patients with follicular lymphoma. Haematologica. 2006; 91: 1252–1256. [PubMed] [Google Scholar]
- 44. Cohen O, Kimchi A. DAP‐kinase: from functional gene cloning to establishment of its role in apoptosis and cancer. Cell Death Differ. 2001; 8: 6–15. [DOI] [PubMed] [Google Scholar]
- 45. Nakamichi I, Tomita Y, Zhang B, Sugiyama H, Kanakura Y, Fukuhara S, Hino M, Knamaru A, Ogawa H, Aozasa K. Correlation between promoter hypermethylation of GSTP1 and response to chemotherapy in diffuse large B cell lymphoma. Ann Hematol. 2007; 86: 557–564. [DOI] [PubMed] [Google Scholar]


