Abstract
Several variants within gene‐encoding endothelial isoform of nitric oxide synthase have been reported to confer prostate cancer (PCa) susceptibility and/or progression. Nevertheless, studies referring to this issue have yielded inconsistent results. In order to elucidate the involvement of these variants in prostate carcinogenesis, we have conducted a meta‐analysis of previously published case‐control and relevant case‐only studies. Eleven studies comprising in total 3,806 cases and 4,466 controls were included in the meta‐analysis which yielded evidence of association of rs2070744 (ORCC = 1.43, 95% CI 1.04–1.97; p = 0.03) and intron 4a/b variant (ORab+aa = 1.47, 95% CI 1.00–2.14; p = 0.05) with PCa risk under recessive and dominant model, respectively. Furthermore, PCa patients carrying 4a/b a allele were found to have an increased risk of cancer progression to a less differentiated form, characterized by a high Gleason score (OR = 2.29, 95% CI 1.51–3.49; p < 0.01) and to higher TNM stage (OR = 2.55, 95% CI 1.71–3.81; p < 0.01). These results support the involvement of NOS3 variants in molecular pathogenesis of PCa.
Keywords: prostate cancer, NOS3, rs1799983, rs2070744, 4a/b, meta‐analysis
Introduction
According to estimates provided by GLOBOCAN project, the number of new cancer cases has increased from about 12.7 million in 2008 to more than 14 million that have occurred in 2012.1, 2 Therefore, as the second most frequently diagnosed cancer among men, prostate cancer (PCa) was recognized as one of the major health issues worldwide. The implementation of screening programs, improvements in treatment efficiency, and actions directed toward raising public awareness of the importance of regular testing have resulted in a downward trend in mortality rates of PCa, mainly in developed countries.3, 4, 5 Nevertheless, this malignancy remained the sixth leading cause of cancer‐related death among men worldwide.1
The striking statistics have led to a large number of studies in PCa epidemiology, pathology, molecular basis of pathogenesis, genetics and, more recently, epigenetics. Among the analyses aiming to identify genetic factors contributing to PCa susceptibility, the major contribution was made by genetic association studies.6 Even though the variants that have been recognized as the most important in determining PCa predisposition had been identified in large genome‐wide association studies,6 several important regions were derived from studies based on candidate genes.7, 8 The findings of these studies have also provided evidence of association between various genetic markers and the parameters of PCa progression, mortality, survival, and recurrence.9, 10, 11
Genes involved in angiogenesis have emerged as suitable candidate genes, based on the importance of this process in tumor biology.12 Variants affecting their function were hypothesized to affect prostate carcinogenesis, due to promotion of tumor growth by vascularization.13 Furthermore, for genes encoding the enzymes that produce nitric oxide (NO), another mechanism of potential involvement in cancer development and progression was proposed, which includes the effect on apoptotic cell death.14 When considering the stimulatory role of NO on this cellular process, but also its influence on promoting angiogenic phenotype, it is obvious that the production of this diffusive messenger could have two opposing roles in malignant growth of prostate cells. Furthermore, excessive NO production can result in formation of reactive nitrogen species with significant DNA damaging potential.15 Another procancerous property of this molecule is the promotion of inflammatory status, since it is considered to be one of the key inflammatory mediators.16
The NO synthases (NOS) family comprises endothelial, inducible, neuronal, and mitochrondrial NOS. The endothelial isoform of NOS (eNOS), encoded by NOS3 gene, produces low amounts of NO, but it seems that, even when synthesized at low levels, this molecule can promote oncogenesis.16 Thus, NOS3 was considered as potential candidate gene in genetic association studies of PCa. These studies focused mainly on the effect of variants located in the coding region, introns, and the promoter of NOS3 on PCa susceptibility and the association of these variants with standard prognostic parameters of PCa progression. Two most commonly analyzed single nucleotide variants are rs1799983 located in the seventh exon and rs2070744 in the promoter of this gene.17, 18, 19, 20, 21, 22, 23, 24 Furthermore, an the insertion–deletion variant 4a/b in an intron of this gene was evaluated as potential PCa‐associated genetic marker.17, 23, 25, 26
These studies have yielded inconsistent results, possibly due to genetic differences between populations of subjects. Also, these differences could reflect the discordances in methodologies used for genotyping, control group selection and recruitment of subjects, matching of controls with PCa patients, as well as in adjustments for possible confounders. Furthermore, since most of these studies had small sample size, it is possible that they could be underpowered to detect variants with relatively small effect. Therefore, in order to elucidate the effect of variants located in NOS3 gene on PCa risk and progression, we have conducted the meta‐analysis of eligible studies. Combining the data from small single studies could result in increased statistical power, thus providing more precise estimations.
Material and Methods
The literature included in this meta‐analysis was selected from PubMed database using the search strategy based on combinations of keywords “SNP” or “variant” or “polymorphism,” “NOS3” or “endothelial NOS,” “association” and “prostate cancer” without language restriction. References cited in retrieved original studies, as well as in review articles, were examined for additional studies suitable for inclusion in this meta‐analysis. Selected articles were published before January 2014.
Eligible studies met the following criteria: (1) analysis of association between variants located in NOS3 gene (including promoter region) and PCa risk and/or prognostic parameters; (2) case‐control or case‐only study design; (3) provided ORs with 95% CIs and p values, or sufficient data about genotype and allele frequencies to calculate risk estimates; (4) the presence of Hardy‐Weinberg equilibrium in control group (for case‐control studies); and (5) provided detailed information about diagnostic protocols, together with criteria for assessment of clinical and pathological characteristics of PCa patients. For our previous study,24 row data were available.
The data extracted from selected studies included: first author's last name, year of publication, country from which participants were recruited, ethnicity, source of controls, methods and study design, sample size, genotype and allele counts or published risk estimates (OR with 95% CIs and p values). Selected studies were classified according to analyzed variants and meta‐analysis was performed if three or more studies corresponded to single variant. Furthermore, stratified analyses were performed according to source of control groups. The criteria used for segregation of PCa patients in the meta‐analyses of association between genetic variants and PCa progression were selected based on their usage in the largest number of included studies. Therefore, two groups of PCa patients were formed according to their Gleason score (GS <7 and GS ≥7) and tumor stage (low stage defined as TNM T1‐T2 without metastases and high stage defined as T3‐T4 or the presence of metastases).
We have used statistical software Open Meta‐Analyst27 for meta‐analysis and heterogeneity tests. Estimates of ORs and its 95% CIs were calculated for each genetic variant using fixed‐effect or random‐effect model based on the results of heterogeneity tests. For assessing heterogeneity of results across studies, Cochran's Q test was used, combined with inconsistency index (I2). Heterogeneity was considered significant at p < 0.1, while I2 = 0–25% suggested no heterogeneity, I2 = 25–50%, moderate heterogeneity, I2 = 50–75%, large heterogeneity and I2 = 75–100% extreme heterogeneity. Random‐effect model was selected for meta‐analysis when heterogeneity tests yielded significant results. For the fixed‐effect model, the inverse variance method of weighting was used, while for pooling results under the random‐effect model, the method proposed by DerSimonian and Laird was applied.28 Since fewer than 10 studies were included in each meta‐analysis, publication bias was not assessed.
Results
The flow chart of study selection for this meta‐analysis is presented in Figure 1 . A total of 30 records were identified through database searching and by reviewing reference lists. After removing overlapping studies, together with one study not relevant to the topic, 12 remaining full‐text articles were assessed for eligibility. One of them was excluded for not meeting the inclusion criteria, since it is a nested case‐control study involving only patients with recurrent PCa.29 Finally, 11 studies, of which 9 were case‐control and 2 were case‐only, comprising 3,806 cases and 4,466 controls were included in the meta‐analysis (Table 1 ). For studies that included multiple control groups, data were extracted separately for each case‐control group comparison. The distributions of genotypes in the controls were consistent with HWE.
Figure 1.
Flow chart of the study selection process.
Table 1.
Characteristics of the studies included in the meta‐analysis
| Authors | Year | Ethnicity | Study design | Type of control group | Variant | Case/control | HWE p value | Ref. |
|---|---|---|---|---|---|---|---|---|
| Medeiros et al. | 2002 | Caucasians | Case‐control | Healthy controls | rs1799983 | 125/153 | 0.63 | 17 |
| 4a/b | 0.42 | |||||||
| Medeiros et al. | 2002 | Caucasians | Case‐only | – | rs1799983 | 161/‐ | – | 18 |
| Medeiros et al. | 2003 | Caucasians | Case‐only | – | 4a/b | 61/‐ | – | 25 |
| Marangoni et al. | 2006 | Caucasians | Case‐control | BPH patients | rs1799983 | 84/65 | 0.79 | 19 |
| Jacobs et al. | 2008 | Approximately 97% Caucasians | Case‐control | Noncancer | rs1799983 | 1,425/1,453 | 0.07 | 13 |
| Marangoni et al. | 2008 | Caucasians | Case‐control | BPH patients | rs2070744 | 83/94 | 0.71 | 20 |
| Lee et al. | 2009 | Non‐Hispanic Caucasians | Case‐control | Non‐PCa | rs1799983 | 1,213/1,433 | 0.94 | 21 |
| African Americans | Case‐control | Non‐PCa | rs1799983 | 107/409 | 0.51 | |||
| Sanli et al. | 2011 | Caucasians | Case‐control | Healthy controls | 4a/b | 132/158 | 0.88 | 26 |
| Ziaei et al. | 2013 | Caucasians | Case‐control | BPH patients | rs1799983 | 95/111 | 0.92 | 22 |
| Safarinejad et al. | 2013 | Caucasians | Case‐control | Healthy controls | rs1799983 | 170/340 | 0.10 | 23 |
| rs2070744 | 0.22 | |||||||
| 4a/b | 0.11 | |||||||
| Branković et al. | 2013 | Caucasians | Case‐control | Healthy controls | rs1799983 | 150/100 | 0.69 | 24 |
| rs2070744 | 0.56 | |||||||
| BPH patients | rs1799983 | 150/150 | 0.70 | |||||
| rs2070744 | 0.66 |
For genetic variant rs1799983, no evidence of association between minor allele T and PCa risk was found (OR = 0.99, 95% CI 0.92–1.06; p = 0.77, P heterogeneity = 0.90). These results did not change even when performing separate subgroup analyses for studies that included healthy controls and those including control group selected from BPH patients (Figure 2 ). Furthermore, neither carriers of GT, nor TT genotype were found to have an altered risk of developing PCa compared to men homozygous for major allele G (ORGT = 1.06, 95% CI 0.96–1.17; ORTT = 0.97, 95% CI 0.82–1.15). Similarly, no significant association with the risk of PCa was found when assuming dominant model of inheritance (OR = 1.03, 95% CI 0.94–1.13), while tests under recessive model were not conducted due to a low number of minor allele homozygotes. In the analysis aiming to evaluate the association of rs1799983 alleles and genotypes with PCa progression, no statistically significant effects were observed (Figure S1).
Figure 2.
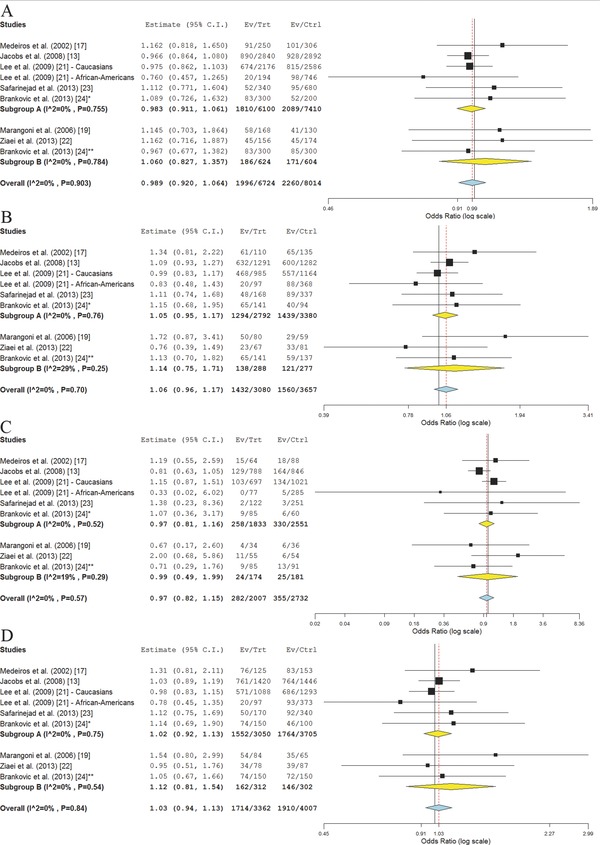
Meta‐analysis of the association between rs1799983 alleles and genotypes and PCa risk across nine study panels from seven studies. (A) rs1799983 T versus G allele; (B) rs1799983 GT versus GG genotype; (C) rs1799983 TT versus GG genotype; (D) rs1799983 GT+TT versus GG genotype. Subgroup A = studies that included healthy controls; Subgroup B = studies that included BPH patients as controls. *Healthy controls; †BPH patients as controls.
This meta‐analysis did not suggest significant association of rs2070744 minor allele C with PCa risk (OR = 1.18, 95% CI 0.95–1.47; p = 0.14, P heterogeneity = 0.18). Individuals carrying TC genotype were not found to have an altered risk of PCa compared to carriers of TT genotype (ORTC = 1.06, 95% CI 0.72–1.56), while for the rs2070744 TT genotype the marginally significant association with the 1.48‐fold increased risk of developing PCa was found (95% CI 1.00–2.20; p = 0.05, P heterogeneity = 0.30). When assuming dominant genetic model, no evidence of association between rs2070744 and PCa risk was obtained (OR = 1.14, 95% CI 0.78–1.66; p = 0.5, P heterogeneity = 0.07). Under recessive model, tests have shown that CC genotype confer 1.43‐fold increased risk of developing PCa, compared to combined TT and TC genotypes (95% CI 1.04–1.97; p = 0.03, P heterogeneity = 0.80; Figure 3 ). For this variant, association with parameters of PCa progression was not assessed, since relevant data from only two studies were available.
Figure 3.
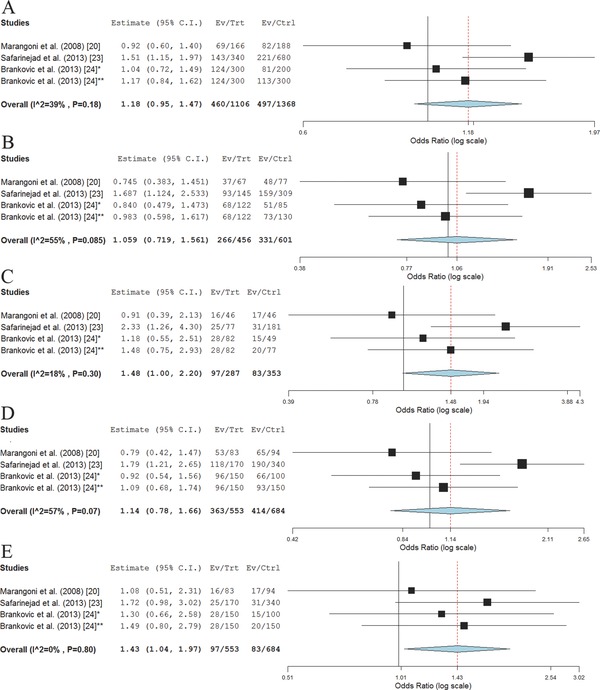
Meta‐analysis of the association between rs2070744 alleles and genotypes and PCa risk across four study panels from three studies. (A) rs2070744 C versus T allele; (B) rs2070744 TC versus TT genotype; (C) rs2070744 CC versus TT genotype; (D) rs2070744 TC+CC versus TT genotype; (E) rs2070744 CC versus TT+TC genotype. *Healthy controls; †BPH patients as controls.
The allelic analysis has shown statistical trend of significance (0.05 < p < 0.1) for associations between the NOS3 intron 4 a/b variant and the risk of developing PCa (ORa = 1.51, 95% CI 0.98–2.35; p = 0.06, P heterogeneity = 0.03). Furthermore, ab genotype was found to be associated with the increased PCa risk (OR = 1.34, 95% CI 1.01–1.77; p = 0.04, P heterogeneity = 0.39), while for the minor allele a homozygote the supposed association was not found to be statistically significant (OR = 3.23, 95% CI 0.73–14.37; p = 0.12, P heterogeneity = 0.02). Similar findings were obtained when assuming dominant genetic model (OR = 1.47, 95% CI 1.00–2.14; p = 0.05, P heterogeneity = 0.14; Figure 4 ). The association under recessive model was not evaluated due to a small number of aa homozygotes in selected studies. Furthermore, PCa patients carrying 4a/b a allele were found to have an increased risk of cancer progression to a less differentiated form characterized with higher Gleason score, when assuming dominant genetic model (OR = 2.29, 95% CI 1.51–3.49; p < 0.01, P heterogeneity = 0.86). Also, this allele was shown to confer a 2.55‐fold increased risk of PCa progression to a higher TNM stage (95% CI 1.71–3.81; p < 0.01, P heterogeneity = 0.51; Figure 5 ).
Figure 4.
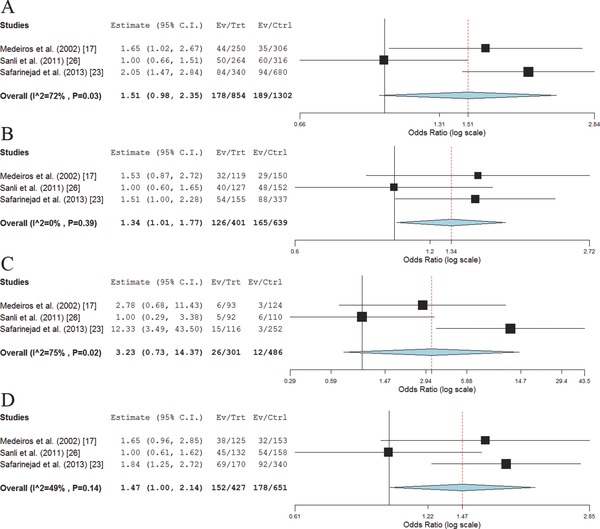
Meta‐analysis of the association between NOS3 intron 4 a/b alleles and genotypes and PCa risk across three studies. (A) 4a/b a versus b allele; (B) 4a/b ab versus bb genotype; (C) 4a/b aa versus bb genotype; (D) 4a/b ab+aa versus bb genotype.
Figure 5.
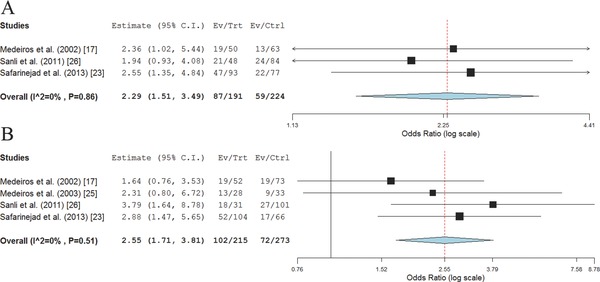
Meta‐analysis of the association between NOS3 intron 4 a/b variant and PCa progression under dominant model. (A) Association with Gleason score; (B) Association with PCa TNM stage.
Discussion
Cancer‐promoting properties of NO and the aberrant expression and/or function of eNOS have been assessed in multiple types of malignant diseases, including PCa.30, 31, 32, 33 Results obtained in these studies suggest involvement of pathogenic levels of NO in cancerous cell proliferation, inhibition of apoptosis, angiogenesis, invasiveness, and metastasis.30, 31 Therefore, genetic variants affecting the function of enzymatic pathways involved in synthesis of NO could potentially contribute to PCa susceptibility, and also stimulate or suppress cancer progression. Based on the accumulating data supporting the involvement of NOS3 in molecular pathogenesis of PCa, variants within gene encoding this enzyme emerged as plausible candidates for involvement in prostate cancerogenesis.25, 32, 33, 34, 35, 36
A variant located in the seventh exon of NOS3 (rs1799983), which causes an amino acid substitution within N‐terminal oxygenase domain of eNOS enzyme (G298E), was previously found to be associated with several pathological entities, such as cardiovascular diseases, including atherosclerosis, essential hypertension, coronary artery disease, and myocardial infarction, but also with male infertility, recurrent miscarriages, and multiple malignant diseases.37, 38, 39, 40, 41, 42, 43, 44, 45 Even though functional analysis in yeast expression system yielded no evidence of altered enzyme function in NOS3 G298E mutants, the replication of association of this variant with several pathological stages in multiple populations clearly qualifies it for a genetic marker of eNOS functionality.46 To date, several studies investigating involvement of this variant in prostate cancerogenesis and/or progression have been published, showing opposing results.13, 17, 18, 19, 21, 22, 23, 24 These inconsistencies could be reflecting differences in ethnic backgrounds of study groups, criteria for recruitment of subjects, classification of patients according to clinical characteristics, but could be also caused by small sample sizes of several studies. In order to clarify the importance of this variant in PCa onset and progression, we have conducted a meta‐analysis of all published case‐control studies referring to this issue, as well as several case‐only studies found to be relevant after assessing their patient classification and recruitment criteria. Our results did not confirm the association of rs1799983 with PCa susceptibility, under various genetic models tested. This variant was also not found to be associated with PCa progression, as assessed by using GS and cancer TNM stage as parameters. Therefore, without further evidence from larger studies conducted in various populations and ethnic groups, including those of African descent, from which only one subpopulation of subjects is derived so far, we cannot consider this variant to be an important genetic factor contributing to PCa development and/or progression to a more aggressive form.
Another potentially functional variant within NOS3 is rs2070744, located in the promoter of this gene. This variant was proposed to cause a decrease in eNOS expression, which promotes cancer onset.20 In a recent meta‐analysis aiming to evaluate the association of this variant with the risk of cancer in general and also with the several types of cancer in subgroup analyses, evidence to support the supposed correlations were obtained.47 Even though this analysis did not show association between rs2070744 and PCa risk, since it did not include a study by Marangoni et al.,20 we decided to perform another assessment. Our results also did not support the association of minor allele with PCa risk in alleleic and dominant model. Conversely, when assuming recessive genetic model, a significant association of CC genotype with PCa susceptibility was shown. Therefore, rs2070744 could affect PCa development, with minor allele homozygote contributing to PCa predisposition. Nevertheless, these results need to be substantiated with evidence from future studies. Also, further studies are needed in order to make conclusions about possible involvement of this variant in PCa progression.
The 4a/b variant in the fourth intron of NOS3 gene was previously found to be associated with development and/or progression of various malignant diseases, including PCa.17, 23, 25, 26, 48, 49, 50, 51 Similarly, as rs2070744, this variant was found to be associated with PCa risk in the current meta‐analysis. The evidence of the supposed association were found when assuming dominant genetic model, and furthermore, when assessing the increase in PCa risk associated with heterozygous genotype, compared to major allele homozygote. The lack of evidence of association between aa homozygote and PCa susceptibility could be explained by a small number of studies referring to this issues, and also by the small size of study groups. This variant was also found to be associated with both GS and the cancer TNM stage, which suggests its involvement in PCa progression. The a allele was found to confer increased risk of developing poorly differentiated PCa, advanced stage of localized cancer, and distant metastases. Therefore, this variant, which was previously shown to affect eNOS promoter activity together with rs2070744,52 could be an important genetic factor in PCa biology, influencing both development and progression of this malignancy.
In order to make further conclusions about the association of NOS3 variants with PCa risk, additional studies in various populations need to be performed. Furthermore, additional variants in NOS3 should be evaluated, beside three of them included in this meta‐analysis. Since minor allele homozygotes of all three variants were found in a small number of participants, larger studies could elucidate the dose‐dependent effect of these variants on PCa risk and progression. Also, with the increase in study numbers, publication bias could be assessed, potentially yielding additional explanations for the observed discordances.
Although this meta‐analysis included a relatively small number of studies, every attempt was made to prevent the potential bias affecting results and to minimize its inherent weakness. Therefore, study heterogeneity tests were performed and the adequate statistical model for meta‐analysis was chosen based on these results. Since heterogeneity was assessed by combining Cochran's Q test, p value, and inconsistency index, the weighting of studies was adjusted to random‐effect model if any of these parameters suggested that included studies were even slightly heterogeneous. The methods of weighting were chosen to be suitable for binary outcomes. Furthermore, for every study potentially suitable for meta‐analysis, an exhaustive review of methodology was performed. This especially refers to recruitment of participants, genotyping methods, as well as ethnic and clinical characteristics of patients involved. By performing subgroup analyses based on control group characteristics, as well as on ethnic backgrounds, we tended to avoid potential bias caused by selection of participants and their demographics. Also, differences in statistical approaches which could lead to discordant results were minimized by extracting row data and submitting them to the uniform tests for assessing ORs and their confidence intervals, which are suitable as effect‐size measures. This meta‐analysis tended not to simply assess the effect of minor alleles on disease susceptibility, but to evaluate the effects of separate genotypes and the potential association under different genetic models that could be misses in allelic test. One of the major limitations of this study is leaving out the results of several studies from disease progression‐based meta‐analysis, since they did not meet the most frequently used criteria for assessment of cancer progression and aggressiveness. Among them are even those with comparably large study groups. Therefore, in addition to analyses of association between these variants and PCa risk, classification of PCa patients according to common, yet clinically and pathologically justifiable criteria should be performed in order to clearly demonstrate the involvement of NOS3 variants in PCa progression, or to reject this hypothesis.
Conclusion
Our study did not confirm the supposed association of rs1799983 with PCa risk and progression. Therefore, we cannot consider this variant to be an important genetic factor contributing to PCa development and/or progression to a more aggressive form. When considering rs2070744, the results show that this variant could affect PCa development, with minor allele homozygote contributing to PCa predisposition. Another variant located in NOS3 gene, 4a/b, was found to be associated with PCa risk in the current meta‐analysis. The 4a/b was also found to be associated with both GS and the cancer TNM stage, which suggests its involvement in PCa progression. Even though this meta‐analysis included a limited number of studies, the obtained results clearly support the involvement of NOS3 variants in molecular pathogenesis of PCa.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Meta‐analysis of the association between rs1799983 and PCa progression under dominant model. (A) association with Gleason score; (B) association with PCa TNM stage.
Acknowledgment
The research was supported by the Ministry of Education, Science and Technological Development of Serbia (Project no. 173016).
References
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010; 127(12): 2893–2917. [DOI] [PubMed] [Google Scholar]
- 2. International Agency for Research on Cancer . GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. [Online]. Available from: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx [Accessed 3rd January 2014].
- 3. Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al.: ERSPC Investigators. Screening and prostate‐cancer mortality in a randomized European study. N Engl J Med. 2009; 360(13): 1320–1328. [DOI] [PubMed] [Google Scholar]
- 4. Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010; 19(8): 1893–1907. [DOI] [PubMed] [Google Scholar]
- 5. Center MM, Jemal A, Lortet‐Tieulent J, Ward E, Ferlay J, Brawley O, Bray F. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012; 61(6): 1079–1092. [DOI] [PubMed] [Google Scholar]
- 6. Xu J, Sun J, Zheng SL. Prostate cancer risk‐associated genetic markers and their potential clinical utility. Asian J Androl. 2013; 15(3): 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burmester JK, Suarez BK, Lin JH, Jin CH, Miller RD, Zhang KQ, Salzman SA, Reding DJ, Catalona WJ. Analysis of candidate genes for prostate cancer. Hum Hered. 2004; 57(4): 172–178. [DOI] [PubMed] [Google Scholar]
- 8. Breyer JP, McReynolds KM, Yaspan BL, Bradley KM, Dupont WD, Smith JR. Genetic variants and prostate cancer risk: candidate replication and exploration of viral restriction genes. Cancer Epidemiol Biomarkers Prev. 2009; 18(7): 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gallagher DJ, Vijai J, Cronin AM, Bhatia J, Vickers AJ, Gaudet MM, Fine S, Reuter V, Scher HI, Halldén C, et al. Susceptibility loci associated with prostate cancer progression and mortality. Clin Cancer Res. 2010; 16(10): 2819–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin DW, FitzGerald LM, Fu R, Kwon EM, Zheng SL, Kolb S, Wiklund F, Stattin P, Isaacs WB, Xu J, et al. Genetic variants in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes are prognostic markers of prostate cancer‐specific mortality. Cancer Epidemiol Biomarkers Prev. 2011; 20(9): 1928–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pomerantz MM, Werner L, Xie W, Regan MM, Lee GS, Sun T, Evan C, Petrozziello G, Nakabayashi M, Oh WK, et al. Association of prostate cancer risk loci with disease aggressiveness and prostate cancer‐specific mortality. Cancer Prev Res. 2011; 4(5): 719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russo G, Mischi M, Scheepens W, De la Rosette JJ, Wijkstra H. Angiogenesis in prostate cancer: onset, progression and imaging. BJU Int. 2012; 110(11 Pt C): E794–E808. [DOI] [PubMed] [Google Scholar]
- 13. Jacobs EJ, Hsing AW, Bain EB, Stevens VL, Wang Y, Chen J, Chanock SJ, Zheng SL, Xu J, Thun MJ, et al. Polymorphisms in angiogenesis‐related genes and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2008; 17(4): 972–977. [DOI] [PubMed] [Google Scholar]
- 14. Tong X, Li H. eNOS protects prostate cancer cells from TRAIL‐induced apoptosis. Cancer Lett. 2004; 210(1): 63–71. [DOI] [PubMed] [Google Scholar]
- 15. Xu W, Liu LZ, Loizidou M, Ahmed M, Charles IG. The role of nitric oxide in cancer. Cell Res. 2002; 12(5–6): 311–320. [DOI] [PubMed] [Google Scholar]
- 16. Ying L, Hofseth LJ. An emerging role for endothelial nitric oxide synthase in chronic inflammation and cancer. Cancer Res. 2007; 67(4): 1407–1410. [DOI] [PubMed] [Google Scholar]
- 17. Medeiros R, Morais A, Vasconcelos A, Costa S, Pinto D, Oliveira J, Lopes C. Endothelial nitric oxide synthase gene polymorphisms and genetic susceptibility to prostate cancer. Eur J Cancer Prev. 2002; 11(4): 343–350. [DOI] [PubMed] [Google Scholar]
- 18. Medeiros RM, Morais A, Vasconcelos A, Costa S, Pinto D, Oliveira J, Ferreira P, Lopes C. Outcome in prostate cancer: association with endothelial nitric oxide synthase Glu‐Asp298 polymorphism at exon 7. Clin Cancer Res. 2002; 8(11): 3433–3437. [PubMed] [Google Scholar]
- 19. Marangoni K, Neves AF, Cardoso AM, Santos WK, Faria PC, Goulart LR. The endothelial nitric oxide synthase Glu‐298‐Asp polymorphism and its mRNA expression in the peripheral blood of patients with prostate cancer and benign prostatic hyperplasia. Cancer Detect Prev. 2006; 30(1): 7–13. [DOI] [PubMed] [Google Scholar]
- 20. Marangoni K, Araújo TG, Neves AF, Goulart LR. The ‐786T>C promoter polymorphism of the NOS3 gene is associated with prostate cancer progression. BMC Cancer. 2008; 8: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee KM, Kang D, Park SK, Berndt SI, Reding D, Chatterjee N, Chanock S, Huang WY, Hayes RB. Nitric oxide synthase gene polymorphisms and prostate cancer risk. Carcinogenesis. 2009; 30(4): 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ziaei SA, Samzadeh M, Jamaldini SH, Afshari M, Haghdoost AA, Hasanzad M. Endothelial nitric oxide synthase Glu298Asp polymorphism as a risk factor for prostate cancer. Int J Biol Markers. 2013; 28(1): 43–48. [DOI] [PubMed] [Google Scholar]
- 23. Safarinejad MR, Safarinejad S, Shafiei N, Safarinejad S. Effects of the T‐786C, G894T, and Intron 4 VNTR (4a/b) polymorphisms of the endothelial nitric oxide synthase gene on the risk of prostate cancer. Urol Oncol. 2013; 31(7): 1132–1140. [DOI] [PubMed] [Google Scholar]
- 24. Branković A, Brajušković G, Nikolić Z, Vukotić V, Cerović S, Savić‐Pavićević D, Romac S. Endothelial nitric oxide synthase gene polymorphisms and prostate cancer risk in Serbian population. Int J Exp Pathol. 2013; 94(6): 355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Medeiros R, Morais A, Vasconcelos A, Costa S, Carrilho S, Oliveira J, Lopes C. Endothelial nitric oxide synthase gene polymorphisms and the shedding of circulating tumour cells in the blood of prostate cancer patients. Cancer Lett. 2003; 189(1): 85–90. [DOI] [PubMed] [Google Scholar]
- 26. Sanli O, Kucukgergin C, Gokpinar M, Tefik T, Nane I, Seckin S. Despite the lack of association between different genotypes and the presence of prostate cancer, endothelial nitric oxide synthase a/b (eNOS4a/b) polymorphism may be associated with advanced clinical stage and bone metastasis. Urol Oncol. 2011; 29(2): 183–188. [DOI] [PubMed] [Google Scholar]
- 27. Wallace BC, Dahabreh IJ, Trikalinos TA, Lau J, Trow P, Schmid CH. Closing the gap between methodologists and end‐users: R as a computational back‐end. J Stat Softw. 2012; 49(5): 1–15. Available at URL: http://www.jstatsoft.org/v49/i05/paper [Google Scholar]
- 28. Der Simonian R, Laird N. Meta‐analysis in clinical trials. Control Clin Trials. 1986; 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 29. Dluzniewski PJ, Wang MH, Zheng SL, De Marzo AM, Drake CG, Fedor HL, Partin AW, Han M, Fallin MD, Xu J, et al. Variation in IL10 and other genes involved in the immune response and in oxidation and prostate cancer recurrence. Cancer Epidemiol Biomarkers Prev. 2012; 21(10): 1774–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choudhari SK, Chaudhary M, Bagde S, Gadbail AR, Joshi V. Nitric oxide and cancer: a review. World J Surg Oncol. 2013; 11: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu W, Liu LZ, Loizidou M, Ahmed M, Charles IG. The role of nitric oxide in cancer. Cell Res. 2002; 12(5–6): 311–320. [DOI] [PubMed] [Google Scholar]
- 32. Cronauer MV, Ince Y, Engers R, Rinnab L, Weidemann W, Suschek CV, Burchardt M, Kleinert H, Wiedenmann J, Sies H, et al. Nitric oxide‐mediated inhibition of androgen receptor activity: possible implications for prostate cancer progression. Oncogene. 2007; 26(13): 1875–1884. [DOI] [PubMed] [Google Scholar]
- 33. Burnett AL, Maguire MP, Chamness SL, Ricker DD, Takeda M, Lepor H, Chang TS. Characterization and localization of nitric oxide synthase in the human prostate. Urology. 1995; 45(3): 435–439. [DOI] [PubMed] [Google Scholar]
- 34. Yu S, Jia L, Zhang Y, Wu D, Xu Z, Ng CF, To KK, Huang Y, Chan FL. Increased expression of activated endothelial nitric oxide synthase contributes to antiandrogen resistance in prostate cancer cells by suppressing androgen receptor transactivation. Cancer Lett. 2013; 328(1): 83–94. [DOI] [PubMed] [Google Scholar]
- 35. Nanni S, Benvenuti V, Grasselli A, Priolo C, Aiello A, Mattiussi S, Colussi C, Lirangi V, Illi B, D'Eletto M, et al. Endothelial NOS, estrogen receptor beta, and HIFs cooperate in the activation of a prognostic transcriptional pattern in aggressive human prostate cancer. J Clin Invest. 2009; 119(5): 1093–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Polytarchou C, Hatziapostolou M, Poimenidi E, Mikelis C, Papadopoulou A, Parthymou A, Papadimitriou E. Nitric oxide stimulates migration of human endothelial and prostate cancer cells through up‐regulation of pleiotrophin expression and its receptor protein tyrosine phosphatase beta/zeta. Int J Cancer. 2009; 124(8): 1785–1793. [DOI] [PubMed] [Google Scholar]
- 37. Casas JP, Cavalleri GL, Bautista LE, Smeeth L, Humphries SE, Hingorani AD. Endothelial nitric oxide synthase gene polymorphisms and cardiovascular disease: a HuGE review. Am J Epidemiol. 2006; 164(10): 921–935. [DOI] [PubMed] [Google Scholar]
- 38. Colombo MG, Paradossi U, Andreassi MG, Botto N, Manfredi S, Masetti S, Biagini A, Clerico A. Endothelial nitric oxide synthase gene polymorphisms and risk of coronary artery disease. Clin Chem. 2003; 49(3): 389–395. [DOI] [PubMed] [Google Scholar]
- 39. Hingorani AD, Liang CF, Fatibene J, Lyon A, Monteith S, Parsons A, Haydock S, Hopper RV, Stephens NG, O'Shaughnessy KM, et al. A common variant of the endothelial nitric oxide synthase (Glu298>Asp) is a major risk factor for coronary artery disease in the UK. Circulation. 1999; 100(14): 1515–1520. [DOI] [PubMed] [Google Scholar]
- 40. Hibi K, Ishigami T, Tamura K, Mizushima S, Nyui N, Fujita T, Ochiai H, Kosuge M, Watanabe Y, Yoshii Y, et al. Endothelial nitric oxide synthase gene polymorphism and acute myocardial infarction. Hypertension. 1998; 32(3): 521–526. [DOI] [PubMed] [Google Scholar]
- 41. Safarinejad MR, Shafiei N, Safarinejad S. The role of endothelial nitric oxide synthase (eNOS) T‐786C, G894T, and 4a/b gene polymorphisms in the risk of idiopathic male infertility. Mol Reprod Dev. 2010; 77(8): 720–727. [DOI] [PubMed] [Google Scholar]
- 42. Tempfer C, Unfried G, Zeillinger R, Hefler L, Nagele F, Huber JC. Endothelial nitric oxide synthase gene polymorphism in women with idiopathic recurrent miscarriage. Hum Reprod. 2001; 16(8): 1644–1647. [DOI] [PubMed] [Google Scholar]
- 43. Hao Y, Montiel R, Huang Y. Endothelial nitric oxide synthase (eNOS) 894 G>T polymorphism is associated with breast cancer risk: a meta‐analysis. Breast Cancer Res Treat. 2010; 124(3): 809–813. [DOI] [PubMed] [Google Scholar]
- 44. Verim L, Toptas B, Ozkan NE, Cacina C, Turan S, Korkmaz G, Yaylim I. Possible relation between the NOS3 gene GLU298ASP polymorphism and bladder cancer in Turkey. Asian Pac J Cancer Prev. 2013; 14(2): 665–668. [DOI] [PubMed] [Google Scholar]
- 45. Jang MJ, Jeon YJ, Kim JW, Chong SY, Hong SP, Oh D, Cho YK, Chung KW, Kim NK. Association of eNOS polymorphisms (‐786T>C, 4a4b, 894G>T) with colorectal cancer susceptibility in the Korean population. Gene. 2013; 512(2): 275–281. [DOI] [PubMed] [Google Scholar]
- 46. Golser R, Gorren AC, Mayer B, Schmidt K. Functional characterization of Glu298Asp mutant human endothelial nitric oxide synthase purified from a yeast expression system. Nitric Oxide. 2003; 8(1): 7–14. [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y, Jia Q, Xue P, Liu Y, Xiong T, Yang J, Song C, He Q, Du L. The ‐786T > C polymorphism in the NOS3 gene is associated with increased cancer risk. Tumour Biol. 2014. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 48. Yeh CC, Santella RM, Hsieh LL, Sung FC, Tang R. An intron 4 VNTR polymorphism of the endothelial nitric oxide synthase gene is associated with early‐onset colorectal cancer. Int J Cancer. 2009; 124(7): 1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramírez‐Patiño R, Figuera LE, Puebla‐Pérez AM, Delgado‐Saucedo JI, Legazpí‐Macias MM, Mariaud‐Schmidt RP, Ramos‐Silva A, Gutiérrez‐Hurtado IA, Gómez Flores‐Ramos L, Zúñiga‐González GM, et al. Intron 4 VNTR (4a/b) polymorphism of the endothelial nitric oxide synthase gene is associated with breast cancer in Mexican women. J Korean Med Sci. 2013; 28(11): 1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hefler LA, Ludwig E, Lampe D, Zeillinger R, Leodolter S, Gitsch G, Koelbl H, Tempfer CB. Polymorphisms of the endothelial nitric oxide synthase gene in ovarian cancer. Gynecol Oncol. 2002; 86(2): 134–137. [DOI] [PubMed] [Google Scholar]
- 51. Riener EK, Hefler LA, Grimm C, Galid A, Zeillinger R, Tong‐Cacsire D, Gitsch G, Leodolter S, Tempfer CB. Polymorphisms of the endothelial nitric oxide synthase gene in women with vulvar cancer. Gynecol Oncol. 2004; 93(3): 686–690. [DOI] [PubMed] [Google Scholar]
- 52. Wang J, Dudley D, Wang XL. Haplotype‐specific effects on endothelial NO synthase promoter efficiency: modifiable by cigarette smoking. Arterioscler Thromb Vasc Biol. 2002; 22(5): e1–e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Meta‐analysis of the association between rs1799983 and PCa progression under dominant model. (A) association with Gleason score; (B) association with PCa TNM stage.


