Abstract
Purpose
Long noncoding RNAs (lncRNAs) constitute an emerging group of noncoding RNAs, which regulate gene expression. Their role in cardiac disease is poorly known. Here, we investigated the association between lncRNAs and left ventricular hypertrophy.
Methods
Wild‐type and adenosine A2A receptor overexpressing mice (A2A‐Tg) were subjected to transverse aortic constriction (TAC) and expression of lncRNAs in the heart was investigated using genome‐wide microarrays and an analytical pipeline specifically developed for lncRNAs.
Results
Microarray analysis identified two lncRNAs up‐regulated and three down‐regulated in the hearts of A2A‐Tg mice subjected to TAC. Quantitative PCR showed that lncRNAs 2900055J20Rik and Gm14005 were decreased in A2A‐Tg mice (3.5‐ and 1.8‐fold, p < 0.01). We found from public microarray dataset that 2900055J20Rik and Gm14005 were increased in TAC mice compared to sham‐operated animals (1.8‐ and 1.4‐fold, after 28 days, p < 0.01). Interestingly, in this public dataset, cardioprotective drug JQ1 decreased 2900055J20Rik and Gm14005 expression by 2.2‐ and 1.6‐fold (p < 0.01).
Conclusions
First, we have shown that data on lncRNAs can be obtained from gene expression microarrays. Second, expression of lncRNAs 2900055J20Rik and Gm14005 is regulated after TAC and can be modulated by cardioprotective molecules. These observations motivate further investigation of the therapeutic value of lncRNAs in the heart.
Keywords: cardiac hypertrophy, long noncoding RNAs, microarrays, adenosine, cardioprotective molecules, experimental disease model
Introduction
Since the completion of the sequencing of the human genome in 2001,1, 2 it has been observed that only a small proportion (less than 2%) of human genome encodes protein‐coding RNAs.3 The vast majority of human genes are transcribed into noncoding RNAs. Multiple types of noncoding RNAs have been characterized and classified depending on their subcellular localization, their size, and their mode of action4 Long noncoding RNAs (lncRNAs) are RNA sequences of more than 200 nucleotides, which are able to regulate gene expression at multiple levels, from modification of epigenetic mechanisms to posttranscriptional regulation.5, 6, 7, 8 While lncRNAs have been shown to be involved in tumor development and may also serve as biomarkers in certain types of cancer,9, 10, 11, 12 the knowledge of the role of lncRNAs in the heart is only at its infancy. Two recent landmark studies revealed that lncRNAs are involved in cardiac development.13, 14 However, the role of lncRNAs in cardiac disease is poorly known.
A few reports (reviewed in Gurha and Marian4 and Schonrock et al.15) support a role for lncRNAs in cardiac disease. For instance, eight single nucleotide polymorphisms in an lncRNA called antisense noncoding RNA in the INK4 locus (ANRIL) have been associated with susceptibility to coronary artery disease.16 Genetic variants in another lncRNA called myocardial infarction‐associated transcript (MIAT) conferred susceptibility to myocardial infarction.17 Antisense transcripts to cardiac troponin I regulate cardiac muscle contraction.18 The involvement of lncRNAs in heart failure (HF) has been suggested by a study showing that transverse aortic constriction (TAC) in mice affected the expression of a subset of lncRNAs.19 However, whether lncRNAs are functionally involved in the progression of HF remains to be determined. HF is a serious health problem, both in sociological and economical terms. It remains a leading cause of mortality in modern countries. A recent epidemiological study enrolling 2.8 millions of North Americans hospitalized for acute myocardial infarction (AMI) revealed that, while the percentage of patients hospitalized for HF in the year following AMI has decreased from 16.1% in 1998 to 14.2% in 2010, the mortality at 1‐year increased from 44.4% in 1998 to 45.5% in 2010.20 These findings indicated improvements in the management of AMI, but also a still poor survival rate of patients developing HF after AMI. This highlights the need for novel treatments of HF. Since lncRNAs can be therapeutically regulated and ongoing studies aim at developing specific inhibitors that may become applicable to the clinic,21 it is believed that lncRNAs might constitute a novel class of therapeutic targets of HF. In this study, we addressed the association between lncRNAs and left ventricular hypertrophy induced by pressure overload. Using an analytical pipeline specifically developed to extract from traditional gene expression microarrays—that is, microarrays initially dedicated to investigation of coding genes—the data related to lncRNAs, we were able to show that lncRNAs are regulated in the hypertrophied heart.
Methods
Animal experiments
Animal experiments were approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University (Philadelphia, PA, USA) and were conducted in accordance with the regulations of the Animal Welfare Act of the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85‐23, revised 1996).
Six male Friend Virus B mice (PolyGene, Zurich, Switzerland) were subjected to TAC: three mice were A2A transgenic (A2A‐Tg) and three mice were wild‐type (Wt) littermates. A2A‐Tg mice carried over an A2A transgene allowing cardiac‐restricted and inducible overexpression of the human adenosine A2A receptor as previously described.22 A2A overexpression was started at 3 weeks of age after removal of doxycycline from mice diet. At 8 weeks of age, mice underwent TAC as described.23 Briefly, an aortic band was created by placing a ligature (7–0 nylon suture) securely between the origin of the right innominate and left common carotid arteries with a 27‐gauge needle as a guide. After 4 weeks, mice were sacrificed, and the heart was harvested and deep frozen. At that time, mice showed signs of left ventricular dysfunction as documented by a decrease of fractional shortening.23
Microarrays
Total RNA was extracted from mice hearts and its quality was verified using a Bioanalyser (Agilent, Santa Clara, CA, USA). cRNAs were generated, labeled, and hybridized to Affymetrix Mouse Gene 1.0 ST Array according to Affymetrix protocol (Affymetrix, Santa Clara, CA, USA). Scanning of the slides was performed using Affymetrix GeneChip Scanner 3000 7G with 2.5‐μm resolution. Raw data were acquired and processed with Partek Genomics Suite (St. Louis, Missouri, USA) using Robust Multiarray Averaging with GC correction and quantile normalization. Probe sets were summarized with mean values for one transcript cluster. Complete microarray dataset is available at Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the accession under GSE45423.
Normalized data were analyzed by using the t‐test procedure within Significance Analysis of Microarrays software 28 (version 3.09, Stanford, CA, USA) to identify differentially expressed transcripts. Transcripts with q‐value <5% were considered as significant. Heat‐maps were generated with Cluster 3 and TreeView softwares (Princteton, NJ, USA). M‐A plot was performed with R Limma package (Melbourne, Australia).24
Extraction of lncRNA sequences from public databases
Transcript sequences were downloaded from three databases: the widely used sequence database NCBI RefSeq, the Ensembl ncRNA database, and the lncRNAdb database, which gathers lncRNAs having, or associated with, biological functions.
All RNA sequences from RefSeq database were downloaded from release 55 (ftp://ftp.ncbi.nih.gov/refseq/M_musculus/mRNA_Prot/mouse.rna.fna.gz). Sequences were divided into two groups: protein‐coding RNAs (i.e., RNA with NM prefix) and noncoding RNAs (i.e., RNA with NR prefix). Then, ribosome RNAs and small noncoding RNAs, that is noncoding RNAs annotated as microRNA, small nucleolar RNA, or small nuclear RNA, were excluded.
Noncoding RNA sequences of Ensembl ncRNA database were obtained from release 68 (ftp://ftp.ensembl.org/pub/release‐68/fasta/mus_musculus/ncrna/). Sequences were removed if their transcript biotype was described as snRNA, Mt‐rRNA, rRNA, snoRNA, miRNA, or Mt_tRNA.
All mouse sequences were downloaded from the lncRNAdb database (http://lncrnadb.com/).
Microarray probe re‐annotation
All transcripts differentially expressed with q‐value < 5% were re‐annotated. Probe sequences were provided by Affymetrix. First, differentially expressed transcripts annotated as mature messenger RNAs by Affymetrix were removed. Then, all the probe sequences of differentially expressed transcripts were aligned to the sequences of the lncRNAs extracted from public databases (see above) using Perl with BioPerl modules (Durham, NC, USA) integrated with BLAST program. Perl scripts are freely available on request from authors. Alignment and filtering were performed as follows. First, probes were aligned to lncRNAs from lncRNAdb, RefSeq, and Ensembl databases. Second, probes with perfect match were aligned to protein‐coding RNAs from RefSeq database. Perfectly matched probes were removed. Third, transcript clusters missing even one probe were removed.
Quantitative RT‐PCR
Mouse heart total RNA (1 μg) was reverse‐transcribed using the Superscript II RT kit (Life Technologies, Merelbeke, Belgium). Controls without reverse transcriptase were performed to ensure the absence of genomic DNA amplification during PCR. Real‐time PCR was performed in a CFX96 apparatus (BioRad, Nazareth, Belgium) with IQ SYBR Green Supermix (BioRad) and primers designed with the Beacon Designer software (Premier Biosoft, Palo Alto, CA, USA; Table 1 ). PCR conditions were as follows: 3 minutes at 95°C, 30 seconds at 95°C, and 1 minute annealing‐extension (40‐fold). Optimal annealing‐extension temperature was determined for each primer pair. The specificity of the PCR was confirmed by a melting curve analysis. GAPDH was chosen as a housekeeping gene for normalization. Expression levels were calculated by the relative quantification method (ΔΔCt) using the CFX Manager 2.1 (BioRad).
Table 1.
Quantitative RT‐PCR primers
| Name | Accession no. | Forward primer | Reverse primer | Annealing temperature (°C) |
|---|---|---|---|---|
| Dancr | NR_015531 | AATGTATCTGGACTTCGTTAG | AGAATTGACACAGGAAGC | 54 |
| Gm10400 | NR_033555 | TCACTCTTGCTTAATCAT | TTAGACCTGTTCAACTAG | 52 |
| Gm14005 | NR_028590 | ACCCACATACCAAGTTCT | AAGTCATCCAGGTAACAAG | 56 |
| 2900055J20Rik | NR_045177 | TAAGTGTGGAATCATTGTT | TTGCGAAGAAATAACCTT | 56 |
| Nppa | NM_008725 | TCTGATGGATTTCAAG | ACCTCATCTTCTACC | 56 |
| Nppb | NM_008726 NM_001287348 | ATGATTCTGTTTCTGCTTT | ATTTCCTCCGACTTTTCT | 56 |
| Acta1 | NM_001272041 NM_009606 | AGGACCTGTACGCCAACAAC | ACATCTGCTGGAAGGTGGAC | 62 |
Public dataset
Microarray dataset consisting of 27 mouse samples was published by Anand et al.25 (GEO accession code: GSE48110). Mice were subjected to TAC or sham‐operation followed after 1.5 days by treatment with the BET bromodomain inhibitor JQ1 or vehicle (DMSO). Cardiac gene expression was investigated in three groups (sham‐vehicle, TAC‐vehicle, and TAC‐JQ1) at three time points after TAC (3, 11, and 28 days) using the Affymetrix Mouse Gene 1.0 ST Array. The full list of differentially expressed transcripts was downloaded from the supplementary files provided by authors.
Statistical analysis
The SigmaPlot v11.0 software (San Jose, CA, USA) was used for statistical analyses. All tests were preceded by the Shapiro–Wilk normality test. Comparisons between two groups were performed using t‐test for Gaussian data and the Mann–Whitney test for non‐Gaussian data. Results are presented as mean ± standard deviation. A p‐value <0.05 was considered significant.
Results
Effect of adenosine A2A receptor overexpression on cardiac transcriptome
The animals included in this study were already used in a previous study and have been described in detail.23 Briefly, TAC induced a decrease of fractional shortening in Wt mice, showing a loss of contractile function. In A2A‐Tg mice however, cardiac function was preserved.
We used the Affymetrix Mouse Gene 1.0 ST microarray to characterize the effects of overexpression of adenosine A2A receptor on gene expression in the heart of TAC mice. This microarray includes 28853 transcript cluster ID covering 26166 total RefSeq (release 36) transcripts. All mice (three A2A‐Tg and three Wt) were subjected to TAC and were sacrificed after 4 weeks. Hearts were harvested and RNA expression was analyzed by microarrays. Adenosine A2A receptor overexpression induced significant changes of gene expression as displayed in the heat‐map and M‐A plot of Figure 1 (a and b). Significance analysis of microarrays revealed that 709 transcripts were differentially expressed between A2A‐Tg mice and Wt littermates. Principal component analysis showed the ability of gene expression data to discriminate A2A‐Tg mice from Wt mice (Figure 1 c). Interestingly, the expression of the canonical hypertrophic markers natriuretic peptide A (Nppa), natriuretic peptide B (Nppb), and actin alpha 1 (Acta1) was strongly decreased following overexpression of adenosine A2A receptor (Figure 1 d). Collectively, these data show that overexpression of adenosine A2A receptor in the heart has significant effects on gene expression, and down‐regulates the expression of hypertrophic markers. This is consistent with the cardioprotective effect of A2A overexpression reported previously.23
Figure 1.
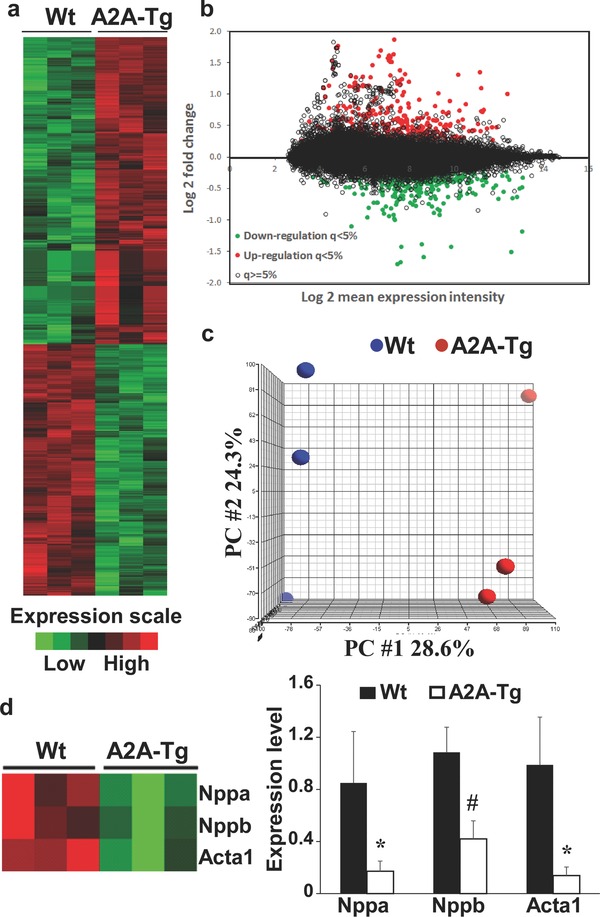
Effect of adenosine A2A receptor overexpression on cardiac transcriptome. Three A2A‐Tg mice and three Wt littermates were subjected to TAC and were sacrificed after 4 weeks. Hearts were harvested and gene expression was profiled using Affymetrix Mouse Gene 1.0 ST microarray. (a and b) Heat‐map and M‐A plot of differentially expressed transcripts as determined by significance analysis of microarrays with a q‐value < 5% as significance threshold. (c) Principal component analysis. (d). Expression of canonical hypertrophic markers Nppa, Nppb, and Acta1. Left panel: heat‐map showing down‐regulation of all three genes in A2A‐Tg mice. Right panel: Assessment of the three genes by quantitative RT‐PCR. *p < 0.05 vs. Wt; #p < 0.01 vs. Wt. Wt = wild‐type; A2A‐Tg = adenosine A2A receptor overexpressing mice; PC = principal component.
Effect of adenosine A2A receptor overexpression on LncRNAs expression
We implemented an analytical pipeline to retrieve lncRNAs data from the microarray dataset generated from three A2A‐Tg mice and three Wt littermates subjected to TAC and shown in Figure 1 . Indeed, the Affymetrix Mouse Gene 1.0 ST microarray used in these experiments has been originally developed to characterize the expression of coding genes, although a significant part of the probes spotted on this microarray recognizes noncoding transcripts. The analytical pipeline is displayed in Figure 2 and included the following steps.
Figure 2.
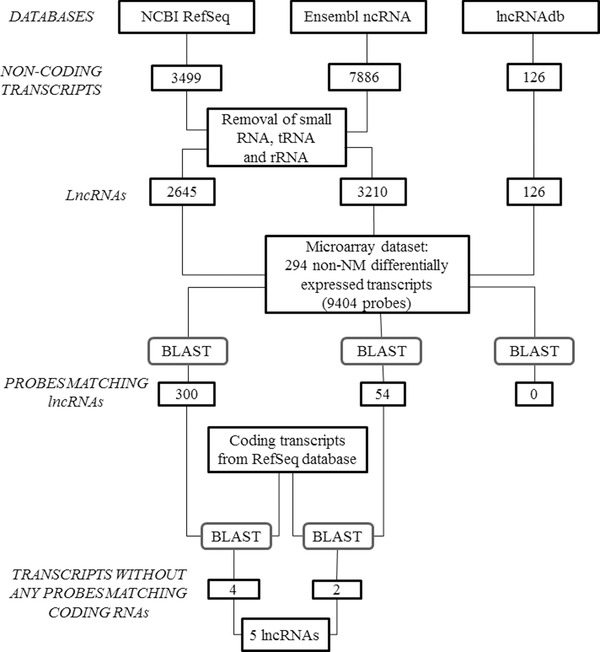
Analytical pipeline to retrieve lncRNAs data from microarray dataset. The Affymetrix Mouse Gene 1.0 ST microarray dataset generated from three A2A‐Tg mice and three Wt littermates subjected to TAC (see Figure 1 ) was used in this analysis. Non‐NM: noncoding transcripts (accession number without NM prefix); rRNA = ribosome RNA; tRNA = transfer RNA.
First, a list of well‐annotated lncRNAs was extracted from public databases. For this purpose, 3499, 7886, and 126 murine noncoding transcripts were downloaded from NCBI RefSeq, Ensembl ncRNA, and lncRNAdb databases, respectively. After removing small noncoding RNAs, transfer RNAs (tRNA) and ribosome RNAs (rRNAs), 2645 transcripts from NCBI RefSeq, 3210 from Ensembl ncRNA, and 126 from lncRNAdb database remained and were considered as lncRNAs for subsequent analyses.
Second, we aimed at identifying differentially expressed lncRNAs in our microarray dataset. Using significance analysis of microarrays with a threshold for significance of 5%, we identified 294 non‐NM differentially expressed transcripts, corresponding to 9404 microarray probes. These probes were aligned (“Blast”) to the lncRNAs selected from RefSeq (2645), Ensembl ncRNA (3210), and lncRNAdb (126) databases: 300, 54, and 0 probes found perfect match with lncRNAs, respectively. To avoid any potential match with protein‐coding sequences, we aligned these probes to sequences from RefSeq database with NM prefix (i.e., coding transcripts). The probes that perfectly matched these coding sequences (80 and 6 sequences from RefSeq and Ensembl databases, respectively) were discarded. Only the transcripts without any probes matching coding RNAs were retained. After this filtering process, four and two lncRNAs from RefSeq and Ensembl ncRNA databases, respectively, were retained. One lncRNAs, named Gm14005, was common between the two databases. Therefore, the analysis ended with five lncRNAs differentially expressed between A2A‐Tg mice and Wt mice. Microarray expression data for these five lncRNAs are presented in Table 2 . Three lncRNAs, 2900055J20Rik, Dancr, and Gm14005, were down‐regulated in A2A‐Tg mice, and two lncRNAs, Gm10400 and BC016548, were up‐regulated in A2A‐Tg mice, compared to Wt littermates.
Table 2.
Differentially expressed lncRNAs in A2A‐Tg mice compared to Wt mice
| Transcript ID | Gene name | Affymetrix transcript ID | Fold change | q‐value (%) |
|---|---|---|---|---|
| NR_045177 | 2900055J20Rik | 10455292 | 0.52 | 0.00 |
| NR_033555 | Gm10400 | 10542592 | 1.30 | 4.00 |
| NR_015531 | Dancr | 10522465 | 0.85 | 4.42 |
| NR_028590 | Gm14005 | 10487506 | 0.81 | 4.42 |
| ENSMUST00000144657 ENSMUST00000153797 | BC016548 | 10485461 | 1.23 | 4.42 |
Data have been obtained from the analytical pipeline displayed in Figure 2 and applied to the microarray data shown in Figure 1 . Microarrays were generated from heart samples of three A2A‐Tg mice and three Wt mice subjected to TAC. Differential expression was determined using significance analysis of microarrays with a q‐value <5% as significance threshold.
Validation of microarray data
Expression of the five lncRNAs identified by microarrays as differentially expressed between A2A‐Tg mice and Wt mice was assessed by quantitative RT‐PCR in heart samples from the six mice used for microarray experiments (Figure 3 ). Gm14005 and 2900055J20Rik were down‐regulated (1.7‐ and 3.5‐fold, respectively) in A2A‐Tg mice compared to wild‐type mice, thus confirming microarray results. Dancr and Gm10400 followed the same trends as in microarray experiments, but differences did not reach statistical significance. BC016548 lncRNA was undetected.
Figure 3.
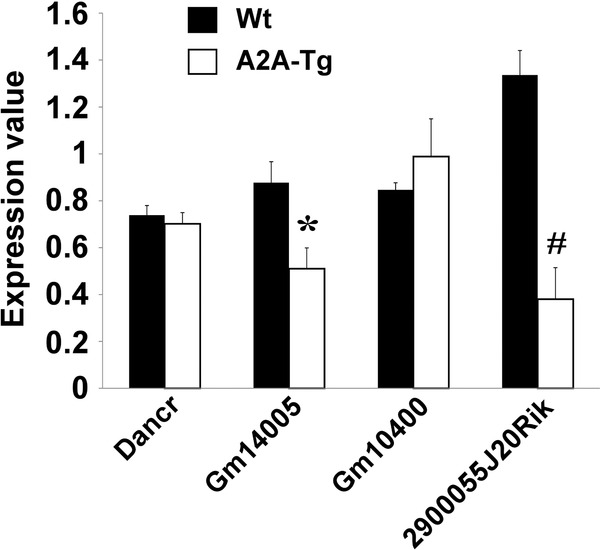
Validation of microarray data. Expression of the five lncRNAs identified by microarrays as differentially expressed between A2A‐Tg mice and Wt mice was measured by quantitative RT‐PCR. Heart samples from the six mice used in microarray experiments were used in these experiments. The lncRNA BC016548 could not be detected. GAPDH was used as housekeeping gene for normalization. *p = 0.007 vs. Wt; #p < 0.001 vs. Wt.
Association between lncRNAs and left ventricular hypertrophy in public data
To further address the association between lncRNAs and left ventricular hypertrophy induced by pressure overload, we used a public dataset from mice subjected to TAC and which used the same Affymetrix Mouse Gene 1.0 ST Arrays as us.25 In addition, this dataset contained data from mice treated with the BET bromodomain inhibitor JQ1, an inhibitor of c‐Myc pathway with anticancer26 and cardioprotective properties.25, 27 From this public dataset, we obtained the expression values of the four lncRNAs 2900055J20Rik, Gm14005, Gm10400, and Dancr (Figure 4 ). Of note, BC016548, the fifth lncRNA found to be regulated in A2A‐Tg mice in our microarray experiments (Table 1 ), was not regulated by TAC or administration of JQ1 in the public dataset. Expression of 2900055J20Rik and Gm14005 was up‐regulated in TAC mice compared to sham‐operated mice. Interestingly, administration of JQ1 resulted in a decrease of expression of both lncRNAs (p < 0.01). In particular, expression of 2900055J20Rik, which was the most down‐regulated lncRNA in our A2A‐Tg mice, showed a 1.8‐fold up‐regulation 28 days after TAC compared to sham‐operated animals (p < 0.01), and a decrease (2.2‐fold, p < 0.01) upon JQ1 treatment. Expression of Gm10400 and Dancr was not reproducibly affected by TAC or JQ1 treatment, which is consistent with our data in A2A‐Tg mice shown in Figure 3 . It has to be noted that a clear left ventricular hypertrophy and an increase of heart/body weight ratio were observed in these mice 28 days after TAC.25 This hypertrophy was reduced upon administration of JQ1. Taken together, our data in A2A‐Tg mice subjected to TAC and the public data in TAC mice subjected to JQ1 treatment show a consistent regulation of the expression of 2900055J20Rik and Gm14005: up‐regulation in the overloaded heart and down‐regulation by cardioprotective strategies aiming at stimulating the adenosine pathway by overexpression of A2A receptor and by the inhibitor of c‐Myc pathway JQ1.
Figure 4.
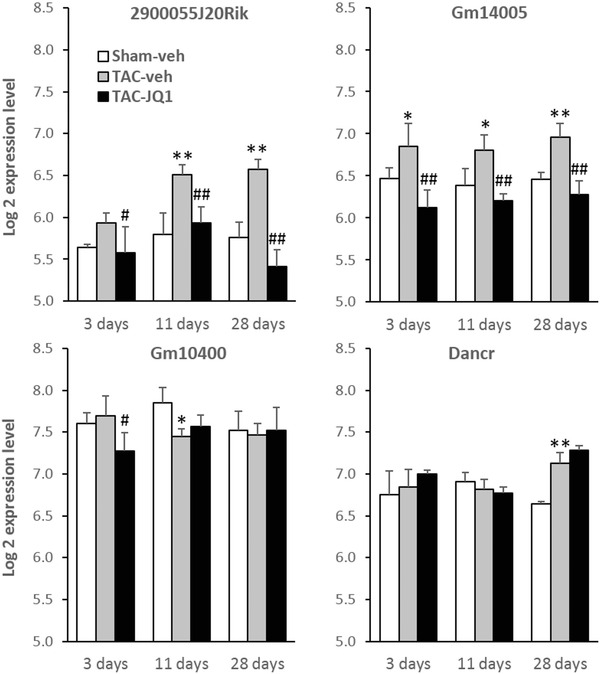
Association between lncRNAs and left ventricular hypertrophy. Data shown are from a public microarray dataset (GSE48110). Mice were subjected to TAC or sham operation and were treated with BET bromodomian inhibitor JQ1 or vehicle (DMSO) for 3, 11, or 28 days. Hearts were harvested and gene expression was analyzed by Affymetrix Mouse Gene 1.0 ST microarray. Graph bars represent the expression values of the four lncRNAs 2900055J20Rik, Gm14005, Gm10400, and Dancr extracted from microarray data and expressed as log2. *p < 0.05 vs. sham, **p < 0.01 vs. sham, #p < 0.05 vs. TAC, ##p < 0.01 vs. TAC (n = 3 per group). Veh = vehicle (DMSO).
Discussion
The main findings of this study are threefold. First, we demonstrated the feasibility of obtaining expression data for lncRNAs in traditional gene expression microarrays. Second, we revealed that expression of lncRNAs is regulated in the heart subjected to pressure overload and can be modulated by cardioprotective strategies. Third, we identified two lncRNAs, which are up‐regulated in the overloaded heart and down‐regulated upon overexpression of adenosine A2A receptor or administration of JQ1.
Genome‐wide microarrays have been widely used to analyze the expression of protein‐coding genes. With the development of new high‐throughput techniques such as next generation sequencing, genomic information has been rapidly updated and stored in public databases. This allowed to re‐annotate previously generated microarray data and realize that these microarrays, such as the widely used Affymetrix Mouse Gene 1.0 ST Array, are a reservoir of information for lncRNAs. So far, microarray re‐annotation for lncRNAs has been performed on different platforms28, 29 using different public RNA databases. Liao et al.28 aligned probes to noncoding transcript sequences from the FANTOM3 (Functional Annotation of Mammalian cDNA) project30 that identified about 35,000 noncoding transcripts. Michelhaugh et al.29 used H‐Invitational version 5 database,31 a comprehensive annotation resource for human transcripts, and RNAdb,32 a database of mammalian noncoding RNAs retracted since 2012. We selected three widely used databases: lncRNAdb,33 a database of lncRNAs with an experimentally validated biological function; RefSeq, a curated nonredundant sequence database34; Ensembl ncRNA, a database of well‐curated noncoding RNAs.35 In RefSeq, we kept only sequences with NR prefix that have well‐established annotations. By combining the three databases, we obtained a comprehensive and well‐annotated set of lncRNA sequences.
The analytical pipeline to mine microarray data has been a useful tool in our study to obtain reliable data for lncRNAs from traditional microarray data. Previous studies already reported similar approaches.36, 37, 38 Genome‐wide expression microarray platforms have been widely used in the past decade to associate gene expression signatures with disease states, and a huge amount of expression data are stored either in servers of research laboratories or are available from public repositories. This constitutes an enormous reservoir of data that can be re‐interpreted in the context of lncRNAs.
The main aim of this study was to investigate the regulation of lncRNAs in the hypertrophied heart. To address this, we used microarrays to compare the expression of lncRNAs in the heart of transgenic mice overexpressing the adenosine A2A receptor and Wt littermates, all subjected to TAC. The rationale beyond this choice was to test a potential link between lncRNAs and the cardioprotection provided by overexpression of the A2A receptor. Indeed, we previously reported beneficial effects of adenosine and its A2A receptor in the heart.23, 39, 40, 41, 42, 43, 44 Cardiac contractile function was preserved at 8 weeks after TAC in A2A receptor transgenic mice, while it was impaired in Wt littermates subjected to TAC.23 After 14 weeks, the end systolic dimension, heart/body ratio, and cardiac fibrosis were markedly attenuated in A2A transgenic mice compared to wild‐type mice. Having identified some lncRNAs regulated following overexpression of the A2A receptor, we used a public microarray dataset from Anand et al.25 to confirm the association between these lncRNAs and left ventricular hypertrophy induced by TAC. It has to be noted that in both our A2A‐Tg mice23 and in mice from the study of Anand, TAC induced a decrease of fractional shortening indicating left ventricular dysfunction with impaired contractility. This phenomenon was reversed by overexpression of A2A receptor and administration of JQ1, which also inhibited the up‐regulation of Nppa, Nppb, and Acta1 induced by TAC. The mechanisms mediating these cardioprotective effects appear to be different and involve a suppression of myocardial inflammation following overexpression of A2A receptor23 and a blockade by JQ1 of the transcription of genes driving pathologic cardiac remodeling.25 Whether adenosine and JQ1 have additive anti‐remodeling effects is an attractive possibility that deserves further investigation.
We identified two lncRNAs associated with left ventricular hypertrophy, 2900055J20Rik and Gm14005. Lee et al.19 developed bioinformatic approaches to analyze the transcriptome of the failing murine heart generated by RNA sequencing. Consistently with findings of the ENCODE consortium for human genome,3 they observed that more than 80% of RNA transcripts had low protein‐coding potential. They were able to characterize 15 and 135 novel lncRNAs differentially expressed between hypertrophied and failing hearts, respectively. Recently, deep sequencing revealed that lncRNAs are regulated in the failing human heart.45 These studies support a role for lncRNAs in cardiac remodeling and in the development of HF. RNA sequencing is unbiased and certainly more sensitive than microarrays to study the regulation of lncRNAs. However, this study had the merit to show that existing microarray datasets can be a useful reservoir of data on lncRNAs to study cardiac pathology. It has to be acknowledged that microarrays lack sensitivity and may lead to false discovery as attested by the fact that only two of the five lncRNAs identified by microarrays could be validated by quantitative PCR. This might also be due to the choice of the 0.05 significance threshold used to select differentially expressed lncRNAs in microarray data. Another option would be to use a fold‐change as threshold instead of a significance threshold. However, the significance threshold (q‐value) has the benefit of taking into account the false discovery rate.
The biological roles of the two lncRNAs, 2900055J20Rik and Gm14005, are unknown. Although their expression pattern follows cardiac remodeling (up‐regulation by TAC and down‐regulation by A2A receptor and JQ1), it remains to be determined whether they play a functional role in cardiac remodeling, and thereby whether they may be considered as potential therapeutic targets. Investigation of the role of lncRNAs in cardiac remodeling may lead to the discovery of novel pathways regulating the development of HF, which may lead to new treatments.
Microarrays have been widely used in different fields of biomedical research such as oncology and cardiology. The majority of microarray data are freely accessible. Mining public microarray data are an economical and time‐saving approach to identify genes regulated in different biological processes and diseases. However, microarrays, compared with more recent techniques such as next generation sequencing, lack sensitivity, and may ignore the expression of new transcripts. Therefore, sequencing is certainly the technology of choice for future studies, particularly to decipher the noncoding transcriptome of complex diseases and to identify novel and sometimes very weakly expressed transcripts.
Conclusion
In conclusion, we have identified two lncRNAs associated with left ventricular hypertrophy. Further studies are required to determine their role in the development of HF, as well as their potential as therapeutic targets.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Authors' Contributions
L.Z. designed the microarray analytical pipeline, acquired microarray data and drafted the manuscript. E.H. and H.F. performed mice experiments. M.V. performed PCR and in vitro experiments. A.M.F. and D.R.W. reviewed the manuscript. Y.D. designed the study, analyzed data and drafted the manuscript.
Acknowledgments
We thank Laurent Vallar, Nathalie Nicot, and Petr Nazarov from the Genomics Research Unit of the Centre de la Recherche Public – Santé for Affymetrix experiments. This work was supported by the Society for Research on Cardiovascular Diseases and the Ministry of Culture, Higher Education and Research of Luxembourg.
Scott A. Waldman acted as the Editor in Chief for this paper.
References
- 1. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. Feb 15 2001; 409(6822): 860–921. [DOI] [PubMed] [Google Scholar]
- 2. Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. Feb 16 2001; 291(5507): 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3. Consortium EP, Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. Sep 6 2012; 489(7414): 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gurha P, Marian AJ. Noncoding RNAs in cardiovascular biology and disease. Circulation research. Dec 6 2013; 113(12): e115–e120. [DOI] [PubMed] [Google Scholar]
- 5. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. Jun 8 2007; 316(5830): 1484–1488. [DOI] [PubMed] [Google Scholar]
- 6. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non‐coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. Nov 15 2012; 491(7424): 454–457. [DOI] [PubMed] [Google Scholar]
- 7. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. Feb 28 2013; 494(7438): 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. Mar 5 2013; 20(3): 300–307. [DOI] [PubMed] [Google Scholar]
- 9. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P et al. Extensive and coordinated transcription of noncoding RNAs within cell‐cycle promoters. Nat Genet. 2011; 43(7): 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011; 21(6): 354–361. [DOI] [PubMed] [Google Scholar]
- 11. Gutschner T, Diederichs S. The hallmarks of cancer: a long non‐coding RNA point of view. RNA Biol. 2012; 9(6): 703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non‐coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013; 339(2): 159–166. [DOI] [PubMed] [Google Scholar]
- 13. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, et al. The tissue‐specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. Jan 28 2013; 24(2): 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. Jan 31 2013; 152(3): 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res. Oct 26 2012; 111(10): 1349–1362. [DOI] [PubMed] [Google Scholar]
- 16. Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. Jun 8 2007; 316(5830): 1491–1493. [DOI] [PubMed] [Google Scholar]
- 17. Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. Identification of a novel non‐coding RNA, MIAT, that confers risk of myocardial infarction. J Human Genet. 2006; 51(12): 1087–1099. [DOI] [PubMed] [Google Scholar]
- 18. Podlowski S, Bramlage P, Baumann G, Morano I, Luther HP. Cardiac troponin I sense‐antisense RNA duplexes in the myocardium. J Cell Biochem. 2002; 85(1): 198–207. [PubMed] [Google Scholar]
- 19. Lee J‐H, Gao C, Peng G, Greer C, Ren S, Wang Y, Xiao X. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. December 9 2011; 109(12): 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J, Hsieh A, Dharmarajan K, Masoudi FA, Krumholz HM. National trends in heart failure hospitalization after acute myocardial infarction for Medicare beneficiaries: 1998–2010. Circulation. Nov 4 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]; 128(24): 2577–2584. [Google Scholar]
- 21. Wahlestedt C. Targeting long non‐coding RNA to therapeutically upregulate gene expression. Nat Rev. Drug Dis. 2013; 12(6): 433–446. [DOI] [PubMed] [Google Scholar]
- 22. Funakoshi H, Chan TO, Good JC, Libonati JR, Piuhola J, Chen X, MacDonnell SM, Lee LL, Herrmann DE, Zhang J, et al. Regulated overexpression of the A1‐adenosine receptor in mice results in adverse but reversible changes in cardiac morphology and function. Circulation. Nov 21 2006; 114(21): 2240–2250. [DOI] [PubMed] [Google Scholar]
- 23. Hamad EA, Zhu W, Chan TO, Myers V, Gao E, Li X, Zhang J, Song J, Zhang X‐Q, Cheung JY, et al. Cardioprotection of controlled and cardiac‐specific over‐expression of A2A‐adenosine receptor in the pressure overload. PLoS ONE. 2012; 7(7): e39919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smyth GK, Michaud J, Scott HS. Use of within‐array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. May 1 2005; 21(9): 2067–2075. [DOI] [PubMed] [Google Scholar]
- 25. Anand P, Brown Jonathan D, Lin Charles Y, Qi J, Zhang R, Artero Pedro C, Alaiti MA, Bullard J, Alazem K, Margulies Kenneth B, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013; 154(3): 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Delmore Jake E, Issa Ghayas C, Lemieux Madeleine E, Rahl Peter B, Shi J, Jacobs Hannah M, Kastritis E, Gilpatrick T, Paranal Ronald M, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c‐Myc. Cell. 2011; 146(6): 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spiltoir JI, Stratton MS, Cavasin MA, Demos‐Davies K, Reid BG, Qi J, Bradner JE, McKinsey TA. BET acetyl‐lysine binding proteins control pathological cardiac hypertrophy. J Mol Cell Cardiol. 2013; 63(0): 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liao Q, Xiao H, Bu D, Xie C, Miao R, Luo H, Zhao G, Yu K, Zhao H, Skogerbo G, et al. ncFANs: a web server for functional annotation of long non‐coding RNAs. Nucleic Acids Res. 2011; 39(Web Server issue): W118–W124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining affymetrix microarray data for long non‐coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem. Feb 2011; 116(3): 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. Sep 2 2005; 309(5740): 1559–1563. [DOI] [PubMed] [Google Scholar]
- 31. Yamasaki C, Murakami K, Fujii Y, Sato Y, Harada E, Takeda J, Taniya T, Sakate R, Kikugawa S, Shimada M, et al. The H‐Invitational Database (H‐InvDB), a comprehensive annotation resource for human genes and transcripts. Nucleic Acids Res. Jan 2008; 36(Database issue): D793–D799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pang KC, Stephen S, Engstrom PG, Tajul‐Arifin K, Chen W, Wahlestedt C, Lenhard B, Hayashizaki Y, Mattick JS. RNAdb—a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. Jan 1 2005; 33(Database issue): D125–D130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. Jan 2011; 39(Database issue): D146–D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non‐redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. Jan 1 2005; 33(Database issue): D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. Jan 1 2002; 30(1): 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gellert P, Ponomareva Y, Braun T, Uchida S. Noncoder: a web interface for exon array‐based detection of long non‐coding RNAs. Nucleic Acids Res. Jan 7 2013; 41(1): e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non‐coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012; 48(1): 1–8. [DOI] [PubMed] [Google Scholar]
- 38. Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. Jul 2013; 20(7): 908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bousquenaud M, Maskali F, Poussier S, Zangrando J, Marie PY, Boutley H, Fay R, Karcher G, Wagner DR, Devaux Y. Cardioprotective effects of adenosine within the border and remote areas of myocardial infarction. EJNMMI Res. 2013; 3(1): 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ernens I, Rouy D, Velot E, Devaux Y, Wagner DR. Adenosine inhibits matrix metalloproteinase‐9 secretion by neutrophils: implication of A2A receptor and cAMP/PKA/Ca2+ pathway. Circ Res. Sep 15 2006; 99(6): 590–597. [DOI] [PubMed] [Google Scholar]
- 41. Haas B, Leonard F, Ernens I, Rodius S, Vausort M, Rolland‐Turner M, Devaux Y, Wagner DR. Adenosine reduces cell surface expression of toll‐like receptor 4 and inflammation in response to lipopolysaccharide and matrix products. J Cardiovasc Transl Res. Dec 2011; 4(6): 790–800. [DOI] [PubMed] [Google Scholar]
- 42. Leonard F, Devaux Y, Vausort M, Ernens I, Rolland‐Turner M, Wagner DR. Adenosine modifies the balance between membrane and soluble forms of Flt‐1. J Leukoc Biol. Mar 29 2011; 90(1): 199–204. [DOI] [PubMed] [Google Scholar]
- 43. Chan TO, Funakoshi H, Song J, Zhang XQ, Wang J, Chung PH, DeGeorge BR, Jr. , Li X, Zhang J, Herrmann DE, et al. Cardiac‐restricted overexpression of the A(2A)‐adenosine receptor in FVB mice transiently increases contractile performance and rescues the heart failure phenotype in mice overexpressing the A(1)‐adenosine receptor. Clin Transl Sci. Sep 2008; 1(2): 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hamad EA, Li X, Song J, Zhang X‐Q, Myers V, Funakoshi H, Zhang J, Wang J, Li J, Swope D, et al. Effects of cardiac‐restricted overexpression of the A2A adenosine receptor on adriamycin‐induced cardiotoxicity. Am J Physiol. June 1, 2010; 298(6): H1738–H1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNA in failing human heart and remodeling with mechanical circulatory support. Circulation. Jan 15 2014; 129(9): 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]


