Abstract
Transcription of bacteriophage T4 late genes requires concomitant DNA replication. T4 late promoters, which consist of a single 8-bp –10 motif, are recognized by a holoenzyme containing Escherichia coli RNA polymerase core and the T4-encoded promoter specificity subunit, gp55. Initiation of transcription at these promoters by gp55-holoenzyme is inefficient, but is greatly activated by the DNA-loaded DNA polymerase sliding clamp, gp45, and the coactivator, gp33. We report that gp33 attaches to the flap domain of the Escherichia coli RNA polymerase β-subunit and that this interaction is essential for activation. The β-flap also mediates recognition of –35 promoter motifs by binding to σ70 domain 4. The results suggest that gp33 is an analogue of σ70 domain 4 and that gp55 and gp33 together constitute two parts of the T4 late σ. We propose a model for the role of the gp45 sliding clamp in activation of T4 late-gene transcription.
Keywords: RNA polymerase structure, sliding clamp, transcription–replication coupling
Transcription of the bacteriophage T4 late genes, which constitute ≈40% of the genome, depends on ongoing DNA replication. The connection between late transcription and genome replication is created by the phage gene 45 protein (gp45), which is the sliding-clamp processivity subunit of T4 DNA polymerase holoenzyme (1), and is also the activator of T4 late transcription (2, 3). The gp45 sliding clamp is a head-to-tail trimer; in crystals, the trimeric ring takes the form of a triangle with bent sides; in solution, the ring is slightly open, like a lock washer (4–7). To activate transcription, gp45 normally must be loaded on DNA by the clamp-loading complex (gp44–62) at primer-template junctions or single-strand breaks. Once loaded, the sliding clamp moves freely along DNA, capable of carrying its ligands, gp43 (the T4 DNA polymerase) and two T4-encoded, Escherichia coli RNA polymerase (RNAP) core-binding proteins, gp55 (the late promoter specificity subunit) and gp33 (the transcriptional coactivator), along with it (8–10).
The ≈40 T4 late promoters contain an 8-bp –10 motif (consensus sequence TATAAATA); there are no other DNA sequence requirements (11). T4 late promoters are specifically recognized by holoenzyme comprising the Escherichia coli RNAP core (subunit composition α2ββ′) and gp55, but the resultant basal transcription is not efficient. Gp33 and gp45 together activate T4 late transcription up to several hundred-fold by strengthening promoter binding and speeding up promoter opening (3).
How the DNA polymerase sliding clamp mediates activation of transcription is not well understood. It has been shown that gp55, gp33, and gp45 are all part of the activated T4 late-promoter complex; gp33 and gp45 are located near DNA, 35–40 bp upstream of the transcriptional start site (12, 13). Whereas gp55 and gp33 bind to RNAP core directly, gp45 interacts with the late-promoter complex through C-terminal hydrophobic motifs of gp55 and gp33. The intact motifs in both gp55 and gp33 are required for gp45-mediated transcriptional activation (3, 14).
Gp55 is a truncated, highly divergent member of the σ70 family [named after the housekeeping E. coli promoter specificity subunit, σ70 (15)]. The very weak similarity of the 185 aa gp55 to the σ70-family subunits is confined to homology segment 2 (15–17), which contains the major core-binding site and the region responsible for recognition of –10 promoter motifs. Gp55 has been shown to bind to the same RNAP core site as σ70 domain 2 (18, 19). It is not known whether other interactions between gp55 and RNAP exist.
The acidic 112 aa gp33 has no similarity to heterologous ORFs in public databases and no predicted functional motifs. Gp33 does not bind DNA; it binds equally well to RNAP core and gp55 holoenzyme, but is displaced from RNAP core by σ70 (20). Although gp33 is required for activation, it represses basal transcription by gp55 holoenzyme in the absence of gp45 (21).
Here, we report that the binding site of gp33 on RNAP is the β-flap domain, the region of RNAP that mediates recognition of –35 promoter motifs by binding to σ70 domain 4 (22). We show that RNAP with a small deletion at the tip of the flap loses the ability to be activated by gp45 and gp33 (together) or repressed by gp33 (alone). We also demonstrate that deleting σ70 domain 4, or disabling its ability to bind to the RNAP flap domain by point mutation, makes the resultant σ70 holoenzyme accessible to gp33. In addition, a peptide scan of gp33 is used to identify the gp33 segment that determines binding to RNAP core. These results are incorporated into a model of T4 late transcription and its activation.
Materials and Methods
Plasmids and Proteins. Preparation of gp45, gp44–62 complex, gp32, gp55, gp33, and E. coli RNA polymerase core C-terminally His6-tagged in the β′-subunit has been described or referenced (23). Plasmids for overproducing individual RNAP core subunits and β-subunit fragments [N-terminal: amino acids 1–643; C-terminal: amino acids 644–1,342, containing the ΩT988 insertion (24)] were kindly provided by K. Severinov (Waksman Institute, Rutgers, The State University of New Jersey, Piscataway). An expression plasmid for σ70R541C/L607P was kindly provided by A. Hochschild (Harvard Medical School, Boston) (25). A plasmid coexpressing RNAP core subunits α, β′, and βΔ900–909 was kindly provided by I. Toulokhonov and R. Landick (University of Wisconsin, Madison) (26). These proteins were purified by following methods specified in the references cited above. Wild-type σ70, σ70Δ4 (containing amino acids 1–528), and σ70Δ3,4 (amino acids 1–448) were prepared as described (27). Plasmids expressing β-flap-chitin-binding domain (CBD) fusion genes were constructed by subcloning the fragment of rpoB corresponding to codons 873–932 from plasmids containing wild-type or Δβ900–909 rpoB into pTYB1 vector. The fusion proteins were overproduced and immobilized on purchased chitin beads according to the manufacturer's instructions. The plasmid expressing gp33 with a C-terminal protein kinase A phosphorylation site and His6 tag was made by adding codons for RRASVH6 to the C terminus of the gp33 ORF in plasmid pET21gp33 (23). T4 AsiA protein was generously provided by M. Ouhammouch (University of California at San Diego).
32P Labeling of gp33. Forty picomoles of C-terminally protein kinase A site- and His6-tagged gp33 were incubated with 60 μCi of [γ32-P]ATP (6000 Ci/mmol; 1 Ci = 37 GBq) and 20 units of bovine heart muscle protein kinase A for 30 min at 37°C in 20 μl of 20 mM Tris·HCl, pH 7.5/100 mM NaCl/10 mM MgCl2/7.5 mM DTT. The reaction mixture was desalted on a spin column equilibrated in 10 mM Tris·HCl, pH 8.0/250 mM NaCl/0.1 mM EDTA. Glycerol was added to 15% (vol/vol) after desalting, and the protein was stored at –80°C.
Transcription in Vitro. Single-round transcription of the partially single-stranded template placO-SK110-rrnB(T1-T2) was performed as described (3). Activated transcription assays were performed at 20°C; repressed basal transcription was assayed at 25°C in the same buffer but without polyethylene glycol 3350.
Peptide Scanning of gp33. Peptides were synthesized on cellulose membrane as described (28). After deprotection by trifluoroacetic acid, the membrane was incubated for 10 h at 20°C in 20 mM Tris·HCl, pH 8/150 mM NaCl/0.1%(wt/vol) sucrose/0.1%(wt/vol) Tween 20 containing 1× blocking buffer (Sigma), and then for 30 min in the same solution (without blocking buffer) containing 100 nM RNAP core. After three washes with the same buffer (without blocking buffer), monoclonal antibody 11D11 (27) covalently linked to horseradish peroxidase was added for 30 min. The membrane was washed three times with the same buffer and developed with enhanced chemiluminescence reagent.
Affinity Chromatography. Nickel-nitrilotriacetic acid agarose beads (20 μl volume) were incubated with 200 pmol of N-terminally His6-tagged gp33 for 20 min at 20°C in 100 μl of 10 mM Tris·HCl, pH 8.0/200 mM NaCl/5% (vol/vol) glycerol/5 mM imidazole/5 mM 2-mercaptoethanol/0.1% (wt/vol) Tween 20 with gentle shaking for 30 min and then washed twice with 1 ml of the same buffer. The beads were then combined with 100 μl of the same buffer containing 50 pmol of the appropriate RNAP subunit and gently shaken for an additional 30 min, followed by three 1-ml washes with the same buffer. Retained proteins were eluted with 30 μl of the same buffer also containing 200 mM imidazole. Samples were analyzed by SDS/PAGE and stained with SYPRO orange dye. Binding of gp33 to CBP–flap fusions was analyzed by incubating 20 μl of chitin beads, containing 0.5 mg CBD-fusion protein per ml of beads, with 50 pmol of [32P]gp33. Samples were processed as above, except that gp33 was eluted from beads by adding SDS to 0.2% (wt/vol) and boiling for 1 min. Proteins were resolved by SDS/PAGE and analyzed by autoradiography.
Native-Gel Electrophoresis of RNAP–gp33 Complexes. One picomole of RNAP core, 5 pmol of σ-subunit (as required), and various amounts (from 0.25 to 10 pmol) of [32P]gp33 were combined in 5 μl of 20 mM Tris·HCl, pH 7.5/200 mM NaCl/10 mM MgCl2/0.1 mM DTT/5% (vol/vol) glycerol/0.1% (wt/vol) Tween-20, and incubated for 5 min at 25°C. For competition experiments, 0.5 pmol of [32P]gp33 combined with 0.5–10 pmol of unlabeled competitor gp33 was used in each reaction. A 1-μl aliquot from each reaction was loaded on a 4–15% gradient PHAST gel. Gels were run in a PHAST apparatus, by using native buffer strips, at 4°C. Protein complexes were visualized by Coomassie R-250 staining, and binding of [32P]gp33 to RNAP was assessed by autoradiography.
Results and Discussion
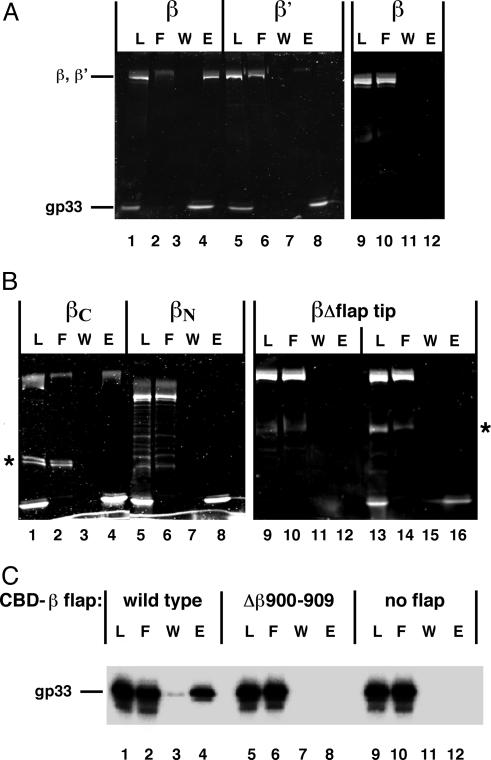
Identification of the gp33-Binding Site on E. coli RNA Polymerase. Binding of individual core subunits to N-His6-gp33 immobilized on nickel-nitrilotriacetic acid beads was analyzed to locate the gp33-binding site on RNAP core. As shown in Fig. 1A, β (lane 4), but not β′ (lane 8), was retained on gp33-containing beads; β was not retained on beads in the absence of gp33 (lane 12). The α-subunit did not bind to gp33 (data not shown). The same experiment with the N- and C-terminal β halves showed only the C-terminal half binding to gp33 (Fig. 1B, compare lanes 4 and 8).
Fig. 1.
Gp33 binds to the RNAP β flap. (A) Gp33 binding to individual RNAP core subunits. RNAP core subunits β (lanes 1–4) and β′ (lanes 5–8) were probed for binding to N-terminally His6-tagged gp33 immobilized on nickel-nitrilotriacetic acid agarose as described in Materials and Methods.L, 1/10th of the material (gp33 combined with β or β') loaded on beads; F, flow-through (1/10th of the unbound fraction); W, 1/10th of the last wash; E, one-half of the fraction eluted with imidazole. Lanes 9–12 show the negative control of binding to beads without gp33. (B) Binding of gp33 to β fragments (lanes 1–8) and Δflap tip β (Δ884–914) (lanes 13–16) (22), lanes 9–12 as in A. Bands indicated by asterisks are unknown polypeptides contaminating partially purified β-fragments. (C) Binding of [32P]gp33 to the isolated flap. Gp33 binding to CBD fused to β-fragments corresponding to amino acids 873–932: wild-type (lanes 1–4), deleted for the flap-tip helix (βΔ900–909) (lanes 5–8), or CBD alone (lanes 9–12).
Analysis of the structure of bacterial RNAP core reveals the flap as the only prominent feature contained completely within the β C-terminal half (29). The β-flap is located at the upstream end of promoter complexes (reviewed in ref. 30) and gp33 is located at the upstream end of the T4 late-promoter complex (13). Thus, it is possible that gp33 might bind to the β-subunit flap domain. β-subunit with a deletion of codons 884–914, that is, lacking the tip of the flap (22), was tested for gp33 binding. As shown in Fig. 1B, deletion of the β-flap tip eliminated gp33 binding (lane 16), specifying that the flap tip is required for binding of gp33 to the β-subunit.
To determine whether the β-flap suffices to bind gp33, the isolated flap domain fragment (amino acids 873–932) was fused to the CBD and immobilized on chitin beads. As shown in Fig. 1C, gp33 was retained on beads containing the wild-type flap–CBD fusion protein (lane 4), but not on beads containing CBD alone (lane 12), or fusion protein deleted for β-amino acids 900–909 in the flap-tip helix (lane 8). Thus, the β-flap domain suffices, and the flap-tip helix is essential for gp33 binding to RNAP.
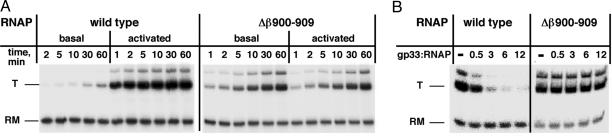
The Flap-Tip Helix Is Required for the Response of RNAP to gp33. To determine whether interaction of gp33 with the β-flap is essential for gp33-mediated activation or repression of basal T4 late transcription, βΔ900–909 RNAP core was used for in vitro transcription from a T4 late promoter. RNAP core, gp55, gp45, gp44–62 complex (for basal transcription), and gp33 (for activated transcription) were incubated with promoter DNA for various periods of time at 20°C, followed by simultaneous addition of rNTPs and heparin to enforce a single round of transcription, thus measuring the accumulation of open promoter complexes over time (3). As shown in Fig. 2A Left, wild-type RNAP demonstrated the expected response to activation: under conditions of basal transcription at 20°C, open promoter complexes accumulated extremely slowly, whereas under activating conditions, the formation of open promoter complexes reached saturation shortly after 1 min. In sharp contrast, transcription by the βΔ900–909 RNAP did not respond to activation (Fig. 2 A Right).
Fig. 2.
Deletion of the flap-tip helix eliminates the response of RNAP to gp33. (A) Basal and activated transcription with wild-type and Δβ900–909 RNAPs. Reactions containing RNAP core, gp55, gp45, and gp44–62, and for activated transcription, gp33, were incubated at 20°C for the indicated time. A single round of transcription was then performed as described in Materials and Methods. An autoradiograph of the denaturing gel is shown. T, transcript; RM, recovery marker. (B) Repression of basal transcription by gp33. RNAP was combined with gp55, DNA template, and increasing amounts of gp33 at 25°C for 15 min (the gp33:RNAP core ratio is indicated). A single round of transcription was then carried out as above.
Repression of basal transcription by gp33 was examined in the same way, except that gp45 was omitted and the effect of increasing gp33 concentration was monitored at a fixed time (15 min of promoter opening) at 25°C. Basal transcription by the wild-type RNAP was strongly repressed by gp33, as expected, whereas βΔ900–909 RNAP was insensitive to gp33 (Fig. 2B). These results clearly show that the β-flap-tip helix is required both for gp33-mediated activation and for repression of basal T4 late transcription.
We note that the basal rate of promoter opening is greater for βΔ900–909 RNAP than for the wild-type enzyme (Fig. 2 A); the difference remains pronounced at higher temperature (30°C; data not shown). It is possible that the flap impedes promoter opening and that removing its tip relieves the block.
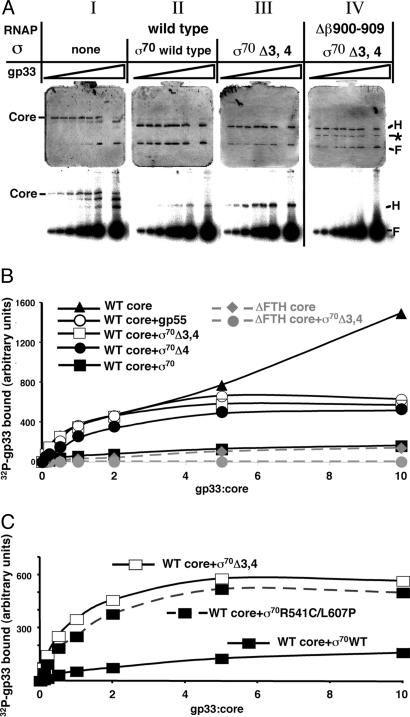
Competition Between gp33 and σ70 Domain 4 for RNAP Binding. The observation that gp33 binds to the gp55 RNAP holoenzyme, but not the σ70 holoenzyme (20), implies that a domain of σ70 without a gp55 counterpart blocks access of gp33 to the σ70 holoenzyme. Domain 4 of σ70, which binds to the flap (22), is the most likely candidate to constitute the block. The next experiment aimed to determine whether removal of σ70 domain 4 would allow binding of gp33 to the σ70-deletion RNAP holoenzyme. RNAP core was combined with σ70 variant proteins; increasing amounts of [32P]gp33 were added, and complexes were separated by native PAGE; the formation of holoenzymes was detected by Coomassie dye staining, and binding of gp33 was revealed by autoradiography. All σ-subunits formed sufficiently stable RNAP holoenzymes to be detected by native PAGE (Fig. 3A and data not shown).
Fig. 3.
Competition between gp33 and σ70 domain 4 for RNAP binding. Reactions containing RNAP core, the appropriate σ (where indicated), and increasing amounts of [32P]gp33 were resolved by native PAGE as described in Materials and Methods.(A) Native gels stained with Coomassie dye (Top) and autoradiographed (Bottom). Bands corresponding to RNAP core, holoenzyme (H) and the dye front (F) containing excess [32P]gp33 and σ are indicated. The band marked by the asterisk is residual α2βΔ900–909 complex in the Δflap-tip helix RNAP (Δβ900–909; ΔFTH) preparation. (B) Quantitative analysis of [32P]gp33 binding to RNAP core and various holoenzymes. The graph shows relative amounts of [32P]gp33 bound to the indicated RNAP complex as a function of the gp33:core ratio. (C) Quantitative analysis of [32P]gp33 binding to σ70 wild-type (WT)-, σ70 Δ3,4-, and σ70(R541C/L607P)-holoenzymes.
Several features of gp33 binding to RNAP should be noted. (i) Binding to the wild-type core did not saturate, even at 10-fold molar excess of gp33 (Fig. 3B and data not shown). Inspection of the same gels stained with Coomassie dye (Fig. 3AI, upper gel) revealed that the core, running as a single band in the absence of gp33 or in the presence of low concentrations of gp33, runs as multiple bands in the presence of high excess of gp33. (ii) Binding of gp33 to gp55 holoenzyme was comparable with binding to core at low gp33 concentrations but reached saturation at higher gp33 concentrations (Fig. 3B). This finding is in agreement with earlier studies (20) and suggests that excess gp33 binding to core is nonspecific. (iii) Binding of gp33 to full-sized σ70 holoenzyme was at least 10-fold weaker than its binding to RNAP core or gp55 holoenzyme, but was reproducibly observed (Fig. 3 AII and B). It had been earlier concluded that gp33 does not bind to the σ70 holoenzyme, but careful examination of the data (20) also shows residual gp33 binding to the σ70 holoenzyme that had been previously discounted. (iv) Removing domains 3 and 4 of σ70 or domain 4 alone allowed binding of gp33 to the resultant holoenzymes to greatly increase and, moreover, saturate at higher gp33 concentrations (Fig. 3 AIII and B).
Gp33 bound to βΔ900–909 RNAP core much more weakly than to wild-type core; binding of gp33 to βΔ900–909 σ70Δ3,4 holoenzyme was not detectable (Fig. 3 AIV and B). These results imply that the sites on the core to which gp33 binds nonspecifically are occluded by σ70 domains 1 and 2, and also confirm that specific binding of gp33 to RNAP is completely eliminated by a deletion of the flap-tip helix.
If deletion of σ70 domain 4 allows specific binding of gp33 to the resultant holoenzyme, might a point mutation that weakens the interaction of σ70 region 4 with the β flap do the same? A recently described σ70 mutant, R541C/L607P in domain 4, is specifically defective in binding to the β-flap (25). The results presented in Fig. 3C show that gp33 binds to RNAP holoenzyme containing the mutant full-length σ70 as tightly as it does to the holoenzymes containing gp55 or σ70Δ3,4. Thus, deleting σ70 domain 4 or weakening its interaction with the β-flap by point mutations allows binding of gp33 to the resultant holoenzyme, specifying that gp33 competes with σ70 domain 4 for RNAP core binding.
Effect of AsiA on Competition Between gp33 and σ70 Domain 4 for RNAP Core Binding. AsiA is an effector of T4 middle transcription that binds to region 4 of σ70 and disrupts its interaction with the β-flap (25, 31). Having established that destabilizing the interaction with of σ70 domain 4 with the flap by mutation increases binding of gp33 to the σ70 holoenzyme, we determined whether destabilizing the domain 4–β-flap interaction with AsiA would do the same. As shown in Fig. 6, which is published as supporting information on the PNAS web site, addition of AsiA resulted in only a marginal (but consistently observed) increase of gp33 binding to the wild-type σ70 holoenzyme. That AsiA does not fully restore gp33 binding to the σ70 holoenzyme suggests that it might sterically hinder gp33 binding. If so, then AsiA should also interfere with gp33 binding to the σ70(R541C/L607P)-holoenzyme, and this is what was observed (Fig. 6). The result specifies that although AsiA and gp33 have different targets on RNAP, their binding sites are located close together.
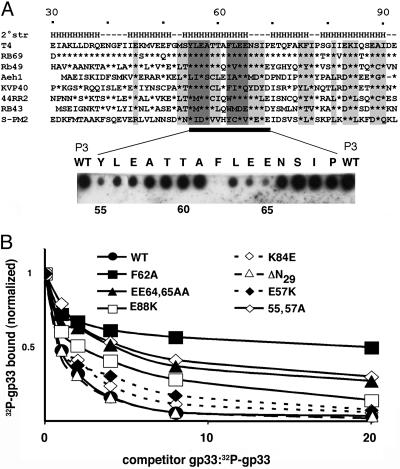
Peptide Scanning of gp33. We have searched for the region of gp33 that binds to RNAP core. A series of 15-mer peptides spanning the entire length of gp33, in two-residue steps, was synthesized on cellulose membrane and probed for RNAP core binding. Three peptides, designated P1-P3 (T4 gp33 amino acids 31–45, 51–65 and 55–69, respectively), yielded positive signals, with P3 displaying the highest affinity (Fig. 7, which is published as supporting information on the PNAS web site). The same peptide scan was also performed with the gp33-like ORF from bacteriophage RB49, a close relative of T4: only one RB49 peptide, corresponding to P3, gave a positive signal, indicating binding to the RNAP core (data not shown). The region of T4 gp33 corresponding to peptide P3 was next subjected to an alanine scan to identify single amino acids important for gp33 binding to RNAP. Fifteen versions of P3, each containing a single alanine substitution (or glycine instead of natural alanine), were tested for binding to the RNAP core, as above. As shown in Fig. 4A, substitutions in two short segments, amino acids 55–57 and 62–65, resulted in the most pronounced defects in core binding.
Fig. 4.
An RNAP core-binding domain of gp33. (A Upper) Amino acid sequence alignment of gp33-like ORFs from T4-group bacteriophages (segments corresponding to T4 gp33 amino acids 30–92 are shown; sequences can be accessed at http://phage.bioc.tulane.edu). Asterisks indicate identity with the corresponding amino acid of T4 gp33. Sequence conservation is highlighted in light gray, and the two acidic/hydrophobic motifs (F/Y)LE(E/A) are dark gray. The predicted secondary structure (a consensus of secondary structure predictions performed at the META PredictProtein Server, which can be accessed at http://cubic.bioc.columbia.edu/pp) is indicated above the T4 gp33 sequence. H, α-helix. (Lower) Alanine scanning of gp33 amino acids 55–69 (with Gly replacing Ala-58 and Ala-61). The peptides were synthesized on a membrane and probed for RNAP core binding, as indicated in Materials and Methods. The replaced amino acid residues are shown above the corresponding spot. (B) Binding defects of gp33 mutants, judged by native PAGE. RNAP core was incubated with σ70Δ3,4 (to block nonspecific gp33 binding). A mixture containing a fixed amount of wild-type [32P]gp33 and increasing amounts of unlabeled competitor wild-type or mutant gp33 was then added, and reaction products were resolved on native PAGE as described in Materials and Methods. The graph shows relative amounts of [32P]gp33 retained by RNAP as a function of the ratio of unlabeled competitor gp33 to wild-type [32P]gp33.
Functional Defects of gp33 Mutants. Mutations identified in the peptide scan, as well as substitutions at several other positions of sequence conservation among gp33-like ORFs from T4-group bacteriophages (Fig. 4A), were introduced into full-length gp33. Binding of these gp33 variants to RNAP was evaluated by testing their ability to compete with 32P-tagged wild-type gp33 for binding RNAP, as judged by native PAGE. As shown in Fig. 4B, wild-type gp33 competed well, as expected, whereas the point mutants were defective to various degrees in their ability to compete with wild-type [32P]gp33. Notably, the F62A substitution, identified as the most severely defective in the peptide scan (Fig. 4A), also showed the most pronounced core-binding defect in the context of full-sized gp33. A deletion of the N-terminal 29 aa of gp33 did not affect its ability to compete for binding to RNAP (Fig. 4B). [This finding is in contradiction with an earlier result, pointing to a severe core-binding defect generated by an internal deletion of N-proximal amino acids 18–27 (32). The sequence of the N-terminal 30 aa of T4 gp33 is not conserved, and the entire segment is absent from some gp33-like ORFs. The prior observation may have reflected misfolding associated with the internal deletion.]
Three of the binding-defective gp33 mutants (F62A, E64, 65AA, and E88K) were assayed for their ability to repress basal transcription (Fig. 8, which is published as supporting information on the PNAS web site): all three were significantly defective.
Despite absence of sequence similarity to σ70, the predicted secondary structure of gp33 shows an alignment of helices that is similar to that of σ70 domain 4 (Fig. 4A and refs. 33 and 34). The peptide scan of gp33 and follow-up mutagenesis identify two short hydrophobic/acidic motifs important for gp33 binding to RNAP. Both motifs are located within a predicted gp33 helix (Fig. 4A), which may bind directly to the flap, possibly to the flap-tip helix. The secondary structure of free gp33 appears to be unstable (35), suggesting that its ligand, the β-flap domain, may be required to stabilize gp33 folding. Finding that gp33 interacts with the isolated flap (Fig. 1C) will facilitate structural analysis of this RNAP core interface.
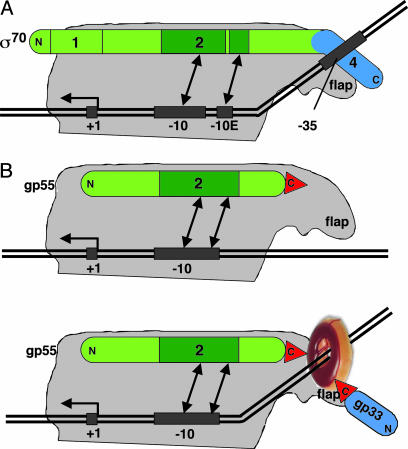
A Model of T4 Late Transcription. The presented findings suggest a model of T4 late transcription and its activation. According to the model, gp33 can be viewed as a structural analogue of σ70 domain 4. Collectively, gp33 and gp55, which provides σ domain 2, can be viewed as two parts of the T4 late σ-subunit. It is plausible to suppose that a role of the DNA-loaded gp45 sliding clamp in T4 late-transcription initiation is to fix these two parts of T4 late σ in an orientation that is favorable for promoter opening (Fig. 5). The model specifies direct stimulation of promoter opening by gp45, in agreement with kinetic analysis and with the observation that promoter opening under activated conditions is robust, even at temperatures as low as 5°C (ref. 3 and S. Kolesky, unpublished observations). Unlike σ70 domain 4, gp33 does not interact with DNA directly, but instead, does so through DNA-loaded gp45. We suggest that this interaction is functionally analogous to DNA sequence-specific interaction of σ70 region 4 with the –35 promoter motif. The provision of a topological link to DNA by the gp45 sliding clamp establishes this interaction independent of DNA sequence (Fig. 5).
Fig. 5.
A model of T4 late-transcription activation. (A) A scheme of the open complex on a σ70-dependent promoter. (B) A model of the T4 late-transcription initiation complex under basal (Upper) and activated (Lower) conditions. DNA is shown as a solid double line; the transcription start site (+1), –35, and –10 motifs (including the extended –10 motif, –10E) are indicated with black rectangles. RNAP core is gray, σ70 and gp55 are green, and gp33 and σ70 domain 4 are blue. Kinking of upstream DNA symbolizes a conformational change facilitating promoter opening. The double arrows show interactions of σ domain 2 (dark green) with –10 and –10 extended promoter motifs. The DNA-loaded gp45 trimer is shown as a ring. Red triangles symbolize the C-terminal hydrophobic motifs of gp55 and gp33 that mediate binding to gp45.
In conclusion, we note that the transcription program of bacteriophage T4 appears to be primarily generated by targeting the interaction between σ domain 4, the RNAP flap, and the –35 motif of T4 promoters. Early promoters contain a –35 motif that is recognized through interaction with σ70 domain 4. At middle promoters, this interaction is replaced with a new interaction between two T4 middle transcription factors, MotA and AsiA, bound to the motA box motif and σ domain 4, respectively (36, 37). At late promoters, gp33 and gp45 constitute the T4 late replacement of σ domain 4, substituting DNA sequence-specific recognition of the –35 motif with topological DNA linkage.
Supplementary Material
Acknowledgments
We thank A. Hochschild, I. Toulokhonov, R. Landick, L. Minakhin (Rutgers, The State University of New Jersey), and K. Severinov for plasmids; L. C. Anthony and R. R. Burgess (University of Wisconsin, Madison) for a generous gift of material that allowed this work to start on the right track; H. Krisch (Centre National de la Recherche Scien-tifique, Toulouse, France) for communicating unpublished phage sequences; and R. Driscoll, R. R. Burgess, G. A. Kassavetis, and K. Severinov for a careful reading of the text. This work was supported by the National Institute of General Medical Sciences.
Author contributions: S.N., M.K.-M., J.-P.L., and E.P.G. designed research; S.N., M.K.-M., E.A., and J.-P.L. performed research; S.N. J.-P.L., and E.P.G. analyzed data; and S.N. and E.P.G. wrote the paper.
Abbreviations: RNAP, Escherichia coli RNA polymerase; CBD, chitin-binding domain.
References
- 1.Nossal, N. G. (1994) in Molecular Biology of Bacteriophage T4, ed. Karam, J. D. (Am. Soc. Microbiol., Washington, DC), pp. 43–54.
- 2.Herendeen, D. R., Kassavetis, G. A. & Geiduschek, E. P. (1992) Science 256, 1298–1303. [DOI] [PubMed] [Google Scholar]
- 3.Kolesky, S. E., Ouhammouch, M. & Geiduschek, E. P. (2002) J. Mol. Biol. 321, 767–784. [DOI] [PubMed] [Google Scholar]
- 4.Hingorani, M. M. & O'Donnell, M. (2000) Curr. Biol. 10, R25–R29. [DOI] [PubMed] [Google Scholar]
- 5.Moarefi, I., Jeruzalmi, D., Turner, J., O'Donnell, M. & Kuriyan, J. (2000) J. Mol. Biol. 296, 1215–1223. [DOI] [PubMed] [Google Scholar]
- 6.Shamoo, Y. & Steitz, T. A. (1999) Cell 99, 155–166. [DOI] [PubMed] [Google Scholar]
- 7.Trakselis, M. A., Alley, S. C., Abel-Santos, E. & Benkovic, S. J. (2001) Proc. Natl. Acad. Sci. USA 98, 8368–8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berdis, A. J., Soumillion, P. & Benkovic, S. J. (1996) Proc. Natl. Acad. Sci. USA 93, 12822–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alley, S. C., Jones, A. D., Soumillion, P. & Benkovic, S. J. (1999) J. Biol. Chem. 274, 24485–24489. [DOI] [PubMed] [Google Scholar]
- 10.Tinker-Kulberg, R. L., Fu, T. J., Geiduschek, E. P. & Kassavetis, G. A. (1996) EMBO J. 15, 5032–5039. [PMC free article] [PubMed] [Google Scholar]
- 11.Kassavetis, G. A., Zentner, P. G. & Geiduschek, E. P. (1986) J. Biol. Chem. 261, 14256–14265. [PubMed] [Google Scholar]
- 12.Tinker, R. L., Kassavetis, G. A. & Geiduschek, E. P. (1994) EMBO J. 13, 5330–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tinker, R. L., Williams, K. P., Kassavetis, G. A. & Geiduschek, E. P. (1994) Cell 77, 225–237. [DOI] [PubMed] [Google Scholar]
- 14.Sanders, G. M., Kassavetis, G. A. & Geiduschek, E. P. (1997) EMBO J. 16, 3124–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribskov, M. & Burgess, R. R. (1986) Nucleic Acids Res. 14, 6745–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmann, J. D. & Chamberlin, M. J. (1988) Annu. Rev. Biochem. 57, 839–872. [DOI] [PubMed] [Google Scholar]
- 17.Lonetto, M., Gribskov, M. & Gross, C. A. (1992) J. Bacteriol. 174, 3843–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonetti, J.-P., Wong, K. & Geiduschek, E. P. (1998) EMBO J. 17, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong, K., Kassavetis, G. A., Leonetti, J. P. & Geiduschek, E. P. (2003) J. Biol. Chem. 278, 7073–7080. [DOI] [PubMed] [Google Scholar]
- 20.Herendeen, D. R., Williams, K. P., Kassavetis, G. A. & Geiduschek, E. P. (1990) Science 248, 573–578. [DOI] [PubMed] [Google Scholar]
- 21.Williams, K. P., Muller, R., Ruger, W. & Geiduschek, E. P. (1989) J. Bacteriol. 171, 3579–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuznedelov, K., Minakhin, L., Niedziela-Majka, A., Dove, S. L., Rogulja, D., Nickels, B. E., Hochschild, A., Heyduk, T. & Severinov, K. (2002) Science 295, 855–857. [DOI] [PubMed] [Google Scholar]
- 23.Kolesky, S., Ouhammouch, M., Brody, E. N. & Geiduschek, E. P. (1999) J. Mol. Biol. 291, 267–281. [DOI] [PubMed] [Google Scholar]
- 24.Wang, Y., Severinov, K., Loizos, N., Fenyo, D., Heyduk, E., Heyduk, T., Chait, B. T. & Darst, S. A. (1997) J. Mol. Biol. 270, 648–662. [DOI] [PubMed] [Google Scholar]
- 25.Gregory, B. D., Nickels, B. E., Garrity, S. J., Severinova, E., Minakhin, L., Urbauer, R. J., Urbauer, J. L., Heyduk, T., Severinov, K. & Hochschild, A. (2004) Proc. Natl. Acad. Sci. USA 101, 4554–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toulokhonov, I., Artsimovitch, I. & Landick, R. (2001) Science 292, 730–733. [DOI] [PubMed] [Google Scholar]
- 27.Rouby, J., Pugniere, M., Mani, J. C., Granier, C., Monmouton, P., Theulier Saint Germain, S. & Leonetti, J.-P. (2002) Biochem. J. 361, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molina, F., Laune, D., Gougat, C., Pau, B. & Granier, C. (1996) Pept. Res. 9, 151–155. [PubMed] [Google Scholar]
- 29.Zhang, G., Campbell, E. A., Minakhin, L., Richter, C., Severinov, K. & Darst, S. A. (1999) Cell 98, 811–824. [DOI] [PubMed] [Google Scholar]
- 30.Murakami, K. S. & Darst, S. A. (2003) Curr. Opin. Struct. Biol. 13, 31–39. [DOI] [PubMed] [Google Scholar]
- 31.Lambert, L. J., Wei, Y., Schirf, V., Demeler, B. & Werner, M. H. (2004) EMBO J. 23, 2952–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkelman, J. W., Kassavetis, G. A. & Geiduschek, E. P. (1994) J. Bacteriol. 176, 1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell, E. A., Muzzin, O., Chlenov, M., Sun, J. L., Olson, C. A., Weinman, O., Trester-Zedlitz, M. L. & Darst, S. A. (2002) Mol. Cell 9, 527–539. [DOI] [PubMed] [Google Scholar]
- 34.Vassylyev, D. G., Sekine, S., Laptenko, O., Lee, J., Vassylyeva, M. N., Borukhov, S. & Yokoyama, S. (2002) Nature 417, 712–719. [DOI] [PubMed] [Google Scholar]
- 35.Shao, W., Kearns, D. R. & Sanders, G. M. (1997) Int. J. Biol. Macromol. 20, 115–121. [DOI] [PubMed] [Google Scholar]
- 36.Ouhammouch, M., Adelman, K., Harvey, S. R., Orsini, G. & Brody, E. N. (1995) Proc. Natl. Acad. Sci. USA 92, 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinton, D. M., March-Amegadzie, R., Gerber, J. S. & Sharma, M. (1996) J. Mol. Biol. 256, 235–248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.