Abstract
Although cisplatin‐based chemotherapy is considered to be the treatment of choice for metastatic bladder cancer, its efficacy and tolerability has proven to be limited. MicroRNAs are small noncoding RNAs, whose genes are frequently organized in clusters. These molecules constitute posttranscriptional regulators of mRNA expression and are claimed to be deregulated in cancer. miR‐143/145 and miR‐183/96/182 clusters have been extensively studied in bladder cancer cells. Herein, we tried to add up to this knowledge by assessing the expression levels of the five mature microRNAs derived from the aforementioned clusters in T24 bladder cancer cells exposed to either cisplatin or paclitaxel. For both compounds, the viability of treated T24 cells was estimated via the MTT colorimetric assay and the Trypan Blue exclusion method, while a fraction of the cells was left to recover. The expression levels of all mature microRNAs were finally quantified both in treated and in recovered cells by performing real‐time PCR. According to our data, cisplatin and paclitaxel strongly decreased T24 cells' viability, showing in parallel the ability to significantly down‐regulate miR‐143 levels, and up‐regulate the expression levels of miR‐145, miR‐183, miR‐96, and miR‐182, which, in their total, demonstrated case‐specific variations after recovery period.
Keywords: cisplatin, microRNAs, paclitaxel, real‐time PCR, T24 bladder cancer cells
Introduction
American Cancer Society has recently estimated that 74,000 new cases of urinary bladder cancer will be diagnosed during 2015 in U.S., while the respective number of deaths expected for the same year will overcome 15,500.1 The most frequently diagnosed histological type of bladder cancer is transitional cell carcinoma which accounts for approximately 90% of all new cases.2 About 25% of bladder malignant tumors belong to muscle‐invasive bladder cancer (MIBC) which represents all tumors classified as T2 or higher according to the current tumor node metastasis (TNM) classification system.3 The therapeutic approach applied for the management of MIBC is mainly determined according to the spread of the disease.4, 5 The use of chemotherapy and especially that of cisplatin‐containing combinations is considered to be the standard of care for patients with metastatic bladder carcinoma. In particular, according to the latest clinical guidelines, metastatic urinary bladder cancer patients with good performance status and renal function should be treated with either the methotrexate, vinblastine, adriamycin, and cisplatin regimen, known as MVAC, or that of gemcitabine/cisplatin (GC) either alone or together with paclitaxel (PGC).4, 5, 6 Cisplatin, also known as cisplatinum or cis‐diamminedichloroplatinum (II), is a DNA‐damaging agent, which demonstrates the ability to bind to purine residues on DNA and accordingly create intrastrand adducts. The latter interfere with the DNA repair mechanisms of the cells leading to lethal damage to DNA and ultimately cell death via various mechanisms.7 The most recent addition to the combinatory therapeutic regimens recommended for the treatment of metastatic bladder cancer is paclitaxel.4, 5, 6 This drug belongs to a group of mitotic inhibitors known as taxanes, which share the capability of inhibiting cell proliferation by binding to microtubules and thus blocking their disassembly reactions during mitosis.8
MicroRNAs (miRNAs) comprise a novel class of small noncoding RNAs implicated in a wide range of physiological and pathological conditions, including cancer.9 These small molecules of approximately 19–25 nucleotides length are processed from longer primary RNA transcripts which frequently contain the sequences of more than one mature miRNA. In the latter case, the primary microRNA transcript resembles the polycistronic messenger RNAs (mRNAs) detected in bacteria and is considered to be the transcription product of several microRNA genes organized in clusters. MicroRNA biogenesis eventually results in the loading of the functional small RNA molecules onto a ribonucleoprotein complex known as RISC (RNA‐induced silencing complex).10 RISC uses the miRNA as a guide seeking mRNAs whose sequence shares partial complementarity with that of the miRNA. Several mRNAs can be recognized as targets per miRNA, while the outcome of this interaction is the suppression of translation or the degradation of the mRNA, which might occur either alone or in combination.11
Upon a search conducted in the literature, the mature miRNAs derived from the miR‐143/145 and miR‐183/96/182 clusters were found to be among the few miRNAs that their role is well documented in bladder cancer cells.12, 13, 14, 15, 16, 17, 18 Thus, we herein sought to determine whether T24 bladder cancer cells respond to treatment with cisplatin and paclitaxel, indicated for bladder cancer therapy, by modulating the expression levels of these miRNAs.
Methods
Cell culture
The human bladder cancer cell line T24 was cultured in McCoy's 5A Medium Modified with 1.5 mM l‐glutamine (PAA Laboratories GmbH, Pasching, Austria) and supplemented with 10% fetal bovine serum (FBS), 100 kU/L penicillin, and 0.1 g/L streptomycin. Cells were grown in an incubator at 37 °C in 5% CO2.
MTT colorimetric assay
The MTT [3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide] (Sigma‐Aldrich, St. Louis, MO, USA) colorimetric assay was performed as follows: 100 μL of cell suspension containing 1,000 cells was transferred to each well of a 96‐well plate and cells were left overnight inside the incubator in order to adhere and adapt to culture environment. Various concentrations of cisplatin (Cisplatin DBL; Faulding Pharmaceuticals, Warwickshire, United Kingdom) and paclitaxel (Taxol, Bristol‐Myers Squibb, Princeton, NJ, USA) were then added to culture medium of T24 cells in quadruplicate and three different incubation periods of 24, 36, and 48 hours followed. At the end of the aforementioned time intervals, 10 μL of MTT solution (5 mg/mL in phosphate‐buffered saline [PBS]) was added to each well and subsequently plates were transferred inside the incubator for 4 hours. Afterwards, the supernatant of each well was removed and replaced by 100 μL of a lysis solution [12.5% (w/v) SDS and 45% (v/v) formamide] containing a suitable organic solvent for the produced formazan. Finally, the absorbance of all cell lysates was measured at 550 nm (reference wavelength set at 630 nm) with the assistance of a microtiter plate reader (HumaReader; Human GmbH, Wiesbaden, Germany).
Drug treatment and Trypan Blue dye exclusion test
In full proportion to MTT assay protocol, 104 cells/mL were seeded in cell culture flasks and cells were left overnight to adapt to culture environment, inside the incubator. Afterwards, selected concentrations of paclitaxel and cisplatin were added to the culture medium of each vessel, in triplicate, and cancer cells were incubated with either drug for 24, 36, and 48 hours. Following the completion of the 48‐hour incubation period, T24 cells were left to recover in the incubator for 24 hours after drug‐containing culture medium was replaced by fresh drug‐free medium. For the 24‐, 36‐, and 48‐hour treated cells, as well for their respective control counterparts, a Trypan Blue exclusion assay was performed. In brief, cancer cells were first trypsinized with (1×) trypsin‐EDTA (PAA Laboratories GmbH) for 5 min at 37 °C and then pelleted via centrifugation. Subsequently, pelleted cells were resuspended in fresh medium and 50 μL of the produced cell suspension was diluted 1:10 in (1×) PBS and then subjected to Trypan Blue exclusion test. For the execution of the latter, a volume of the diluted cell suspension was mixed with Trypan Blue (Sigma‐Aldrich, St. Louis, MO, USA) following manufacturer's recommendations and sequentially loaded onto a hemocytometer so as to perform the cell count.
RNA extraction
Total RNA from cultured cancer cells was extracted using TRI Reagent (Ambion Ltd, Huntingdon, United Kingdom) according to the manufacturer's instructions. A spectrophotometric analysis at 260 and 280 nm followed the extraction of total RNA so that its concentration and purity to be assessed.
microRNA levels quantification via real‐time PCR
miRNA quantification was achieved by developing a highly sensitive quantitative real‐time PCR protocol for each miRNA included in our study based on the basic principles of the methodology described by Shi and Chiang.19 In particular, 1 μg of total RNA was initially subjected to polyadenylation in a 10 μL‐reaction mixture composed of 2 U of Escherichia coli Poly(A) polymerase (PAP; New England Biolabs, Hertfordshire, United Kingdom), 1 mM ATP, and (1×) PAP buffer, which was incubated at 37 °C for 1 hour. Polyadenylated RNA was then reverse transcribed to cDNA within the same tube (final reaction volume of 20 μL) by adding 0.5 μM of a poly(T) adapter (5'‐GCGAGCACAG AATTAATACGACTCACTATAGGTTTTTTTTTTTTVN‐3',
V = A, G, C; N = A, T, G, C) and M‐MuLV Reverse Transcriptase RNase H– (Finnzymes, Espoo, Finland) according to manufacturer's instructions. Regarding real‐time PCR reactions, SYBR Green I was used as the chemical detection system in a 10 μL‐reaction mixture consisted of 0.1 μL of cDNA, (2×) KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, Charlestown, MA, USA), 200 nM of a universal reverse primer, and 200 nM of a miRNA‐specific forward primer. All sequences of primers used for the execution of PCR reactions can be found in Table 1. The thermal protocol applied for all reactions included one step at 95 °C for 3 min and 40 sequential cycles of two steps performed at 95 °C for 15 s and at 60 °C for 1 min. Each sample was assayed in triplicate, while all PCR reactions ran in an ABI 7500 thermal cycler (Applied Biosystems, Foster City, CA, USA). PCR products specificity was confirmed by conducting melting curve analysis at the end of each reaction. For miRNA levels assessment, a relative quantification (RQ) approach was performed by applying the comparative threshold cycle (C T) (2‐ΔΔC T) method after all prerequisite conditions have been met.20 In brief, the relative expression levels of all mature miRNAs analyzed per sample were measured by firstly determining C T of each miRNA and then normalizing its value to that corresponding to the C T of the small nucleolar RNA, C/D box 48 (SNORD48; also known as RNU48) selected as the appropriate reference gene. RNU48 expression was analyzed by using the same experimental conditions mentioned above. Afterwards, the RQ units of each miRNA per sample, expressed in copies of target gene to copies of reference gene (miRNA copies/RNU48 copies), were calculated by applying the 2‐ΔΔC T formula, in which ΔΔCT stands for the difference between the above normalized C T value (ΔC T = C T miRNA − C T RNU48) and the respective ΔC T value calculated for the same miRNA in the sample set as the calibrator. The untreated (control) cells included in our study per incubation period served as the calibrator in all drug treatment experiments, whereas regarding recovery experiments two distinct RQ units were measured per miRNA either by using the 48‐hour treated cells as a calibrator or by taking the average ΔC T of the three control samples and applying their value to the 2‐ΔΔC T formula. In parallel, in order to verify the validity of our data, the amplification efficiencies for the five miRNAs and RNU48 were estimated by constructing a standard curve per molecule and afterwards applying the formula E(%) = [−1 + 10(‐1 ⁄ α)]×100, where E is the PCR amplification efficiency and α is the slope of the RNA‐specific standard curve constructed.20
Table 1.
Sequences of primers used in real‐time PCR reactions
| Small RNA‐specific forward primers | Accession Number (Database) | |
|---|---|---|
| RNU48 | 5'‐TGATGATGACCCCAGGTAACTCT‐3' | NR_002745.1 (GenBank) |
| miR‐183 | 5'‐TATGGCACTGGTAGAATTCACT‐3' | MIMAT0000261 (miRBase) |
| miR‐96 | 5'‐TGGCACTAGCACATTTTTGCTAAA‐3' | MIMAT0000095 (miRBase) |
| miR‐182 | 5'‐TTTGGCAATGGTAGAACTCACA‐3' | MIMAT0000259 (miRBase) |
| miR‐143 | 5'‐TGAGATGAAGCACTGTAGCTCAAA ‐3' | MIMAT0000435 (miRBase) |
| miR‐145 | 5'‐CCAGTTTTCCCAGGAATCCCTAA‐3' | MIMAT0000437 (miRBase) |
| Universal reverse primer | 5'‐GCGAGCACAGAATTAATACGAC‐3' |
Statistical analysis
The statistical significance of all variations observed upon the pairwise comparisons performed between the differentially treated cells was estimated using a 2‐tailed Student's t‐test (p < 0.05).
Results
Cytotoxic activity of cisplatin and paclitaxel against T24 cells
Exposure of T24 cells to different concentrations of either cisplatin or paclitaxel for 24, 36, and 48 hours revealed the capability of both drugs to decrease the viability of these cells in a time‐ and dose‐dependent manner (Figure 1). Upon the same process the maximal inhibitory concentration (IC50) of each drug was calculated and found to be equal to 2 μM for cisplatin and 3 nM for paclitaxel, respectively. By adding the aforementioned IC50 doses of these drugs to the culture medium of T24 cells and sequentially performing Trypan Blue assay, we found that the decrease noticed in cell viability was accompanied by almost no disruption of T24 cells' membrane as regardless of the incubation period that preceded, the percentage of Trypan Blue‐positively stained cancer cells did not exceed 3.5% in either case.
Figure 1.
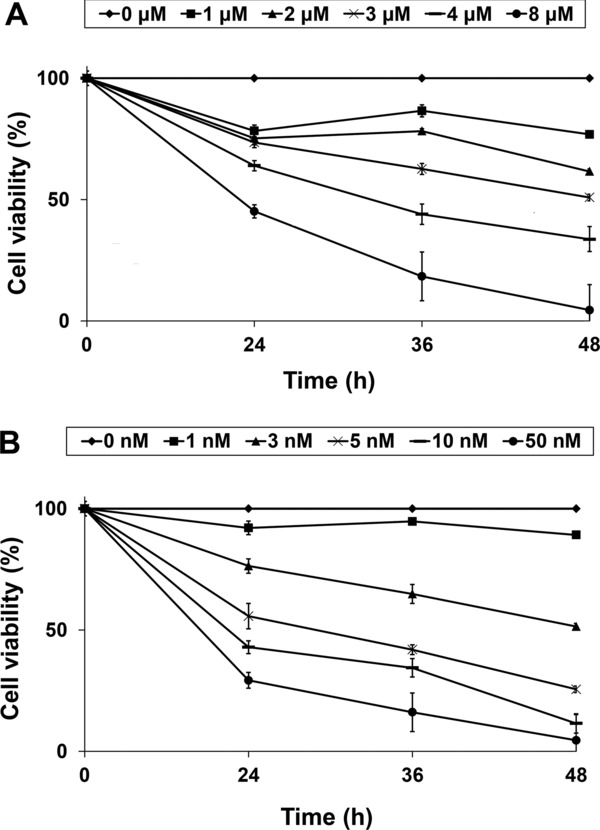
Impact of chemotherapy agents on cell viability of T24 cancer cells. The effect of (A) cisplatin and (B) paclitaxel on the viability of T24 cells was assessed via the colorimetric MTT assay. Each point represents the mean value (± standard error) of the (%) cell viability relative to the untreated cells (0 nM) as assessed from four biological replicates.
Cisplatin‐ and paclitaxel‐induced modulations in miR‐143/145 and miR‐183/96/182 clusters expression of T24 cells
Alterations in the expression of miR‐143/145 and miR‐183/96/182 clusters of T24 cells upon treatment with cisplatin or paclitaxel, as well as following their removal from the culture medium of the cells were estimated by analyzing the relative levels of the corresponding mature miRNAs via quantitative real‐time PCR. The amplification plots generated per assay, as well the analytical specificity of each reaction, as attested by melting curve analysis, are illustrated in Figure 2, while the individual PCR efficiency of each amplicon was estimated and found to range from 96.0% to 97.9%, ensuring the validity of the variations recorded. In particular, cisplatin treatment had a negative impact on miR‐143 expression leading to a significant down‐regulation of 2.8‐fold at 36 hours of treatment and another one of 1.8‐fold in 48‐hour treated cells (Figure 3A). Upon recovery though, the latter was reversed as miR‐143 levels were found to be elevated by 2.4‐fold compared with those of the 48‐hour treated cells, reaching miR‐143 expression levels detected in control cells (Table 2). Contrary to miR‐143, cisplatin had a positive influence on miR‐145 levels, whose expression was found to be up‐regulated by 3.4‐fold at 24 hours of incubation and by 2.2‐fold at the respective time interval of 48 hours (Figure 3B). However, withdrawal of cisplatin from the culture medium of the cells further augmented miR‐145 expression as illustrated in Table 2. Moving to the analysis performed for miR‐183/96/182, cisplatin showed the ability to up‐regulate miR‐183 expression levels, which were found to be elevated at the end of all three indicated incubation periods by 2.6‐, 2.1‐, and 3.0‐fold at 24, 36, and 48 hours, respectively (Figure 3C), while they were even more enhanced following recovery period (Table 2). As for miR‐96 (Figure 3D) and miR‐182 (Figure 3E), they exhibited a similar expression pattern, which consisted of lack of variation at 24 hours, an initial up‐regulation by 2.4‐fold for miR‐96 and 2.9‐fold for miR‐182 at 36 hours, and a further increase at 48 hours of exposure equal to 3.8‐ and 5.0‐fold for miR‐96 and miR‐182, correspondingly. Likewise, upon recovery both miRNAs were above the levels of control but exhibited the propensity to decline (Table 2). Analysis of paclitaxel‐induced modulations in the expression of the aforementioned miRNAs disclosed an analogous expression pattern. In particular, miR‐143 levels exhibited a 1.6‐fold down‐regulation at 24 and 48 hours of incubation but only the latter reached a statistically significant level (Figure 4A). Interestingly, the decline observed in miR‐143 levels was not stopped even when paclitaxel was removed from the culture medium and T24 cells were left to recover (Table 2). The expression pattern of miR‐145, though, was quite the opposite as it was found to be up‐regulated by 2.2‐, 3.0‐, and 2.0‐fold at 24, 36, and 48 hours of exposure (Figure 4B), respectively, whereas recovery process did not manage to further impact on its expression levels (Table 2). Likewise, all mature miRNAs of the miR‐183/96/182 cluster were overexpressed upon treatment (Figure 4 C–E) but their miRNA levels dropped at the level of control cells after recovery period (Table 2). In detail, miR‐183 levels were found be elevated at all three indicated time periods (24, 36, and 48 hours) by 1.7‐, 4.5‐, and 4.9‐fold, correspondingly. Regarding miR‐96, a 2.0‐fold increase was noticed for both 36 and 48 hours of exposure, while miR‐182 expression was found to be enhanced by 2.3‐ and 2.5‐fold, at the respective time points.
Figure 2.
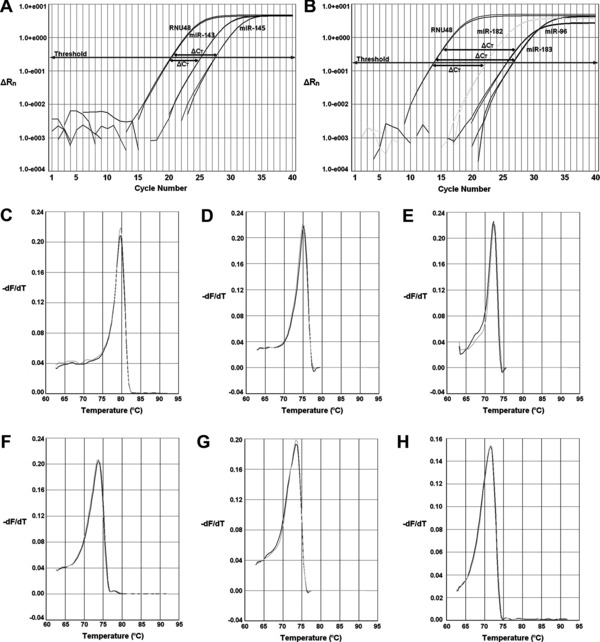
Ilustration of the amplification plots produced by real‐time PCR for the reference gene RNU48 and the mature miRNAs of the (A) miR‐143/145 and (B) miR‐183/96/182 clusters of T24 cells, along with the respective dissociation curves generated for (C) RNU48, (D) miR‐143, (E) miR‐145, (F) miR‐183, (G) miR‐96 and (H) miR‐182. ΔRn displayed in amplification plots represents the normalized and baseline‐corrected fluorescence intensity, while ΔCT stands for the difference between the threshold cycles (C T) of target and reference genes. In dissociation curves, the term ‐dF/dT, shown in y axis of the plot, corresponds to the first negative derivative of fluorescence over temperature.
Figure 3.
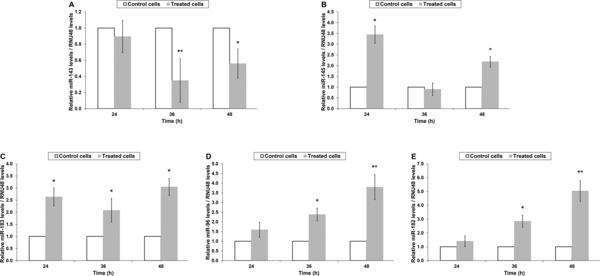
Cisplatin‐induced modulations in microRNAs expression. The expression levels of the mature miRNAs derived from the (A, B) miR‐143/145 cluster, as well as those of the (C, D, and E) miR‐183/96/182 cluster of T24 cancer cells were estimated via real‐time PCR. Each column represents the mean relative levels of each miRNA normalized to the expression levels of RNU48 ± standard error according to the results of three biological replicates. Statistically significant variations have been marked by stars. (*p < 0.05; **p < 0.01).
Table 2.
Fold changes in the expression levels of microRNAs after recovery period
| Cisplatin | Paclitaxel | |||
|---|---|---|---|---|
| Fold change* (p‐value+) | Fold change* (p‐value+) | |||
| Treated cells‡ | Untreated cells§ | Treated cells‡ | Untreated cells§ | |
| miR‐143 | 2.4 (0.031) | 1.4 (n.s.) | −1.4 (n.s.) | −2.2 (0.019) |
| miR‐145 | 3.5(0.028) | 7.6 (0.009) | 1.0 (n.s.) | 2.0 (0.044) |
| miR‐183 | 3.5 (0.013) | 10.5 (0.001) | −3.5 (0.013) | 1.4 (n.s.) |
| miR‐96 | −1.5 (n.s.) | 2.5 (0.021) | −1.9 (0.048) | 1.3 (n.s.) |
| miR‐182 | −1.9(0.039) | 2.6 (0.008) | −2.0 (0.034) | 1.0 (n.s.) |
Positive and negative values reflect increase and decrease in miRNA expression, respectively.
n.s. stands for not significant variations (p‐value > 0.05).
Fold changes in comparison with the relative levels of miRNAs detected in the 48‐hour treated cells.
Compared with the average of the relative miRNA levels measured in untreated (control) cells.
Figure 4.
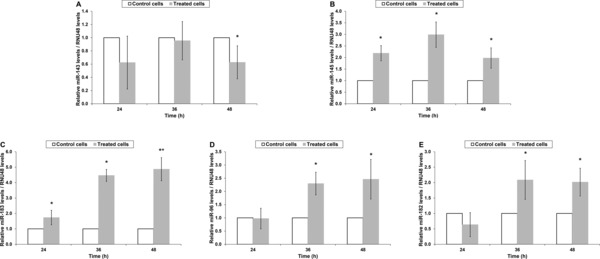
Alterations in microRNA levels of T24 cells upon treatment with paclitaxel. Relative levels (mean± standard error) of (A) miR‐143, (B) miR‐145, (C) miR‐183, (D) miR‐96, and (E) miR‐182 of T24 cells were estimated by performing three independent biological experiments. Observed variations were accordingly subjected to statistical analysis and those demonstrating significance are illustrated as follows: *p < 0.05; **p < 0.01.
Discussion
Metastatic bladder cancer is currently treated via the administration of chemotherapy regimens, which in their vast majority are platinum‐based.4, 5, 6 These combination chemotherapy schemes have managed to increase survival of bladder cancer patients but still their efficacy and tolerability require significant improvement.21 Toward this direction, several studies have sought to identify the molecular mechanisms implicated in the action of these agents in order to locate therapeutic targets that could enhance their antineoplastic activity.22, 23 Upon a search conducted in the literature, we found that a lot of research has been performed, at a preclinical level, regarding the potential impact of synthetic microRNA sequences on bladder cancer cells' viability. Even more, lots of these studies have especially focused on the mature miRNAs derived from the miR‐143/145 and miR‐183/96/182 clusters, whose function has been also extensively studied in bladder cancer cells.12, 13, 14, 15, 16, 17, 18, 24, 25, 26 Specifically, several protein‐coding genes implicated in the process of carcinogenesis in urinary bladder have been identified as targets of the above miRNAs, as shown in Table 3. Nevertheless, the research conducted so far does not provide any information with regard to the effect that chemotherapy agents indicated for the management of bladder cancer exert on the expression levels of these miRNAs. Thus, aiming to cover this missing link, we herein estimated how cisplatin and paclitaxel impact on the expression levels of the mature miRNAs generated from the above clusters both upon manifestation of cytotoxicity and following removal of these drugs from bladder cancer cells' environment.
Table 3.
Identified gene targets of miR‐143/145 and miR‐183/96/182 clusters reported to be deregulated in bladder cancer
| miR‐143 | miR‐145 | miR‐183 | miR‐96 | miR‐182 |
|---|---|---|---|---|
| AKT 12, 14 | ILK 14 | IRS1 26 | SMAD4 17 | |
| PAI‐1 31 | PAI‐1 31 | NR* | MAP4K1 26 | RECK 17 |
| COX‐2 13 | CBFB 15 | FOXO1 18 | ||
| ERK5 12 | CLINT1 15 | |||
| PPP3CA 15 | ||||
| FSCN1 32 | ||||
| SOCS7 25 | ||||
| PAK1 33 | ||||
| IGF‐IR 34 |
NR = none retrieved.
So far, several studies have been conducted regarding the implication of miR‐143 and miR‐145 in different malignancies. Among them, the role of mi‐143 and miR‐145 has been extensively studied in colorectal cancer, prostate cancer, and esophageal squamous cell carcinoma, where both miRNAs are claimed to act as tumor suppressors, while their expression levels are found to be down‐regulated.35, 36, 37, 38 The expression analysis performed herein disclosed a number of interesting and in some cases unexpected variations. The latter refers especially to the alterations noticed in miR‐143 expression levels that, despite its also well documented antitumor activity in bladder cancer cells12, 13, 14, 24 and its proven down‐regulation in bladder cancer tissue specimens,39 demonstrated significant decrease upon treatment of T24 cells with either cisplatin or paclitaxel. This paradox is even more enhanced if we consider that cisplatin,40 paclitaxel,41 and miR‐14312, 13 have been, separately, reported to be associated with the induction of apoptosis in bladder cancer cells. Data from previous preclinical studies in established bladder cancer cell lines suggest that the synthetic sequence of miR‐143 should be considered for clinical investigation in bladder cancer.12, 13, 24 Interestingly, in one of the aforementioned studies researchers subjected bladder cancer cells to co‐treatment with cisplatin and synthetic miR‐143 sequence concluding that this combination may exert additive effect on suppressing cells' growth.12 Hence, our results seem to substantiate this concept as the tumor suppressor miR‐143 appears to be down‐regulated in T24 bladder cancer cells exposed to chemotherapy and thus restoring its expression levels might be a promising therapeutic approach. Furthermore, this might be even more interesting in case of paclitaxel, as our data show that the down‐regulation noticed in paclitaxel‐treated cells was not reversed upon recovery. Instead, the other tumor suppressive miRNA generated from the miR‐143/145 cluster12, 15, 25 behaved as expected, as miR‐145 levels were found to be up‐regulated in T24 cells regardless of the chemotherapy drug added to their culture medium. Moreover, this increase was not affected upon withdrawal of the antineoplastic agents. Therefore, our results imply that miR‐145 might be a crucial mediator of cisplatin and paclitaxel cytotoxic activity against bladder cancer cells. Within the frame of these observations, it is worthwhile mentioning that cisplatin and paclitaxel have been reported to trigger apoptotic cell death in bladder cancer cells, in which, likewise, miR‐145 has been implicated in the induction of both caspase‐dependent and caspase‐independent apoptosis.15, 40, 41
Contrary to the mature miRNAs derived from miR‐143/145 cluster of T24 cells, those of miR‐183/96/182 cluster presented a more consistent expression pattern as they were all found to be up‐regulated upon treatment with either drug and, with the exception of miR‐183 in cisplatin‐treated T24 cells, all miRNAs dropped at the level of control cells following the removal of the antineoplastic agents from cells' environment. Alike miR‐143, though, these results were not in agreement with the suggested role of miR‐183, miR‐96, and miR‐182 in bladder cancer, which has proven to be oncogenic.16, 17, 18, 26 Evidence toward this direction has been provided from different studies conducted by transfecting bladder cancer cells with either the synthetic sequences of the above mature miRNAs or with the antisense inhibitors of their action.16, 17, 26 Within the same frame, the expression levels of miR‐183, miR‐96, and miR‐182 have been reported to be up‐regulated in bladder cancer upon analysis performed in fresh cancerous and adjacent noncancerous bladder tissue specimens.39 In addition, the same expression pattern for all three mature miRNAs has been also well documented in colorectal cancer, prostate cancer, lung cancer, and hepatocellular carcinoma,42 whereas miR‐183/96/182 cluster has been claimed to demonstrate oncogenic properties in various malignancies.27, 28, 29, 30 Therefore, the fact that their expression levels are herein found to be elevated as a result of drug treatment might reflect the propensity of T24 bladder cancer cells to resist to the above chemotherapy agents. Although further experimentation is required in order to determine whether miR‐183, miR‐96, and miR‐182 confer chemoresistance to bladder cancer cells, our data add up to current suggestions and moreover present a different perspective on the idea of considering the use of synthetic devices against the miR‐183/96/182 cluster16 for clinical evaluation in bladder cancer, either alone or in combination with conventional chemotherapy.
Conclusion
Overall, we herein present a number of significant modulations in the expression levels of the mature miRNAs derived from the miR‐143/145 and miR‐183/96/182 clusters of T24 bladder cancer cells upon their treatment with cisplatin or paclitaxel, as well as following the removal of the aforementioned drugs from cells' environment. In their vast majority, the alterations noticed in the expression of the five mature miRNAs of T24 cells were found to be inconsistent with their either oncogenic or tumor suppressive role, as defined by previous studies, reflecting the complexity of the molecular events that occur upon exposure of bladder cancer cells to these anticancer agents. In addition, our data along with those from other preclinical studies argue in favor of considering synthetic miRNA sequences as investigational products for clinical evaluation in bladder cancer.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Acknowledgments
Part of this work was supported by the Hellenic Society of Medical Oncology.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015; 65: 5–29. [DOI] [PubMed] [Google Scholar]
- 2. Chalasani V, Chin JL, Izawa JI. Histologic variants of urothelial bladder cancer and nonurothelial histology in bladder cancer. Can Urol Assoc J. 2009; 3(Suppl 4): S193–S198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013; 63: 234–241. [DOI] [PubMed] [Google Scholar]
- 4. Bellmunt J, Orsola A, Leow JJ, Wiegel T, De Santis M, Horwich A. ESMO Guidelines Working Group: Bladder cancer: ESMO Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2014; 25(Suppl 3): iii40–8. [DOI] [PubMed] [Google Scholar]
- 5. Clark PE, Agarwal N, Biagioli MC, Eisenberger MA, Greenberg RE, Herr HW, Inman BA, Kuban DA, Kuzel TM, Lele SM, et al. National Comprehensive Cancer Network (NCCN): bladder cancer. J Natl Compr Canc Netw. 2013; 11: 446–475. [DOI] [PubMed] [Google Scholar]
- 6. Witjes JA, Compérat E, Cowan NC, De Santis M, Gakis G, Lebrét T, Ribal MJ, Sherif A, van der Heijden AG. Guidelines on bladder cancer—muscle invasive and metastatic. Eur Assoc Urol. 2014. Available at: http://uroweb.org/guideline/bladder‐cancer‐muscle‐invasive‐and‐metastatic/. Accessed, March 2015. [DOI] [PubMed] [Google Scholar]
- 7. Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014; 740: 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Weger VA, Beijnen JH, Schellens JH. Cellular and clinical pharmacology of the taxanes docetaxel and paclitaxel–a review. Anticancer Drugs. 2014; 25: 488–494. [DOI] [PubMed] [Google Scholar]
- 9. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014; 20: 460–469. [DOI] [PubMed] [Google Scholar]
- 10. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014; 15: 509–524. [DOI] [PubMed] [Google Scholar]
- 11. Hu W, Coller J. What comes first: translational repression or mRNA degradation? The deepening mystery of microRNA function. Cell Res. 2012; 22: 1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Noguchi S, Mori T, Hoshino Y, Maruo K, Yamada N, Kitade Y, Naoe T, Akao Y. MicroRNA‐143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Lett. 2011; 307: 211–220. [DOI] [PubMed] [Google Scholar]
- 13. Song T, Zhang X, Wang C, Wu Y, Dong J, Gao J, Cai W, Hong B. Expression of miR‐143 reduces growth and migration of human bladder carcinoma cells by targeting cyclooxygenase‐2. Asian Pac J Cancer Prev. 2011; 12: 929–933. [PubMed] [Google Scholar]
- 14. Noguchi S, Yasui Y, Iwasaki J, Kumazaki M, Yamada N, Naito S, Akao Y. Replacement treatment with microRNA‐143 and ‐145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013; 328: 353–361. [DOI] [PubMed] [Google Scholar]
- 15. Ostenfeld MS, Bramsen JB, Lamy P, Villadsen SB, Fristrup N, Sørensen KD, Ulhøi B, Borre M, Kjems J, Dyrskjøt L, et al. miR‐145 induces caspase‐dependent and ‐independent cell death in urothelial cancer cell lines with targeting of an expression signature present in Ta bladder tumors. Oncogene. 2010; 29: 1073–1084. [DOI] [PubMed] [Google Scholar]
- 16. Liu Y, Han Y, Zhang H, Nie L, Jiang Z, Fa P, Gui Y, Cai Z. Synthetic miRNA‐mowers targeting miR‐183‐96‐182 cluster or miR‐210 inhibit growth and migration and induce apoptosis in bladder cancer cells. PLoS One. 2012; 7: e52280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. Oncogenic miRNA‐182‐5p targets Smad4 and RECK in human bladder cancer. PLoS One. 2012; 7: e51056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo Y, Liu H, Zhang H, Shang C, Song Y. miR‐96 regulates FOXO1‐mediated cell apoptosis in bladder cancer. Oncol Lett. 2012; 4: 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi R, Chiang VL. Facile means for quantifying microRNA expression by real‐time PCR. Biotechniques. 2005; 39: 519–525. [DOI] [PubMed] [Google Scholar]
- 20. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods. 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 21. Sio TT, Ko J, Gudena VK, Verma N, Chaudhary UB. Chemotherapeutic and targeted biological agents for metastatic bladder cancer: a comprehensive review. Int J Urol 2014; 21: 630–637. [DOI] [PubMed] [Google Scholar]
- 22. Galluzzi L, Vitale I, Michels J, Brenner C, Szabadkai G, Harel‐Bellan A, Castedo M, Kroemer G. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis. 2014; 5: e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Ling MT, Guan XY, Tsao SW, Cheung HW, Lee DT, Wong YC. Identification of a novel function of TWIST, a bHLH protein, in the development of acquired taxol resistance in human cancer cells. Oncogene. 2004; 23: 474–482. [DOI] [PubMed] [Google Scholar]
- 24. Kitamura Y, Masegi Y, Ogawa S, Nakashima R, Akao Y, Ueno Y, Kitade Y. Chemically modified siRNAs and miRNAs bearing urea/thiourea‐bridged aromatic compounds at their 3'‐end for RNAi therapy. Bioorg Med Chem. 2013; 21: 4494–4501. [DOI] [PubMed] [Google Scholar]
- 25. Noguchi S, Yamada N, Kumazaki M, Yasui Y, Iwasaki J, Naito S, Akao Y. socs7, a target gene of microRNA‐145, regulates interferon‐β induction through STAT3 nuclear translocation in bladder cancer cells. Cell Death Dis. 2013; 4: e482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Luo H, Li Y, Chen T, Wu S, Yang L. hsa‐miR‐96 up‐regulates MAP4K1 and IRS1 and may function as a promising diagnostic marker in human bladder urothelial carcinomas. Mol Med Rep. 2012; 5: 260–265. [DOI] [PubMed] [Google Scholar]
- 27. Villadsen SB, Bramsen JB, Ostenfeld MS, Wiklund ED, Fristrup N, Gao S, Hansen TB, Jensen TI, Borre M, Ørntoft TF, et al. The miR‐143/‐145 cluster regulates plasminogen activator inhibitor‐1 in bladder cancer. Br J Cancer. 2012; 106: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M. miR‐145 and miR‐133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010; 102: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kou B, Gao Y, Du C, Shi Q, Xu S, Wang CQ, Wang X, He D, Guo P. miR‐145 inhibits invasion of bladder cancer cells by targeting PAK1. Urol Oncol. 2014; 32: 846–854. [DOI] [PubMed] [Google Scholar]
- 30. Zhu Z, Xu T, Wang L, Wang X, Zhong S, Xu C, Shen Z. MicroRNA‐145 directly targets the insulin‐like growth factor receptor I in human bladder cancer cells. FEBS Lett. 2014; 588: 3180–3185. [DOI] [PubMed] [Google Scholar]
- 31. Kamatani A, Nakagawa Y, Akao Y, Maruyama N, Nagasaka M, Shibata T, Tahara T, Hirata I. Downregulation of anti‐oncomirs miR‐143/145 cluster occurs before APC gene aberration in the development of colorectal tumors. Med Mol Morphol. 2013; 46: 166–171. [DOI] [PubMed] [Google Scholar]
- 32. Goto Y, Kurozumi A, Enokida H, Ichikawa T, Seki N. Functional significance of aberrantly expressed microRNAs in prostate cancer. Int J Urol. 2015; 22: 242–252. [DOI] [PubMed] [Google Scholar]
- 33. Liu R, Liao J, Yang M, Sheng J, Yang H, Wang Y, Pan E, Guo W, Pu Y, Kim SJ, et al. The cluster of miR‐143 and miR‐145 affects the risk for esophageal squamous cell carcinoma through co‐regulating fascin homolog 1. PLoS One. 2012; 7: e33987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu BL, Xu LY, Du ZP, Liao LD, Zhang HF, Huang Q, Fang GQ, Li EM. MiRNA profile in esophageal squamous cell carcinoma: downregulation of miR‐143 and miR‐145. World J Gastroenterol. 2011; 17: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013; 10: 396–404. [DOI] [PubMed] [Google Scholar]
- 36. Konstantakou EG, Voutsinas GE, Karkoulis PK, Aravantinos G, Margaritis LH, Stravopodis DJ. Human bladder cancer cells undergo cisplatin‐induced apoptosis that is associated with p53‐dependent and p53‐independent responses. Int J Oncol. 2009; 35: 401–416. [PubMed] [Google Scholar]
- 37. Au JL, Kalns J, Gan Y, Wientjes MG. Pharmacologic effects of paclitaxel in human bladder tumors. Cancer Chemother Pharmacol. 1997; 41: 69–74. [DOI] [PubMed] [Google Scholar]
- 38. Zhang QH, Sun HM, Zheng RZ, Li YC, Zhang Q, Cheng P, Tang ZH, Huang F. Meta‐analysis of microRNA‐183 family expression in human cancer studies comparing cancer tissues with noncancerous tissues. Gene. 2013; 527: 26–32. [DOI] [PubMed] [Google Scholar]
- 39. Zhang Q, Ren W, Huang B, Yi L, Zhu H. MicroRNA‐183/182/96 cooperatively regulates the proliferation of colon cancer cells. Mol Med Rep. 2015; 12: 668–674. [DOI] [PubMed] [Google Scholar]
- 40. Leung WK, He M, Chan AW, Law PT, Wong N. Wnt/β‐Catenin activates MiR‐183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett. 2015; 362: 97–105. [DOI] [PubMed] [Google Scholar]
- 41. Li P, Sheng C, Huang L, Zhang H, Huang L, Cheng Z, Zhu Q. MiR‐183/‐96/‐182 cluster is up‐regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014; 16: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tang H, Bian Y, Tu C, Wang Z, Yu Z, Liu Q, Xu G, Wu M, Li G. The miR‐183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr Cancer Drug Targets. 2013; 13: 221–231. [DOI] [PubMed] [Google Scholar]


