Abstract
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related mortality in the world. Late diagnosis is one of the most significant reasons for the high mortality rate of lung cancer. The identification of microRNAs (miRNAs) has opened a new field for molecular diagnosis of cancer. The purpose of the present study was to investigate whether plasma miRNAs may be used as biomarkers for early-stage NSCLC. A total of 232 participants, including 149 NSCLC patients and 83 healthy controls, were recruited between July 2012 and May 2014. We measured the levels of 10 miRNAs (miR-30d, miR-383, miR-20a, miR-145, miR-221, miR-25, miR-223, miR-21, miR-126 and miR-210) in plasma samples of 40 individuals (20 patients and 20 matched healthy controls) at the point of identification of disease, and 129 NSCLC patients and 83 healthy controls at the validation stage using reverse transcription-quantitative polymerase chain reaction. Receiver operating characteristics (ROC) curves were generated for each possible combination of the miRNAs. We observed that the expression of plasma miR-145, miR-20a, miR-21 and miR-223 was significantly increased in the early-stage NSCLC samples compared with controls. miRNAs have significant diagnostic value for early-stage NSCLC. Combined ROC analyses using these four miRNAs revealed an elevated area under the ROC curve (AUC) of 0.897, with a sensitivity and specificity of 81.8 and 90.1%, respectively. This AUC helped in distinguishing early-stage NSCLC. Furthermore, the levels of the four plasma miRNAs were significantly decreased following surgery (P<0.05). Altered expression of miR-145, miR-20a, miR-21 and miR-223 in plasma are of tumor origin, and the four miRNAs may represent potential novel non-invasive biomarkers for early-stage NSCLC.
Keywords: plasma, microRNA, non-small cell lung cancer
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide (1). Approximately 85% of lung cancers are classified as non-small cell lung cancer (NSCLC) (2), and the majority of patients present with unresectable advanced disease. Despite the advances over the past decades in terms of cancer treatments, the overall five-year survival rate for NSCLC patients remains less than 15% (3). Late diagnosis of NSCLC is one of the significant contributing factors to the poor clinical outcome, since the tumor has often spread to distant organs at the time of diagnosis. The five-year survival rate following surgical resection is ~80% for early-stage NSCLC, but that rate drops to just 30% in patients with advanced stages (4). Thus, earlier detection of NSCLC would greatly facilitate more effective management of the disease.
Currently, histological examination is still the gold standard for the diagnosis of NSCLC. However, in this approach it is necessary to obtain tissues or cells from patients, and invasive examination methods are required, including bronchoscopy, lung puncture, endobronchial ultrasound or thoracotomy. Although chest X-ray and computed tomography examinations are capable of detecting early-stage NSCLC, certain studies have demonstrated that 50% of pulmonary nodules detected by these approaches are benign (5), and that these methods have limited function in reducing lung cancer mortality (6). Moreover, the limited sensitivity and specificity of previously known lung cancer biomarkers, including cytokeratin fragment 21-1, tumor polysaccharides and carcinoembryonic antigen, hampered their further application and development (7). Therefore, a biomarker with non-invasiveness, high sensitivity and high specificity for the early diagnosis of lung cancer is required.
microRNAs (miRNAs) are a class of highly conserved non-coding small RNAs, consisting of 20–24 nt and existing widely in eukaryotic cells. The first miRNA was identified in the mutant of Caenorhabditis elegans by Lee et al in 1993 (8). Following that, a large number of miRNAs were identified in Drosophila, Arabidopsis, zebra fish, rice and human cells. According to the latest version of the miRBase (http://www.mirbase.org/), there are more than 2600 miRNAs in humans, and ~60% of human genes are regulated by miRNAs. Previous studies have demonstrated that there are a large number of miRNAs existing in plasma (9), where they have different expression profiles between lung cancer patients and normal healthy controls (10). Other studies have demonstrated that the abnormally expressed miRNAs may be used as diagnostic markers for NSCLC (7,11). However, studies using the abnormal expression of miRNAs in plasma for early diagnosis of NSCLC are still lacking. In this study, following a large number of relevant studies which have reported the diagnostic value of miRNAs for NSCLC (7,11–14), we investigated the plasma levels of 10 miRNAs in the early stage of NSCLC patients using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The valuable diagnostic miRNAs were identified by receiver operating characteristics (ROC) curve analysis, and are likely to improve the diagnostic ability in early-stage NSCLC patients.
Materials and methods
Patients and ethics
All 232 participants were recruited from Shanghai Chest Hospital, China, between July 2012 and May 2014. In the training set, we selected 20 early-stage NSCLC patients and 20 age- and gender-matched healthy controls to compare the expression profile of candidate plasma miRNAs between NSCLC patients and healthy controls. In the validation set, 109 early-stage NSCLC patients and 63 healthy controls were recruited. To increase the number of samples for validation, we merged the 40 cases of samples in the screening stage with those in the validation stage. To investigate whether the altered expression of the valuable diagnostic miRNAs in plasma was of tumor origin, their expression levels were measured in an independent set of 20 cases with early-stage NSCLC. The inclusion criteria for NSCLC patients included: i) no previous history of cancer-related diseases; ii) did not receive radiotherapy or chemotherapy prior to surgery; iii) diagnosed as NSCLC pathologically following surgery; iv) I and II stages of NSCLC according to the tumor-node-metastasis (TNM) staging guidelines of the American Joint Committee on Cancer (7th version) (15). The inclusion criterion for the healthy control group was that the individuals were without tumor-associated lesions confirmed by chest CT, blood test and other full body examinations. Samples were collected two days after admission and 7 to 9 days after surgery. The collection of samples was approved by the Medical Ethics Committee of Shanghai Chest Hospital, Shanghai Jiaotong University. All patients and healthy controls signed an informed consent form.
Sample processing and RNA isolation
Whole blood (4 ml) was added to an ethylenediamine tetraacetic acid (EDTA)-treated anticoagulant tube, and then plasma was isolated by centrifugation at 1,200 rpm for 10 min and subsequently at 12,000 rpm for 10 min at 4°C. A total of 400 µl plasma was added to an equal volume of TRIzol. After putting on ice for 5 min, 800 µl chloroform was added and incubated on ice for 5 min. Following centrifugation at 1,200 rpm for 10 min at 4°C, the supernatant was collected. In order to obtain a suitable internal control following the isolation of miRNAs, we added cel-miR-39 (Takara Biological Engineering Co., Ltd., Dalian, China) to the supernatant as reported previously (9,12). The synthetic sequences were from the miRBase database. Total RNA was isolated using the mirVana PARIS kit following the manufacturer's instructions (Ambion Life Technologies, Carlsbad, CA, USA). Total RNA (100 µl) was collected from the filter by washing with enzyme-free water (Shanghai Biological Engineering Co., Ltd., Shanghai, China), and then RNA concentration and purity were measured using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA).
Evaluation of internal controls for quantification of plasma miRNAs
To select an appropriate internal control, we examined the expression levels of miR-16 and RNU6B in the plasma of 20 cases of NSCLC patients and 20 age- and gender-matched healthy individuals, as described previously (16–20). Using the fixed volume approach, we added cel-miR-39 to each sample as a control. In order to investigate the stability of the two potential internal controls at room temperature, we randomly selected three copies of plasma samples, and each one was divided into four parts. These sample aliquots were maintained at room temperature for 0, 2, 4 and 8 h in nuclease-free tubes before being processed for RNA isolation. Following quantification by RT-qPCR, PCR products were randomly selected to analyze the sequence integrity by electrophoresis.
RT-qPCR for miRNAs
Reverse transcription was performed using a TaqMan microRNA reverse transcription kit (Ambion Life Technologies), and qPCR was performed using a Brilliant III Ultra-Fast SYBR-Green qPCR master mix kit (Ambion Life Technologies). RT-PCR was performed as described previously by Kroh et al (18). PCR primers with a stem-loop structure of each miRNA were designed based on the miRNA sequences obtained from the miRBase database. The primers were synthesized by Shanghai Biological Engineering Co., Ltd. (Table I). We used the fixed volume approach for detection in the reaction systems of reverse transcription and qPCR since the total RNA concentration detected by the microspectrophotometer was very low (~10 ng/µl). A total of 1.5 µl 10X RT-PCR buffer, 0.15 µl 100X dNTP mixture, 1 µl 50 U/µl Multiscribe RT enzyme, 0.19 µl 20 U/µl RNase inhibitor, 1 µl 10 µmol/l primer and 10 µl total RNA were added to the reverse transcription system and made up to 15 µl volume with diethylpyrocarbonate. The reaction conditions for reverse transcription were 16°C for 30 min, 42°C for 30 min, 85°C for 5 min and terminated at 4°C. The qPCR system (20 l) included TaqMan 2X 10 µl Universal PCR master mix, 1 µl primers (final concentration 200 nM) and 9 µl cDNA. The reaction was performed in an ABI 7500 Real-Time PCR system (Ambion Life Technologies), and the reaction conditions for qPCR were 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. Three parallel samples and a negative control were set. To check the integrity of the amplification product, we used 3% agarose gel electrophoresis to analyze and verify the specificity of the PCR product. Relative expression levels were calculated using the 2−ΔΔCq method (21), and miR-16 was used as an internal control.
Table I.
miRNA-specific primers.
| Primer | Sequences (5′-3′) |
|---|---|
| Cel-miR-39 | TCACCGGGTGTAAATCAG |
| miR-30d | CGCTGTAAACATCCCCGAC |
| miR-383 | CGCAGATCAGAAGGTGATT |
| miR-16 | GTAGCAGCACGTAAATATTGG |
| miR-20a | CGCTAAAGTGCTTATAGTGC |
| miR-145 | TGAACTTCGCAACTACCGTTTG |
| miR-21 | CGCTAGCTTATCAGACTGA |
| miR-221 | CGAGCTACATTGTCTGCTGGGT |
| miR-126 | CGCTCGTACCGTGAGTAAT |
| miR-223 | GCGGGTGTCAGTTTGTCAAATA |
| miR-25 | CATTGCACTTGTCTCGGTCTG |
| miR-210 | CGCAGCCCCTGCCCACCGC |
| RNU6B | ACGCAAATTCGTGAAGCGTT |
Selection and validation of plasma microRNA
In accordance with previous studies, we selected 10 miRNAs (miR-30d, miR-383, miR-20a, miR-145, miR-221, miR-25, miR-223, miR-21, miR126 and miR-210) (7,11–14) and examined their expression using RT-qPCR in a small set of plasma samples (20 NSCLC patients and 20 gender- and age-matched healthy controls). The upregulated markers in NSCLC plasma were further validated in an independent large-scale set of plasma from 109 NSCLC patients and 63 healthy controls using RT-qPCR. The inclusion criteria were as mentioned above. The effect of miRNAs in the early diagnosis of NSCLC was analyzed by ROC curve. In order to observe whether the abnormally expressed miRNAs are derived from tumor tissues, we collected the plasma from 20 cases of early-stage NSCLC before and after surgery, then examined the expression of miRNAs using RT-PCR.
Statistical analysis
Differences in miRNA levels between cases and controls were assessed by the Mann-Whitney U test or the Kruskall-Wallis test. The Chi-square test and one-way analysis of variance were used to assess the difference in clinicopathological characteristics and association between miRNA levels and clinicopathological characteristics between cases and controls. The multivariate logistic regression model was used to establish the optimum regression equation and calculate the odds ratio and 95% confidence interval for each variable. An ROC curve was established to interpret the ability of miRNA in discriminating patients from healthy controls. The area under the curve (AUC), sensitivity and specificity at the optimal cut-off were computed in order to validate the diagnostic application of these effective miRNAs as cancer biomarkers. All P-values were shown bilaterally, and a value less than 0.05 was considered to indicate a statistically significant difference. Statistical analysis of the data was performed using SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) and graphs were generated using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Clinical characteristics of study population
There were 149 NSCLC patients and 83 healthy controls (Table II). In the training set, there were 20 patients in the NSCLC group and 20 healthy individuals in the control group (gender and age were matched in the two groups). At the verification stage, there were 109 NSCLC patients and 63 healthy controls. There were no significant differences in the mean age, gender and smoking history between the two groups (P>0.05). The peripheral blood was collected from 20 early-stage NSCLC patients and the expression levels of miRNAs were investigated before and after surgery (Table II).
Table II.
Clinical characteristics of NSCLC patients and healthy controls (cases, %).
| Training set | Validation set | ||||||
|---|---|---|---|---|---|---|---|
| Category | Control (n=20) | NSCLC (n=20) | P | Control (n=63) | NSCLC (n=109) | P | Pre- and post-surgery (n=20) |
| Gender | 1 | 0.525 | |||||
| Male | 12 (60) | 12 (60) | 36 (57.1) | 69 (63.3) | 13 (65) | ||
| Female | 8 (40) | 8 (40) | 27 (42.9) | 40 (36.7) | 7 (45) | ||
| Age (year) | 1 | 0.539 | |||||
| <60 | 9 (45) | 9 (45) | 31 (49.2) | 47 (43.1) | 8 (40) | ||
| >60 | 11 (55) | 11 (55) | 32 (40.8) | 62 (56.9) | 12 (60) | ||
| Mean age (year) | 61.7+8.8 | 61.0+8.1 | 0.749 | 59.7+8.0 | 59.3+9.0 | 0.783 | 61.4+8.3 |
| Smoking statusa | 0.747 | 0.064 | |||||
| Yes | 11 (55) | 13 (65) | 27 (42.9) | 64 (58.7) | 13 (65) | ||
| No | 9 (45) | 7 (35) | 36 (57.1) | 45 (41.3) | 7 (45) | ||
| TNM stage | |||||||
| I | 7 (35) | 48 (44.0) | 6 (30) | ||||
| II | 13 (65) | 61 (56.0) | 14 (70) | ||||
| Pathological type | |||||||
| Adenocarcinoma | 9 (45) | 49 (45.0) | 11 (55) | ||||
| Squamous cell carcinoma | 11 (55) | 42 (38.5) | 9 (45) | ||||
| Other | 18 (16.5) | ||||||
NSCLC, non-small cell lung cancer; P, P-value; TNM, tumor-node-metastasis.
Individuals with smoking index more than 400 are classed as smokers.
Evaluation of potential internal control for quantification of plasma miRNA
To identify an internal control that is capable of reliably quantifying the expression of the target miRNAs in plasma, we examined the levels of miR-16 and RNU6B using RT-qPCR in plasma samples of 20 NSCLC patients and 20 healthy controls. To normalize the difference in extraction efficiency and reverse transcription efficiency among the different samples, the plasma levels of miR-16 and RNU6B were compared with spiked-in cel-miR-39. No significant difference was observed in the levels of miR-16 (P=0.158) and RNU6B (P=0.557) between the NSCLC patients and healthy controls (Fig. 1). To examine the stability of the two internal controls, we measured the levels in the samples prepared at various time points. We randomly selected three copies of plasma from the 20 NSCLC cases and 20 controls (each was divided into four parts). Total RNA was isolated and quantified after the samples had been kept at room temperature for 0, 2, 4 and 8 h. We observed that miR-16 was relatively stable at room temperature, and there were no significant differences in the expression of miR-16 among the samples kept at room temperature for 0, 2, 4 and 8 h (P>0.05, Fig. 2). However, the expression levels of RNU6B were significantly different when the samples were kept at room temperature for 4 and 8 h (P<0.05, Fig. 2). Together, the observations indicated that miR-16 exhibited higher stability and abundance than RNU6B in plasma. Therefore, we selected miR-16 as the normalization control to determine the expression of the 10 miRNAs in the present study.
Figure 1.
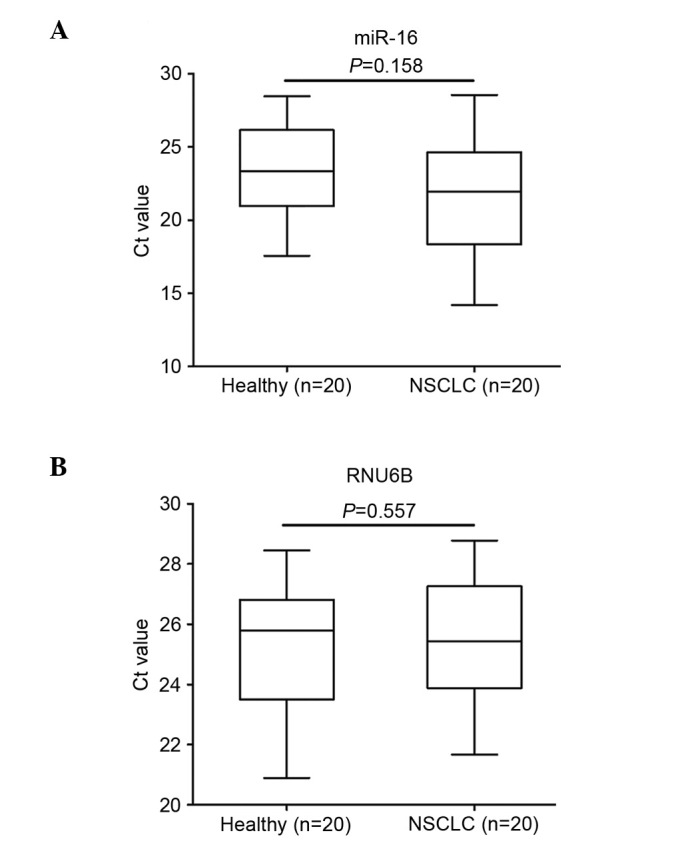
Expression levels of miR-16 and RNU6B.
Figure 2.
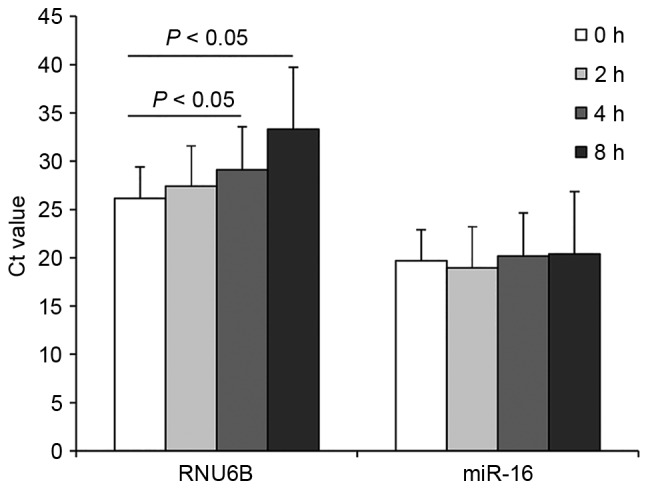
Stability of miR-16 and RNU6B at room temperature.
Evaluation of 10 candidate miRNAs as biomarkers for NSCLC screening in training set
To screen the potential upregulated miRNAs, we first examined the expression levels of 10 candidate miRNAs (miR-30d, miR-383, miR-20a, miR-145, miR-221, miR-25, miR-223, miR-21, miR-126 and miR-210) based on previous studies (7,11–14) using RT-qPCR in 40 plasma samples (20 NSCLC cases and 20 controls). We observed that the expression of four miRNAs in the plasma of NSCLC patients was more than two-fold higher than that in the healthy control group (Table III, P<0.05). miRNA-383 was not detectable in the plasma (Ct value>35). There were no significant differences in the expression of miR-221, miR-25 and miR-30d in the plasma of NSCLC patients and healthy controls (P>0.05). The expression levels of miR-126 and miR-210 were slightly decreased in the plasma of NSCLC patients, but there was no significant difference between the cases and controls (P>0.05).
Table III.
Expression levels of 10 plasma miRNAs between 20 NSCLC patients and 20 healthy controls (mean ± SD).
| miRNAs | Expression | NSCLC/healthy (fold) | P-value |
|---|---|---|---|
| miR-145 | ↑ | 21.67+0.89 | 2.04×10−4 |
| miR-20a | ↑ | 13.39+1.02 | 9.22×10−5 |
| miR-21 | ↑ | 6.15+0.49 | 3.72×10−4 |
| miR-223 | ↑ | 2.64+0.39 | 1.48×10−3 |
| miR-221 | ↑ | 1.37+0.31 | 0.0612 |
| miR-25 | ↑ | 1.23+0.28 | 0.7510 |
| miR-30d | ↑ | 1.13+0.29 | 0.3326 |
| miR-126 | ↓ | 0.93+0.27 | 0.3942 |
| miR-210 | ↓ | 0.62+0.16 | 0.1195 |
| miR-383 | / | / | / |
NSCLC, non-small cell lung cancer.
miRNA validation and ROC curve analysis
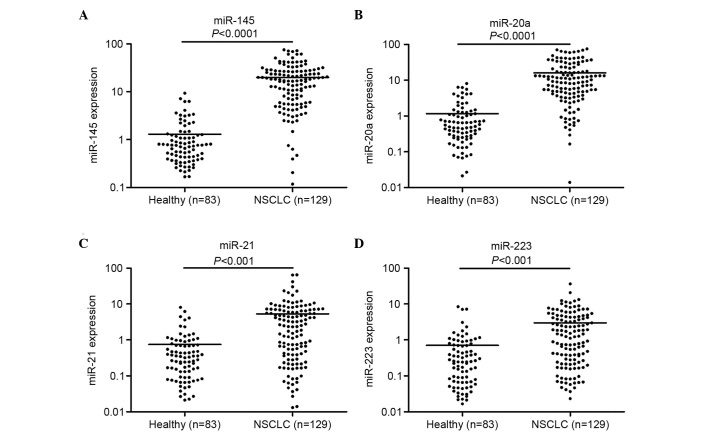
In order to increase the number of samples for validation, we merged the 40 cases of samples in the screening stage with those in the validation stage, making 129 cases of NSCLC patients and 83 healthy individuals. Following detection by RT-PCT, we observed that the expression of miR-145, miR-20a, miR-21 and miR-223 in the plasma of NSCLC patients was significantly enhanced compared with that of the healthy controls (Fig. 3). ROC curve analysis revealed that these four miRNAs distinguished NSCLC patients from healthy individuals (Fig. 4). The AUCs of miR-145, miR-20a, miR-21 and miR-223 were 0.886 (95% CI, 0.835–0.925), 0.889 (95% CI, 0.839–0.928), 0.838 (95% CI, 0.782–0.885) and 0.809 (95% CI, 0.749–0.860), respectively. Their optimum cut-off point was 4.086, 2.428, 1.101 and 1.020, respectively, and the sensitivity and specificity at this cut-off point were 80.6 and 89.2%; 79.8 and 88.0%; 77.5 and 85.5%; and 69.8 and 84.3%, respectively. Multivariate logistic regression analyses on variables including gender, age, smoking history and plasma miRNAs revealed that plasma miR-145, miR-20a, miR-21 and miR-223 were potential biomarkers for early-stage NSCLC diagnosis. The odds ratios of miR-145, miR-20a, miR-21 and miR-223 were 18.5 (95% CI, 7.410–45.649), 16.0 (95% CI, 6.516–43.810), 13.0 (95% CI, 5.570–36.219) and 11.0 (95% xCI, 4.516–30.629), respectively. The panel of miR-145, miR-20a, miR-21 and miR-223 had the highest predictive accuracy in early-stage NSCLC screening (Fig. 5). The AUC, optimum cut-off point, sensitivity and specificity of this combination were 0.897 (95% CI, 0.875–0.917), 1.485, 81.8 and 90.1%, respectively.
Figure 3.
Large-scale validation of (A) miR-145, (B) miR-20a, (C) miR-21 and (D) miR-223 in plasma samples. Expression levels of the miRNAs (Log10 scale for y-axis) are normalized to miR-16. The line represents the median value. The Mann-Whitney U test was used to determine statistical significance.
Figure 4.
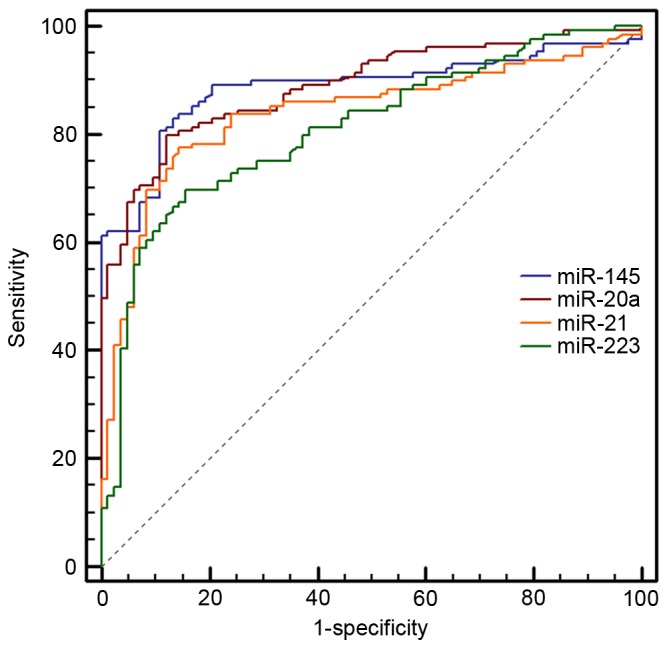
Receiver operating characteristic curve analysis of miR-145, miR-20a, miR-21 and miR-223.
Figure 5.
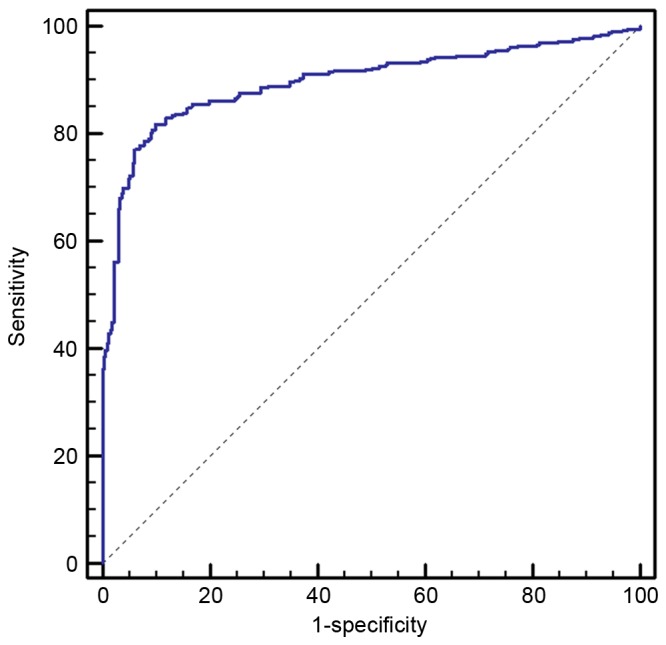
Combination receiver operating characteristic curve analysis of miR-145 + miR-20a + miR-21 + miR-223.
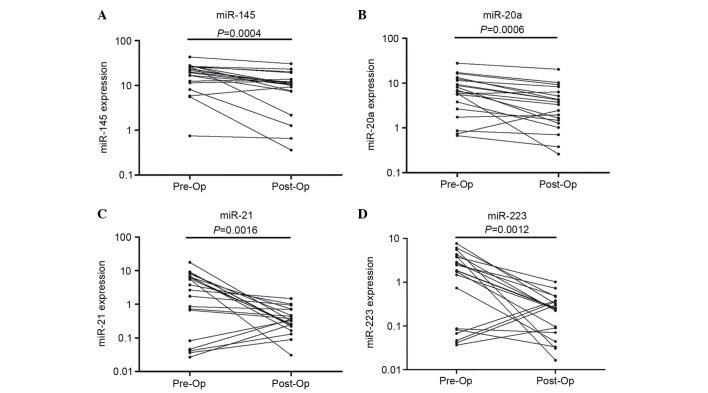
Expression levels of plasma miRNAs before and after surgery
To investigate whether the plasma miR-145, miR-20a, miR-21 and miR-223 were of tumor origin, we collected plasma samples from an independent set of 20 early-stage NSCLC patients, and then examined the plasma expression levels of these four miRNAs before and after surgery with RT-qPCR. We observed that the expression of these four miRNAs was significantly decreased in the post-operative patients compared with the pre-operative patients (Fig. 6).
Figure 6.
Changes in miRNA expression levels: (A) miR-145, (B) miR-20a, (C) miR-21 and (D) miR-223, before and 7–10 days after surgery. Expression levels of the miRNAs (Log10 scale for y-axis) are normalized to miR-16.
Discussion
Lung cancer is a major global health problem. Due to the high level of incidence and low therapeutic efficacy, it has become the leading cause of malignancy-related mortality in numerous countries (22). Late diagnosis of lung cancer is one of the significant contributing factors to the high mortality of this disease since the tumor has often spread to distant organs at the time of diagnosis (23). Conventional diagnostic methods, including CT, positron emission tomography and X-ray, have several limitations. CT screening has a relatively high false positive rate, and benign lung nodules may be misdiagnosed as malignant tumors (24). Several protein biomarkers have been identified as non-invasive and cost-effective diagnostic tools for early-stage NSCLC, which have limited sensitivity and specificity, including cytokeratin fragment 21-1 (sensitivity≈0.5, specificity≈0.95), tissue polypeptide-specific antigen (sensitivity≈0.34, specificity≈0.95) and carcinoembryonic antigen (sensitivity≈0.53, specificity≈0.95) (25,26). Therefore, developing novel non-invasive biomarkers by taking advantage of recent developments in molecular genetics for the screening of early-stage NSCLC is of clinical significance.
Accumulating studies suggest that circulating miRNAs may be used as potential molecular biomarkers for several disease conditions, including human malignancies (27–29). Chen et al reported that the expression profiles of plasma miRNAs in lung cancer, colorectal cancer and diabetic patients are different from those of healthy individuals, and proposed that miR-25 and miR-223 may be used as diagnostic markers for NSCLC (12). In 2011, Chen et al further studied the function of plasma miRNAs as diagnostic markers for NSCLC, and identified that 10 miRNAs may potentially be used as diagnostic markers, with the sensitivity and specificity of the combined use of these 10 miRNAs reaching 93 and 90%, respectively (7). Shen et al revealed that miR-21, miR-126, miR-210 and miR-486-5p may be used as diagnostic markers for stage I NSCLC, and the sensitivity and specificity of the combined use of these four miRNAs were 73.33 and 96.55%, respectively (11). Foss et al demonstrated that plasma miR-1254 and miR-574-5p serve as non-invasive screening tools in the early detection of NSCLC with relatively high accuracy (30). Subsequently, an increasing number of studies have investigated the diagnostic value of miRNAs for early-stage lung cancer (7). Compared with these previous studies, the present study has several advantages. Firstly, we concentrated on the detection of early-stage NSCLC by using miRNAs as biomarkers, and observed that plasma miR-145, miR-20a, miR-21 and miR-223 may be used as biomarkers for the early detection of NSCLC with relatively high sensitivity and specificity. Secondly, normalization is a key step for the accurate quantification of RNA levels with RT-qPCR. Our results revealed that RNU6B is unstable at room temperature. miR-16 exhibited a higher stability and abundance than RNU6B in plasma. Finally, we demonstrated that the expression of plasma miR-145, miR-20a, miR-21 and miR-223 was significantly decreased in the postoperative plasma samples when compared with the preoperative samples. To our knowledge, the present study is the first to evaluate the expression levels of plasma miRNAs before and after surgery.
In the present study, we observed that miR-145, miR-20a, miR-21 and miR-223 were significantly dysregulated in NSCLC patients compared with healthy controls. The data from our study demonstrated that each single miRNA presents high sensitivity and specificity in the detection process. Despite the different expression levels, all four of these miRNAs were validated to have the potential to discriminate early-stage NSCLC patients from healthy controls. The panel of four candidate miRNAs demonstrated the highest predictive accuracy in NSCLC detection (AUC=0.897). We also analyzed the expression levels of plasma miRNAs before and after surgery, and observed that the levels of the four candidate miRNAs decreased rapidly following surgical removal of the tumors. Collectively, miR-145, miR-20a, miR-21 and miR-223 presented great clinical value in NSCLC preliminary screening, and further studies in a large population are required to validate the feasibility of these miRNAs as novel non-invasive biomarkers.
Upregulation of miR-21 has been observed in numerous human cancers (31). Capodanno et al previously revealed that miR-21 expression was significantly increased in NSCLC tissues, and may be used to distinguish NSCLC from non-cancerous lung tissues (32). Furthermore, high expression of miR-21 predicts recurrence and unfavorable survival in non-small cell lung cancer (33). The data produced from the present study imply that plasma miR-21 may serve as a biomarker for the diagnosis of lung cancer. miR-20a inhibits E2F1, which is a transcription factor associated with lung cancer cell growth, and it serves as a non-invasive screening tool for early detection of lung cancer (34). miR-145 inhibits proliferation of NSCLC cells by targeting c-Myc (35) and plays an inhibitory role in tumor angiogenesis, cell growth and invasion and tumor growth through post-transcriptional regulation through N-RAS and vascular endothelial growth factor-A in breast cancer (36). However, conclusions from several studies which focused on the serum expression of miR-145 were inconsistent. These included studies in breast cancer (37,38). As for miR-223, scientists have proven that miR-223 functions as a tumor suppressor in lung cancer cells at multiple steps of tumorigenesis and progression (39,40), which serves as reasonable explanation for the function of miR-223 as an NSCLC biomarker.
In our study, we also noted that the expression levels of these four miRNAs were significantly different before and 7–10 days after the surgery in early-stage NSCLC patients. However, it is not known what causes this change, and additional studies are required to clarify this.
In this study, we have demonstrated that these four miRNAs in plasma had dysregulated expression in NSCLC, suggesting that they may serve as biomarkers in precise clinical diagnosis of early-stage NSCLC. However, certain limitations in our tests need to be addressed. In the training set, there were only 10 miRNAs included in our study. Further studies are required to expand the investigation number of miRNAs. Secondly, the changes in the miRNA expression level before and after surgery need to be verified using a larger sample. The selection of reference gene is a crucial step for accurate quantification by RT-PCR. However, there is still no well-recognized reference gene for miRNA quantification in plasma. In this study, we selected miR-16 and RNU6B as candidate reference genes and observed no significant difference between them in early-stage NSCLC patients and healthy individuals. However, RNU6B was not stable at room temperature; therefore, we selected miR-16 as the internal reference, and this was also supported by previous studies (16,19,41,42). Further studies are still required to identify and validate more suitable reference genes to study circulating miRNAs in early-stage NSCLC patients, and thus provide more accurate results for RT-PCR.
In summary, miR-145, miR-20a, miR-21 and miR-223 appear to be novel biomarkers for early detection of early-stage NSCLC. However, other miRNAs that could function as biomarkers for the early diagnosis of NSCLC may also exist in plasma, therefore further studies are required to screen more suitable miRNAs, and thus improve the diagnostic ability for early-stage NSCLC patients.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Ettinger DS, Akerley W, Bepler G, Blum MG, Chang A, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Ganti AK, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8:740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 3.Crinò L, Weder W, van Meerbeeck J, Felip E. ESMO Guidelines Working Group: Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21:v103–v115. doi: 10.1093/annonc/mdq207. (Suppl 5) [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 5.Swensen SJ, Jett JR, Hartman TE, Midthun DE, Mandrekar SJ, Hillman SL, Sykes AM, Aughenbaugh GL, Bungum AO, Allen KL. CT screening for lung cancer: five-year prospective experience. Radiology. 2005;235:259–265. doi: 10.1148/radiol.2351041662. [DOI] [PubMed] [Google Scholar]
- 6.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabrò E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel non-invasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller A, Leidinger P, Gislefoss R, Haugen A, Langseth H, Staehler P, Lenhof HP, Meese E. Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 2011;8:506–516. doi: 10.4161/rna.8.3.14994. [DOI] [PubMed] [Google Scholar]
- 11.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabrò E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva J, Garcia V, Zaballos Á, Provencio M, Lombardía L, Almonacid L, García JM, Domínguez G, Peña C, Diaz R, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. 2011;37:617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 15.Tsim S, O'Dowd CA, Milroy R, Davidson S. Staging of non-small cell lung cancer (NSCLC): a review. Respir Med. 2010;104:1767–1774. doi: 10.1016/j.rmed.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 17.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 18.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16:360–364. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 20.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Russ R, Slack FJ. Cigarette-smoke-induced dysregulation of MicroRNA expression and its role in lung carcinogenesis. Pulm Med. 2012;2012:791234. doi: 10.1155/2012/791234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Piper-Hunter MG, Crawford M, Nuovo GJ, Marsh CB, Otterson GA, Nana-Sinkam SP. MicroRNAs in the pathogenesis of lung cancer. J Thorac Oncol. 2009;4:1028–1034. doi: 10.1097/JTO.0b013e3181a99c77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swensen SJ, Jett JR, Sloan JA, Midthun DE, Hartman TE, Sykes AM, Aughenbaugh GL, Zink FE, Hillman SL, Noetzel GR, et al. Screening for lung cancer with low-dose spiral computed tomography. Am J Respir Crit Care Med. 2002;165:508–513. doi: 10.1164/ajrccm.165.4.2107006. [DOI] [PubMed] [Google Scholar]
- 25.Wieskopf B, Demangeat C, Purohit A, Stenger R, Gries P, Kreisman H, Quoix E. Cyfra 21-1 as a biologic marker of non-small cell lung cancer. Evaluation of sensitivity, specificity, and prognostic role. Chest. 1995;108:163–169. doi: 10.1378/chest.108.1.163. [DOI] [PubMed] [Google Scholar]
- 26.Nisman B, Lafair J, Heching N, Lyass O, Baras M, Peretz T, Barak V. Evaluation of tissue polypeptide specific antigen, CYFRA 21-1, and carcinoembryonic antigen in nonsmall cell lung carcinoma: does the combined use of cytokeratin markers give any additional information? Cancer. 1998;82:1850–1859. doi: 10.1002/(SICI)1097-0142(19980515)82:10<1850::AID-CNCR6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Andersen M, Grauslund M, Ravn J, Sørensen JB, Andersen CB, Santoni-Rugiu E. Diagnostic potential of miR-126, miR-143, miR-145, and miR-652 in malignant pleural mesothelioma. J Mol Diagn. 2014;16:418–430. doi: 10.1016/j.jmoldx.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Zheng H, Zhang L, Zhao Y, Yang D, Song F, Wen Y, Hao Q, Hu Z, Zhang W, Chen K. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS One. 2013;8:e77853. doi: 10.1371/journal.pone.0077853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang RJ, Zheng YH, Wang P, Zhang JZ. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:765–771. [PMC free article] [PubMed] [Google Scholar]
- 30.Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6:482–488. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 31.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capodanno A, Boldrini L, Proietti A, Alì G, Pelliccioni S, Niccoli C, D'Incecco A, Cappuzzo F, Chella A, Lucchi M, et al. Let-7g and miR-21 expression in non-small cell lung cancer: correlation with clinicopathological and molecular features. Int J Oncol. 2013;43:765–774. doi: 10.3892/ijo.2013.2003. [DOI] [PubMed] [Google Scholar]
- 33.Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong W, Liao Y, Du J. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2013;49:604–615. doi: 10.1016/j.ejca.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 34.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng A, Hu J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res. 2010;29:151. doi: 10.1186/1756-9966-29-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou C, Xu Q, Mao F, Li D, Bian C, Liu LZ, Jiang Y, Chen X, Qi Y, Zhang X, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle. 2012;11:2137–2145. doi: 10.4161/cc.20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mar-Aguilar F, Mendoza-Ramirez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodríguez-Padilla C, Reséndez-Pérez D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.1155/2013/259454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL, et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haneklaus M, Gerlic M, O'Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. 2013;274:215–226. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nian W, Ao X, Wu Y, Huang Y, Shao J, Wang Y, Chen Z, Chen F, Wang D. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclin-dependent kinase 2. Oncol Lett. 2013;6:359–366. doi: 10.3892/ol.2013.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 42.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]