Abstract
The purpose of the present study was to investigate the significance of C-X-C motif chemokine receptor type 4 (CXCR4) and epidermal growth factor receptors (EGFRs) in triple-negative breast cancer (TNBC). CXCR4 and EGFR expression levels were immunohistochemically determined in 207 primary breast cancer specimens. The associations between receptor expression and clinicopathological characteristics were analyzed, and receptor expression was also assessed as a prognostic factor. In the human MDA-MB-231 TNBC cell line, CXCR4 or EGFR was stably knocked down by short hairpin RNA, and the biological behavior of the cells, including migration, invasion and tumorigenesis, was investigated. The results revealed that TNBC was associated with younger age, higher histological grade and an aggressive phenotype. CXCR4 and EGFR were highly expressed in patients with TNBC, and those with high CXCR4 or EGFR expression exhibited an unfavorable prognosis in terms of disease-free survival and overall survival. In MDA-MB-231 cells, the expression of CXCR4 protein was decreased following EGFR silencing, while CXCR4 knockdown also caused a decrease in EGFR protein levels. The migratory and invasive capabilities of MDA-MB-231 cells were decreased following the knockdown of CXCR4 or EGFR expression. A strong correlation between CXCR4 and EGFR expression was identified in patients with TNBC. The results suggest that elevated expression levels of these two receptors may serve as predictive factors for poor prognosis in patients with TNBC. In addition, tumor proliferation, migration, invasion and tumorigenesis are weakened in MDA-MB-231 cells following suppression of CXCR4 or EGFR expression. Therefore, EGFR and CXCR4 may be potential therapeutic targets for TNBC.
Keywords: triple-negative breast cancer, epidermal growth factor receptor, C-X-C motif chemokine receptor type 4, prognosis
Introduction
Triple-negative breast cancer (TNBC) is characterized by the absence of estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 (HER2) expression. TNBC accounts for ~15% of all invasive breast cancers, typically occurs in younger women, and is associated with high histological grade and poor prognosis (1,2).
Although numerous molecules are involved in breast cancer metastasis, the precise mechanism of tumor cell migration to specific organs is not well established. In the past two decades, C-X-C motif chemokine receptor type 4 (CXCR4) has been identified to be important in cancer metastasis (3). Chemokines belong to a superfamily of small, cytokine-like proteins that induce cytoskeletal rearrangement and adhesion to endothelial cells, and direct migration through their interaction with G-protein-coupled receptors (GPCRs). Among the chemokines, CXCR4 is consistently expressed in human breast cancer cell lines and human primary and metastatic breast cancer tissues (4). Retrospective studies have reported that the expression of CXCR4 is associated with higher rates of recurrence and cancer-associated mortality, larger tumor size, advanced tumor-node-metastasis stage and shorter survival, and predicts poor prognosis in cancer patients (5), particularly those with TNBC (6). However, the conflicting results of previous studies have indicated that the expression levels of CXCR4 and its localization in malignant cells remain to be established (7).
Epidermal growth factor receptor (EGFR) and HER2 belong to the ErbB family of receptor tyrosine kinases, and their overexpression predicts poor prognosis for patients with breast cancer (8,9). HER2 has been reported to be overexpressed in 20–25% of patients with breast cancer, and has been validated as a therapeutic target. Additionally, EGFR is frequently overexpressed in TNBC (10), and may be used as a potential drug target. Several inhibitors of EGFR, including gefitinib, lapatinib and cetuximab, have been developed as targeted therapies (11). However, EGFR inhibition for the treatment of TNBC has yielded little clinical benefit (12).
Previously, studies have demonstrated crosstalk between CXCR4 and EGFR in human breast tumor tissues, which is important in the metastasis and prognosis of breast cancer, particularly for TNBC (13,14). Another study reported that C-X-C motif chemokine 12 (CXCL12)/CXCR4 signaling transactivates EGFR through the activation of proto-oncogene c-Src in breast cancer cells (15); the activation of EGFR also upregulates CXCR4 transcriptionally via the increased expression and activity of hypoxia-inducible factor 1α (HIF-1α) (16,17). However, the specific mechanisms underlying the involvement of EGFR and CXCR4 in breast cancer remain to be studied in depth, and EGFR and CXCR4 may be double targets for treatment of TNBC. The purpose of the present study was to investigate the significance of CXCR4 and EGFR in TNBC.
Materials and methods
Patients and immunohistochemical (IHC) analysis
A total of 207 cases of female patients with primary breast cancer, which had been diagnosed as invasive ductal carcinoma between January 2002 and December 2012, were collected at the Cancer Hospital of Shantou University Medical College (Shantou, China). Of these, 109 cases were non-TNBC and 98 were TNBC. Clinicopathological parameters, including tumor size, lymph node status and pathological features, were recorded from the patients' medical records. The use of patient tissues was approved by the Ethics Committee of Shantou University Medical College, and informed consent was obtained from all enrolled patients.
IHC staining was used to determine the expression of EGFR and CXCR4 in human breast cancer tissues. Tissue slices were embedded in melted wax at a high temperature prior to sectioning at a thickness of 5 µm. The tissue sections were then deparaffinized in xylene and rehydrated in graded alcohol. For antigen retrieval, sections were treated with proteinase A (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) for 10 min at 37°C in a constant temperature cabinet. Endogenous peroxidase activity was blocked by 0.3% hydrogen peroxide for 10 min. Sections were then blocked with 4% goat serum (OriGene Technologies, Inc., Beijing, China) for 30–40 min at room temperature, and incubated with mouse monoclonal anti-EGFR (OriGene Technologies, Inc.; ready-to-use; #ZM-0093) or mouse monoclonal anti-CXCR4 (Abcam, Cambridge, UK; dilution 1:200; #ab58176) primary antibodies at 4°C overnight. Tissue sections were rinsed three times with PBS (10 min per rinse), and then incubated with a horseradish peroxidase-conjugated anti-mouse secondary antibody (OriGene Technologies, Inc.; dilution 1:500; #ZB-2305) for 1 h at room temperature. Antibody staining was visualized with 3,3′-diaminobenzidine, and all tissue sections were incubated for 20–30 sec. The sections were counterstained with hematoxylin (Baso Diagnostics Inc., Zhuhai, China) for 3 min. Finally, the sections were gently cleaned in water for 10 min and then sealed with a cover slide.
The immunostaining intensity and the percentage of positive cells were assessed and scored. With regard to CXCR4 and EGFR staining, if the cytoplasm and/or membrane of the breast cancer cells were colored brown, the cells were considered positive. The staining intensity score was based on four classes: None 0; weak brown 1; moderate brown 2; intense brown 3. The percentage of positive tumor cells was classified into five grades, as follows: Negative 0; 1–25% 1; 26–50% 2; 51–75% 3; ≥76% 4. The total score was obtained by multiplying the staining intensity and percentage scores. Sections with a total score ≥6 were considered to have high expression, and scores <6 were considered low expression (18).
Cell culture
The MDA-MB-231 TNBC cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). This cell line is known to overexpress CXCR4 and EGFR. Cells were grown in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). Stable EGFR-silenced and CXCR4-silenced MDA-MB-231 cells were also cultured in the same way. All cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Knockdown of CXCR4 or EGFR by short hairpin RNA (shRNA)
shRNAs targeting CXCR4 or EGFR (Shanghai GenePharma Co., Ltd., Shanghai, China), driven by a U6 promoter, were cloned into pGPU6/GFP/Neo plasmids (Shanghai GenePharma Co., Ltd.). The sense sequences used for the targeted silencing of CXCR4 were as follows: shRNA #1, 5′-GTATGACAACAGCCTCAAG-3′; shRNA #2, 5′-GAAGCATGACGGACAAGTA-3′. The sense sequences for targeting EGFR were as follows: shRNA #1, 5′-CGCAAAGTGTGTAACGGAATA-3′; shRNA #2, 5′-CTGACTCCGTCCAGTATTGAT-3′. Empty vector-transfected MDA-MB-231 cells were used as the negative control. The day prior to transfection, 5×105 MDA-MB-231 cells were inoculated into each well of a 6-well culture plate. For transfection, 8 µg shRNA plasmid was combined with 10 µl Lipofectamineä 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol, and added to the cells (19). After 6 h, cells were supplied with 3 ml/well RPMI-1640 supplemented with 10% FBS. Selection commenced at 48 h following infection; shCXCR4 cells were treated with 2 µg/ml puromycin (Sangon Biotech, Shanghai, China) and shEGFR cells were treated with 1,000 µg/ml geneticin (G418; Sangon Biotech) (3). The cells that were infected with shRNAs targeting CXCR4 and EGFR were selected with puromycin or G418 simultaneously for ~2 weeks.
MTT assays
An MTT Cell Proliferation assay kit (Beyotime Institute of Biotechnology, Shanghai, China) was used to assess cell proliferation. Firstly, the cells were seeded into 96-well plates at a density of 2×103 cells/well and incubated for 1–5 days (20). Every 24 h, one plate was removed for MTT analysis: 10 µl MTT solution was added to each well and the cells were incubated at 37°C in a humidified atmosphere containing 5% CO2 for 4 h. Subsequently, 100 µl formazan was added into each well, and the cells were incubated at 37°C with 5% CO2 until the formazan crystals were solubilized in 150 µl dimethyl sulfoxide (DMSO). The optical density (OD) of each sample was determined by measuring the absorbance at 570 nm with a microplate reader (BioTek Instruments, Inc., Winooski, VT, USA) (21,22).
Cell migration and invasion assay
Cellular migration was determined using 24-well Transwell Chambers with an 8-µm pore size and a track-etched membrane (Corning, Inc., New York, CA, USA) (4,23). Briefly, a chamber insert was placed in each well of a 24-well dish containing 600 µl RPMI-1640 supplemented with 20% FBS in the bottom chamber. According to previous protocol, the cells were then suspended at a concentration of 1×105 cells/ml in serum-free RPMI-1640 medium supplemented with 0.2% bovine serum albumin (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany), and 100 µl cell suspension was added into the upper chamber (19,21,22). Cells were incubated at 37°C with 5% CO2 for 24 h, and then the cells were fixed with 100% methanol for 30 min at room temperature and stained with 0.1% crystal violet for 20 min at room temperature. Following each step, the cells were washed three times for 3 min/time with PBS at room temperature. Subsequently, the non-migrated cells on the upper side of the membranes were removed and the migrated cells on the underside of the membranes were observed under a TS100 inverted fluorescence microscope (Nikon, Corporation, Tokyo, Japan) in five randomized fields. A cell invasion assay was also conducted in the same manner (13,17), but with Matrigel™ Invasion Chamber 24-well plates with 8.0 µm pores (BD Biosciences, San Jose, USA) and an incubation time of 48 h. The number of migrated or invasive cells was counted in five randomly chosen fields under a microscope.
Western blotting
Western blot analysis was performed as reported previously (24). Cells were lysed in radioimmunoprecipitation assay lysis buffer (EMD Millipore, Billerica, MA, USA). Total protein concentration was determined using a Bradford Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA) (25). Extracted protein (~30 µg) was subjected to electrophoresis on a 10% SDS polyacrylamide gel, and was then transferred to a polyvinylidene difluoride membrane (EMD Millipore Corporation, Billerica, MA, USA) at 80 V for 2 h at 4°C. The membrane was blocked in skim milk for 1 h, and subsequently incubated overnight at 4°C with a mouse monoclonal anti-CXCR4 (Abcam; dilution 1:2,000; #ab58176) or a rabbit monoclonal anti-EGFR (Cell Signaling Technology, Inc., Danvers, MA, USA; dilution 1:3,000; #4405) antibody. The following day, the membrane was gently washed in Tris-buffered saline with 0.1% Tween-20 (TBST), and then incubated with goat anti-mouse (#ZB-2305) or goat anti-rabbit (#ZB-2301) horseradish peroxidase-conjugated secondary antibodies (OriGene Technologies, Inc.; dilution 1:2,000) for 2 h at room temperature. Subsequent to washing in TBST, the immunocomplexes were detected using ECL Plus reagent (Applygen Technologies, Inc., Beijing, China) (16,26).
Tumor xenografts
All animal studies were approved by the Institutional Animal Care and Use Committees of Shantou University Medical College and were performed with 8–10-week-old female nude mice (n=12; weights 16–19 g; Vital River Laboratory Animal Technology Co., Ltd., Beijing, China). Nude mice were bred in laminar air chambers, and their food and water were sterilized at a high temperature (121°C). Room ventilation was performed 10–15 times per hour. Using artificial lighting, the light-dark cycle was as follows: 10 h of light and 14 h of dark each day. A total of 2×106 stably transduced MDA-MB-231 cells were suspended in 100 µl PBS and injected into the right leg of 3 female nude mice in the control, shEGFR, shCXCR4, shEGFR and shCXCR4 groups (27). Tumor growth was determined, along the longest diameter and shortest width, every week with digital calipers. The tumor volume was calculated according to the following formula: Volume = 0.5 × width2 × length (28,29). All animals were sacrificed through overdose of CO2 at four weeks following injection with the cells. Finally, tumors were removed, measured and weighed.
Statistical analysis
Analysis was performed using SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA). The associations between patient characteristics and the expression of CXCR4 and EGFR between the groups were examined using Pearson's χ2 test. Correlations between variables were evaluated via the Spearman's rank correlation co-efficient test. Multivariate survival analysis was performed using a Cox proportional hazards regression analysis model, including age at diagnosis, histological grade, tumor size, lymph node status, distant metastasis and family history. Disease-free survival (DFS) and overall survival (OS) rates were also described (30). P<0.05 was considered to indicate a statistically significant difference.
Results
Clinicopathological characteristics of the patients
A total of 207 patients with breast cancer were enrolled in the present study, including 98 patients with TNBC and 109 patients with non-TNBC. The mean age was 51.2 years, and the mean follow-up time was 60 months. The clinicopathological characteristics are summarized in Table I. Compared with non-TNBC cases, patients with TNBC were younger at the time of initial diagnosis (χ2=5.898; P=0.015), and more frequently had a family history of breast cancer (χ2=4.382; P=0.036). A tumor diameter of <2 cm accounted for ~20.4% of TNBC cases, but only 6.4% of non-TNBC cases (χ2=7.709; P=0.005). The percentage of patients with a histological grade of III was 59.2% in the TNBC group, compared with 29.4% in the non-TNBC group (χ2=17.486; P<0.001). In total, 34/98 patients with TNBC (34.7%) had distant metastasis, exhibiting a significantly higher rate of distant metastasis, compared with patients in the non-TNBC group (20/109; 18.3%; χ2=7.150, P=0.007). However, no significant differences in lymph node status or clinical stage were identified between the TNBC and non-TNBC groups.
Table I.
Clinicopathological characteristics of the breast cancer patients.
| Characteristic | TNBC patients, n (%) | Non-TNBC patients, n (%) | χ2 | P-value |
|---|---|---|---|---|
| Age, years | 5.898 | 0.015a | ||
| ≤40 | 29 (29.6) | 16 (14.7) | ||
| >40 | 67 (70.4) | 93 (85.3) | ||
| Histological grade | 17.486 | <0.001a | ||
| I–II | 40 (40.8) | 77 (70.6) | ||
| III | 58 (59.2) | 32 (29.4) | ||
| Clinical stage | 0.194 | 0.660 | ||
| I–II | 50 (51.0) | 60 (55.0) | ||
| III | 48 (49.0) | 49 (45.0) | ||
| Tumor size | 7.709 | 0.005a | ||
| T0-T1 (<2 cm) | 20 (20.4) | 7 (6.4) | ||
| T2-T4 (≥2 cm) | 78 (79.6) | 102 (93.6) | ||
| LN status | 0.080 | 0.777 | ||
| Negative | 35 (35.7) | 41 (37.6) | ||
| Positive | 63 (64.3) | 68 (62.4) | ||
| Number of metastatic LNs | 5.570 | 0.062 | ||
| ≤3 | 44 (44.9) | 62 (56.9) | ||
| 4–9 | 35 (35.7) | 23 (21.1) | ||
| ≥10 | 19 (19.4) | 24 (22.0) | ||
| Distant metastasis | 7.150 | 0.007a | ||
| Negative | 64 (65.3) | 89 (81.7) | ||
| Positive | 34 (34.7) | 20 (18.3) | ||
| Family history of breast cancer | 4.382 | 0.036a | ||
| Negative | 87 (88.8) | 105 (96.3) | ||
| Positive | 11 (11.2) | 4 (3.7) |
P<0.05. TNBC, triple-negative breast cancer; T, tumor stage; LN, lymph node.
Expression of CXCR4 and EGFR in various subtypes of human breast cancers
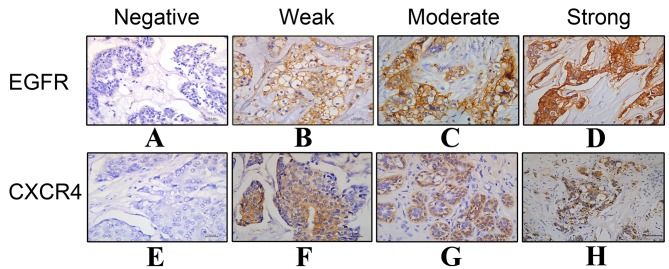
The results of IHC staining for EGFR and CXCR4 in breast cancer tissues are presented in Fig. 1 and Table II. The results revealed that 69/98 (70.4%) TNBC tissues highly or moderately expressed CXCR4, whereas only 43/109 (39.4%) non-TNBC tissues expressed CXCR4 (χ2=18.691; P<0.001). For EGFR, high expression levels were observed in 60 patients with TNBC (61.2%), exhibiting a significantly higher rate of positive expression compared with the non-TNBC group, in which in 25/109 patients (22.9%) had high EGFR expression (χ2=29.698; P<0.001). Furthermore, among the 98 patients with TNBC, EGFR and CXCR4 were co-expressed in 44 (44.9%) patients, 41 (41.8%) patients expressed either EGFR or CXCR4, and 13 (13.3%) patients expressed neither EGFR nor CXCR4. In the non-TNBC group, 11/109 patients (10.1%) co-expressed EGFR and CXCR4, 46 (42.2%) patients expressed either EGFR or CXCR4, and 52 (47.7%) patients expressed neither EGFR nor CXCR4. Therefore, TNBC patients exhibited significant co-overexpression of the two receptors (χ2=43.024; P<0.001). Spearman's rank correlation analysis revealed that EGFR expression was associated with that of CXCR4 (r=0.188; P=0.007).
Figure 1.
Immunohistochemical staining of (A-D) EGFR and (E-H) CXCR4 expression in breast cancer. (A) Negative expression (−) of EGFR; (B) weak expression (+) of EGFR; (C) moderate expression (++) of EGFR; (D) intense expression (+++) of EGFR; (E) negative expression (−) of CXCR4; (F) weak expression (+) of CXCR4; (G) moderate expression (++) of CXCR4; (H) intense expression (+++) of CXCR4. Magnification, 400x; scale bar, 100 µm. EGFR, epidermal growth factor receptor; CXCR4, C-X-C motif chemokine receptor type 4.
Table II.
Expression levels of CXCR4 and EGFR in human TNBC and non-TNBC tissues and their correlations.
| Characteristic | TNBC patients, n (%) | Non-TNBC patients, n (%) | χ2 | P-value |
|---|---|---|---|---|
| EGFR expression | 29.698 | <0.001a | ||
| High | 60 (61.2) | 25 (22.9) | ||
| Low | 38 (38.8) | 84 (77.1) | ||
| CXCR4 expression | 18.691 | <0.001a | ||
| High | 69 (70.4) | 43 (39.4) | ||
| Low | 29 (29.6) | 66 (60.6) | ||
| Co-expression | 43.024 | <0.001a | ||
| Both high | 44 (44.9) | 11 (10.1) | ||
| Either high | 41 (41.8) | 46 (42.2) | ||
| Both low | 13 (13.3) | 52 (47.7) |
P<0.05. Differences in high and low expression levels of the two biomarkers between TNBC and non-TNBC were assessed by Pearson's χ2 analysis. EGFR, epidermal growth factor receptor; CXCR4, C-X-C chemokine receptor type 4; TNBC, triple-negative breast cancer.
The 5-year DFS rates for patients with TNBC and without TNBC were 41 and 52%, respectively (P=0.041). The OS rates for TNBC and non-TNBC were 63 and 75%, respectively (P=0.044). These results suggest that patients with TNBC have a poorer prognosis, compared with patients without TNBC.
CXCR4 suppression reduces EGFR expression and EGFR suppression reduces CXCR4 expression in MDA-MB-231 cells
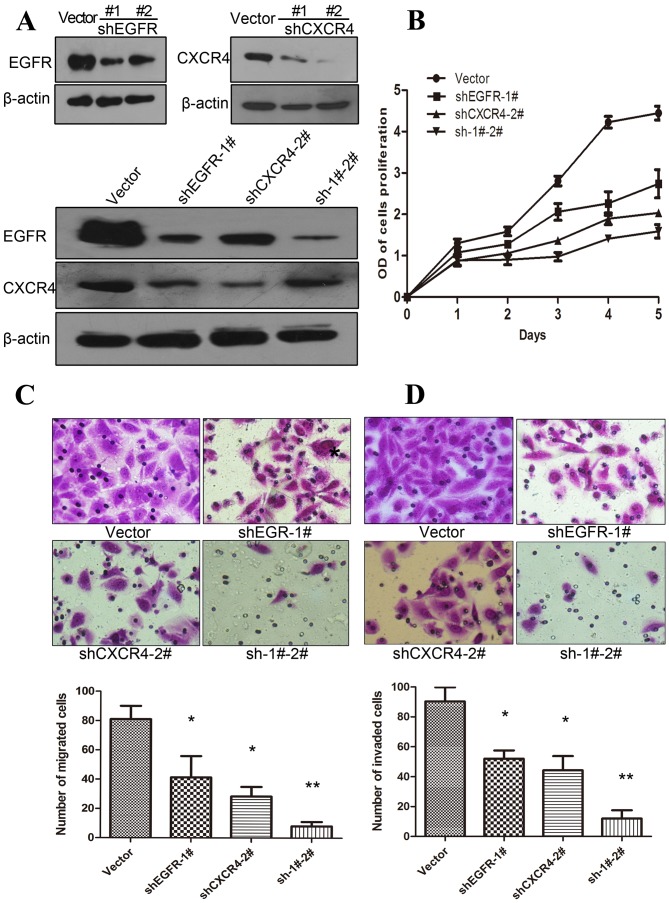
Following individual receptor knockdown by shRNA in MDA-MB-231 tumor cells, western blot analysis was used to investigate whether the expression levels of CXCR4 and EGFR were associated or independent. MDA-MD-231 cells expressed a high level of CXCR4 and EGFR, as described previously (4,31,32), and EGFR and CXCR4 protein were effectively silenced by shRNA (Fig. 2A). The results revealed that CXCR4 expression levels were reduced when EGFR expression was suppressed by shRNA. Similarly, the expression levels of EGFR were lower when CXCR4 was suppressed by shRNA in MDA-MB-231 cells. However, when EGFR was suppressed in cells expressing a low level of CXCR4, the protein expression levels of CXCR4 were increased marginally (Fig. 2A). These results suggest that the expression levels of EGFR and CXCR4 are co-dependent.
Figure 2.
EGFR and CXCR4 expression levels are reciprocally affected in MDA-MB-231 cells, and alter the biological behavior of cells. (A) Western blot analysis results: (a) Transfection with shEGFR #1 effectively blocked the expression of EGFR protein; (b) shCXCR4 #2 blocked the expression of CXCR4 protein almost completely; (c) when EGFR was suppressed, CXCR4 was reduced. Each experiment was repeated ≥3 times. (B) The MTT assay results revealed that MDA-MB-231 cells with suppressed CXCR4 or EGFR exhibited a significant reduction in proliferation, compared with those transfected with the empty vector control, and the greatest inhibition was observed in cells in which EGFR and CXCR4 were both suppressed. (C) Transwell migration and (D) Transwell invasion assays were performed in MDA-MB-231 cells with EGFR and/or CXCR4 silenced by shRNA, and the results were quantified by comparison with the empty vector-transfected MDA-MB-231 cells. *P<0.05 and **P<0.01, compared with control vectors. Error bars correspond to the mean ± standard deviation of triplicate experiments. EGFR, epidermal growth factor receptor; CXCR4, C-X-C motif chemokine receptor type 4; shRNA, short hairpin RNA; shEGFR, shRNA targeting EGFR; shCXCR4, shRNA targeting CXCR4; sh-1#-2#, shRNAs targeting EGFR and CXCR4.
Suppression of CXCR4 and EGFR inhibits the proliferation, migration and invasion of MDA-MB-231 cells
In order to delineate the role of the EGFR and CXCR4 in cell proliferation, MTT experiments were performed in MDA-MB-231 cell lines, in which EGFR and/or CXCR4 were stably knocked down. The results revealed that the suppression of EGFR or CXCR4 inhibited the proliferation of breast cancer cells by 40–50%, compared with untransfected cells. Furthermore, MDA-MB-231 cells with double knockdown of CXCR4 and EGFR expression exhibited a greater reduction in proliferation of almost 67%, compared with the vector control (Fig. 2B).
To further investigate the biological effects of MDA-MB-231 cells following CXCR4-silencing and/or EGFR-silencing, Transwell migration and invasion assays were performed. The results revealed that suppression of either EGFR or CXCR4 substantially reduced the migratory ability of MDA-MB-231 cells by 50 and 69%, respectively. In addition, it was observed that migration was further reduced by >90%, when EGFR and CXCR4 were suppressed (Fig. 2C).
Notably, similar results were obtained in the Transwell invasion assays in the CXCR4-silenced and/or EGFR-silenced MDA-MB-231 cells: EGFR or CXCR4 inhibition alone reduced invasion ability, and the maximum reduction occurred when the two proteins were suppressed simultaneously (Fig. 2D).
Downregulation of CXCR4 and EGFR in MDA-MB-231 cells inhibits tumor growth in mouse models
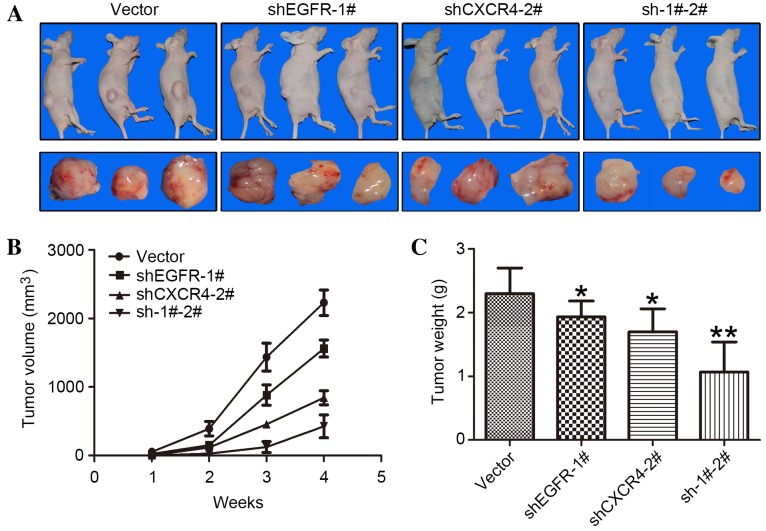
To study the effects of CXCR4 and EGFR expression in tumorigenesis, MDA-MB-231 cells, in which EGFR and/or CXCR4 had been stably knocked down, were subcutaneously implanted into nude mice. MDA-MB-231 cells transfected with the empty vector were used as a control. Tumor growth was determined with a caliper, and tumor volume was calculated as aforementioned. At 4 weeks following implantation, a 25% reduction in the tumor volume of EGFR-silenced MDA-MB-231 cells, a 58% reduction in the tumor volume of CXCR4-silenced cells and an 80% reduction in tumor volume of double CXCR4- and EGFR-silenced MDA-MB-231 cells were observed, compared with the control (Fig. 3A and B).
Figure 3.
Downregulation of CXCR4 and/or EGFR by shRNA in MDA-MB-231 cells inhibits xenograft tumor growth in mouse models. (A) A total of 2×106 stably transduced MDA-MB-231 cells were implanted into the right legs of nude mice. The mice (n=3 per group) were observed for 4 weeks. (B) Tumors were measured every week with external calipers and tumor volume was calculated according to the following formula: Volume = 0.5 × a2 × b, where ‘a’ is the smallest superficial diameter and ‘b’ is the largest superficial diameter. (C) After 4 weeks, the tumors were excised and weighed. *P<0.05 and **P<0.01, compared with control empty vector cells. Error bars correspond to the mean ± standard deviation of three mice in each group. EGFR, epidermal growth factor receptor; CXCR4, C-X-C motif chemokine receptor type 4; shRNA, short hairpin RNA; shEGFR, shRNA targeting EGFR; shCXCR4, shRNA targeting CXCR4; sh-1#-2#, shRNAs targeting EGFR and CXCR4.
At the end of the observation period, subcutaneous tumors were dissected and weighed (Fig. 3A). In concordance with the previous results, EGFR or CXCR4 knockdown alone resulted in a lower tumor weight, compared with the control, and the greatest reduction in tumor weight occurred in tumors in which the two proteins were knocked down simultaneously, with a 60% decrease observed (Fig. 3C).
Discussion
Due to its lack of hormone receptors and HER2 expression, TNBC is not suitable for hormone therapy or anti-HER2-targeted therapy, making traditional chemotherapy the only treatment option (33,34). Studies have reported that the DFS and OS times of patients with TNBC are significantly shorter, compared with those with non-TNBC (7,8,35,36). In the present study, the role of the EGFR tyrosine kinase receptor and CXCR4 chemokine receptor were investigated to determine whether they may serve as novel therapeutic targets for TNBC (37).
Previous studies have reported that CXCR4 is important in visceral metastasis in breast cancer (38,39,6). The current study also demonstrated that CXCR4 was highly expressed in TNBC, concordant with the results of a previous study (6). The mechanism underlying cancer metastasis mediated by CXCR4 is considered to involve the binding of CXCR4 to its ligand, CXCL12, which in turn causes the chemokine receptor to translocate to specific targeted organs (5). Therefore, CXCL12 and CXCR4 compose a signal transduction pathway, leading to tumor growth, invasion and metastasis (4,39). By contrast, EGFR is a transmembrane glycoprotein, and belongs to the tyrosine kinase receptor family. The EGFR-mediated signaling pathway consists of mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (11,16). EGFR has been implicated in cell growth and proliferation, differentiation and other physiological processes of tumor cells (40,41).
In another study, EGFR was demonstrated to be a prognostic factor for TNBC, which is associated with aggressive clinicopathological characteristics (42). However, the correlation between EGFR and CXCR4 in TNBC remains to be elucidated. In the present study, IHC revealed 73.47% CXCR4 positivity and 61.22% EGFR positivity in TNBC tissues, and the CXCR4 and EGFR expression levels were significantly higher, compared with those in non-TNBC. Furthermore, 44.9% of TNBC tumors co-expressed CXCR4 and EGFR, which was also higher compared with those in non-TNBC.
Based on the aforementioned findings, additional experiments were conducted in the present study using MDA-MB-231 cells. Suppression of CXCR4 using shRNA led to the downregulation of EGFR expression, which may occur through increased proteasome-mediated degradation and altered protein trafficking (17). Additionally, it was demonstrated that knockdown of EGFR by shRNA in MDA-MB-231 cells similarly resulted in a reduction in CXCR4 expression levels. Increased levels of CXCR4 transcripts have been associated with increased HIF-1α levels in EGFR-expressing breast cancer (43), and the degradation of CXCR4 protein is also associated with itchy E3 ubiquitin protein ligase and β-arrestin 1/2 in EGFR-expressing breast cancer (13). These studies may explain why CXCR4 decreases when EGFR is silenced in MDA-MB-231 cells, and vice versa. Taken together, the present results suggest the mutual regulation of CXCR4 and EGFR in TNBC cells. However, the functional analysis in the present study may not exclude the possibility that CXCR4 status affects EGFR status (or vice versa) in the neighboring cells.
Suppression of CXCR4 in breast cancer cells has been demonstrated to decrease cellular proliferation, migration and invasive potential (13). Notably, in the present study, the proliferative potential and invasive ability of the TNBC MDA-MB-231 cells were markedly reduced when CXCR4 or EGFR was silenced. In the animal models, tumor growth was significantly decreased in mice injected with MDA-MB-231 cells when CXCR4 or EGFR were knocked down. Previous studies demonstrated the enhancement of CXCR4 by EGFR overexpression in breast cancer cells (44). The current study also confirmed the presence of crosstalk between the two growth factors. A previous study demonstrated that the 38/MAPK signaling pathway in EGFR-expressing breast cancer is essential in the regulation of CXCR4 expression (16). However, the specific underlying mechanism requires additional investigation, particularly regarding the signaling pathways downstream of growth factor receptors.
In conclusion, the present results demonstrate significantly higher expression levels of CXCR4 and EGFR in TNBC, in comparison with non-TNBC, and reveal a positive correlation between the levels of these two receptors. In addition, the findings identified mutual regulation between EGFR and CXCR4 in MDA-MB-231 breast cancer cells. Furthermore, cellular proliferative potential and invasive ability are significantly reduced following the silencing of CXCR4 or EGFR in vitro, and the knockdown of either molecule leads to the inhibition of tumorigenesis in an animal model. These results suggest the possibility of a novel therapeutic strategy involving the dual inhibition of EGFR and CXCR4 in the treatment of TNBC.
Acknowledgements
The present study was supported by The Key International Collaborative Project of The National Natural Science Fund Committee (grant no. 81320108015), The National Natural Science Fund project (grant no. 31271068) and The Special Research Fund for the Doctoral Program of Higher Education (grant no. 20124402110003).
References
- 1.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nassar A, Sussman ZM, Lawson D, Cohen C. Inference of the Basal epithelial phenotype in breast carcinoma from differential marker expression, using tissue microarrays in triple negative breast cancer and women younger than 35. Breast J. 2012;18:399–405. doi: 10.1111/j.1524-4741.2012.01279.x. [DOI] [PubMed] [Google Scholar]
- 3.Choi WT, Yang Y, Xu Y, An J. Targeting chemokine receptor CXCR4 for treatment of HIV-1 infection, tumor progression and metastasis. Curr Top Med Chem. 2014;14:1574–1589. doi: 10.2174/1568026614666140827143541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin F, Brockmeier U, Otterbach F, Metzen E. New insight into the SDF-1/CXCR4 axis in a breast carcinoma model: Hypoxia-induced endothelial SDF-1 and tumor cell CXCR4 are required for tumor cell intravasation. Mol Cancer Res. 2012;10:1021–1031. doi: 10.1158/1541-7786.MCR-11-0498. [DOI] [PubMed] [Google Scholar]
- 5.Luker KE, Lewin SA, Mihalko LA, Schmidt BT, Winkler JS, Coggins NL, Thomas DG, Luker GD. Scavenging of CXCL12 by CXCR7 promotes tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene. 2012;31:4750–4758. doi: 10.1038/onc.2011.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu QD, Panu L, Holm NT, Li BD, Johnson LW, Zhang S. High chemokine receptor CXCR4 level in triple negative breast cancer specimens predicts poor clinical outcome. J Surg Res. 2010;159:689–95. doi: 10.1016/j.jss.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Petersen ER, Sørensen PD, Jakobsen EH, Madsen JS, Brandslund I. Serum HER-2 predicts response and resistance to trastuzumab treatment in breast cancer. Clin Chem Lab Med. 2013;51:1483–1492. doi: 10.1515/cclm-2012-0558. [DOI] [PubMed] [Google Scholar]
- 8.Tang Y, Zhu L, Li Y, Ji J, Li J, Yuan F, Wang D, Chen W, Huang O, Chen X, et al. Overexpression of epithelial growth factor receptor (EGFR) predicts better response to neo-adjuvant chemotherapy in patients with triple-negative breast cancer. J Transl Med. 2012;10:S4. doi: 10.1186/1479-5876-10-S1-S4. (Suppl 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skedgel C, Rayson D, Younis T. Is adjuvant trastuzumab a cost-effective therapy for HER-2/neu-positive T1bN0 breast cancer? Ann Oncol. 2013;24:1834–1840. doi: 10.1093/annonc/mdt069. [DOI] [PubMed] [Google Scholar]
- 10.Burness ML, Grushko TA, Olopade OI. Epidermal growth factor receptor in triple-negative and basal-like breast cancer: Promising clinical target or only a marker? Cancer J. 2010;16:23–32. doi: 10.1097/PPO.0b013e3181d24fc1. [DOI] [PubMed] [Google Scholar]
- 11.Brand TM, lida M, Luthar N, Starr MM, Huppert EJ, Wheeler DL. Nuclear EGFR as a molecular target in cancer. Radiother Oncol. 2013;108:370–377. doi: 10.1016/j.radonc.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 12.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahimi M, George J, Tang C. EGFR variant-mediated invasion by enhanced CXCR4 expression through transcriptional and post-translational mechanisms. Int J Cancer. 2010;126:1850–1860. doi: 10.1002/ijc.24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salvucci O, Bouchard A, Baccarelli A, Deschênes J, Sauter G, Simon R, Bianchi R, Basik M. The role of CXCR4 receptor expression in breast cancer: A large tissue microarray study. Breast Cancer Res Treat. 2006;97:275–283. doi: 10.1007/s10549-005-9121-8. [DOI] [PubMed] [Google Scholar]
- 15.Cabioglu N, Summy J, Miller C, Parikh NU, Sahin AA, Tuzlali S, Pumiglia K, Gallick GE, Price JE. CXCL-12/stromal cell-derived factor-1alpha transactivates HER2-neu in breast cancer cells by a novel pathway involving Src kinase activation. Cancer Res. 2005;65:6493–6497. doi: 10.1158/0008-5472.CAN-04-1303. [DOI] [PubMed] [Google Scholar]
- 16.Phillips RJ, Mestas J, Gharaee-Kermani M, Burdick MD, Sica A, Belperio JA, Keane MP, Strieter RM. Epidermal growth factor and hypoxia-induced expression of CXC chemokine receptor 4 on non-small cell lung cancer cells is regulated by the phosphatidylinositol 3-kinase/PTEN/AKT/mammalian target of rapamycin signaling pathway and activation of hypoxia inducible factor-1alpha. J Biol Chem. 2005;280:22473–22481. doi: 10.1074/jbc.M500963200. [DOI] [PubMed] [Google Scholar]
- 17.Rahimi M, Toth TA, Tang CK. CXCR4 suppression attenuates EGFRvIII-mediated invasion and induces p38 MAPK-dependent protein trafficking and degradation of EGFRvIII in breast cancer cells. Cancer Lett. 2011;306:43–51. doi: 10.1016/j.canlet.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Ji R, Li J, Gu Q, Zhao X, Sun T, Wang J, Li J, Du Q, Sun B. Correlation effect of EGFR and CXCR4 and CCR7 chemokine receptors in predicting breast cancer metastasis and prognosis. J Exp Clin Cancer Res. 2010;29:16. doi: 10.1186/1756-9966-29-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Z, Zhao C, Han X, Han Y. Wnt5a promotes ewing sarcoma cell migration through upregulating CXCR4 expression. BMC Cancer. 2012;12:480. doi: 10.1186/1471-2407-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Zhang B, Wu M, Li H, Niu R, Ying G, Zhang N. Screening of a PKC zeta-specific kinase inhibitor PKCzI257.3 which inhibits EGF-induced breast cancer cell chemotaxis. Invest New Drugs. 2010;28:268–275. doi: 10.1007/s10637-009-9242-8. [DOI] [PubMed] [Google Scholar]
- 21.Lapteva N, Yang AG, Sanders DE, Strube RW, Chen SY. CXCR4 knockdown by small interfering RNA abrogates breast tumor growth in vivo. Cancer Gene Ther. 2005;12:84–89. doi: 10.1038/sj.cgt.7700770. [DOI] [PubMed] [Google Scholar]
- 22.Spina A, Di Maiolo F, Esposito A, Sapio L, Chiosi E, Sorvillo L, Naviglio S. cAMP elevation down-regulates β3 integrin and focal adhesion kinase and inhibits leptin-induced migration of MDA-MB-231 breast cancer cells. Biores Open Access. 2012;1:324–332. doi: 10.1089/biores.2012.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Q, Kantonen S, Gomez-Cambronero J. Serum deprivation confers the MDA-MB-231 breast cancer line with an EGFR/JAK3/PLD2 system that maximizes cancer cell invasion. J Mol Biol. 2013;425:755–766. doi: 10.1016/j.jmb.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murillo G, Peng X, Torres KE, Mehta RG. Deguelin inhibits growth of breast cancer cells by modulating the expression of key members of the Wnt signaling pathway. Cancer Prev Res (Phila) 2009;2:942–950. doi: 10.1158/1940-6207.CAPR-08-0232. [DOI] [PubMed] [Google Scholar]
- 25.Pattarozzi A, Gatti M, Barbieri F, Würth R, Porcile C, Lunardi G, Ratto A, Favoni R, Bajetto A, Ferrari A, Florio T. 17beta-estradiol promotes breast cancer cell proliferation-inducing stromal cell-derived factor-1-mediated epidermal growth factor receptor transactivation: Reversal by gefitinib pretreatment. Mol Pharmacol. 2008;73:191–202. doi: 10.1124/mol.107.039974. [DOI] [PubMed] [Google Scholar]
- 26.Sales KJ, Grant V, Catalano RD, Jabbour HN. Chorionic gonadotrophin regulates CXCR4 expression in human endometrium via E-series prostanoid receptor 2 signalling to PI3K-ERK1/2: Implications for fetal-maternal crosstalk for embryo implantation. Mol Hum Reprod. 2011;17:22–32. doi: 10.1093/molehr/gaq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallett MA, Teng B, Hasegawa H, Schwab LP, Seagroves TN, Pourmotabbed T. Anti-matrix metalloproteinase-9 DNAzyme decreases tumor growth in the MMTV-PyMT mouse model of breast cancer. Breast Cancer Res. 2013;15:R12. doi: 10.1186/bcr3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasser MW, Qamri Z, Deol YS, Smith D, Shilo K, Zou X, Ganju RK. Crosstalk between chemokine receptor CXCR4 and cannabinoid receptor CB2 in modulating breast cancer growth and invasion. PLoS One. 2011;6:e23901. doi: 10.1371/journal.pone.0023901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liedtke C, Hess KR, Karn T, Rody A, Kiesel L, Hortobagyi GN, Pusztai L, Gonzalez-Angulo AM. The prognostic impact of age in patients with triple-negative breast cancer. Breast Cancer Res Treat. 2013;138:591–599. doi: 10.1007/s10549-013-2461-x. [DOI] [PubMed] [Google Scholar]
- 31.Wendel C, Hemping-Bovenkerk A, Krasnyanska J, Mees ST, Kochetkova M, Stoeppeler S, Haier J. CXCR4/CXCL12 participate in extravasation of metastasizing breast cancer cells within the liver in a rat model. PLoS One. 2012;7:e30046. doi: 10.1371/journal.pone.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MF, Pan MH, Chiou YS, Cheng AC, Huang H. Resveratrol modulates MED28 (Magicin/EG-1) expression and inhibits epidermal growth factor (EGF)-induced migration in MDA-MB-231 human breast cancer cells. J Agric Food Chem. 2011;59:11853–11861. doi: 10.1021/jf202426k. [DOI] [PubMed] [Google Scholar]
- 33.Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH. Triple-negative breast cancer: Clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol. 2010;23:123–133. doi: 10.1038/modpathol.2009.145. [DOI] [PubMed] [Google Scholar]
- 34.de Ruijter TC, Veeck J, de Hoon JP, van Engeland M, Tjan-Heijnen VC. Characteristics of triple-negative breast cancer. J Cancer Res Clin Oncol. 2011;137:183–192. doi: 10.1007/s00432-010-0957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae SY, Lee SK, Koo MY, Hur SM, Choi MY, Cho DH, Kim S, Choe JH, Lee JE, Kim JH, et al. The prognoses of metaplastic breast cancer patients compared to those of triple-negative breast cancer patients. Breast Cancer Res Treat. 2011;126:471–478. doi: 10.1007/s10549-011-1359-8. [DOI] [PubMed] [Google Scholar]
- 36.Choi YL, Oh E, Park S, Kim Y, Park YH, Song K, Cho EY, Hong YC, Choi JS, Lee JE, et al. Triple-negative, basal-like, and quintuple-negative breast cancers: Better prediction model for survival. BMC Cancer. 2010;10:507. doi: 10.1186/1471-2407-10-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chacón RD, Costanzo MV. Triple-negative breast cancer. Breast Cancer Res. 2010;12:S3. doi: 10.1186/bcr2574. (Suppl 2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boudot A, Kerdivel G, Habauzit D, Eeckhoute J, Le Dily F, Flouriot G, Samson M, Pakdel F. Differential estrogen-regulation of CXCL12 chemokine receptors, CXCR4 and CXCR7, contributes to the growth effect of estrogens in breast cancer cells. PLoS One. 2011;6:e20898. doi: 10.1371/journal.pone.0020898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hawkins OE, Richmond A. The dynamic yin-yang interaction of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res. 2012;14:103. doi: 10.1186/bcr3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lill NL, Sever NI. Where EGF receptors transmit their signals. Sci Signal. 2012;5:pe41. doi: 10.1126/scisignal.2003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai PC, Hsieh CY, Chiu CC, Wang CK, Chang LS, Lin SR. Cardiotoxin III suppresses MDA-MB-231 cell metastasis through the inhibition of EGF/EGFR-mediated signaling pathway. Toxicon. 2012;60:734–743. doi: 10.1016/j.toxicon.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 42.Liu D, He J, Yuan Z, Wang S, Peng R, Shi Y, Teng X, Qin T. EGFR expression correlates with decreased disease-free survival in triple-negative breast cancer: A retrospective analysis based on a tissue microarray. Med Oncol. 2012;29:401–405. doi: 10.1007/s12032-011-9827-x. [DOI] [PubMed] [Google Scholar]
- 43.Billadeau DD, Chatterjee S, Bramati P, Sreekumar R, Shah V, Hedin K, Urrutia R. Characterization of the CXCR4 signaling in pancreatic cancer cells. Int J Gastrointest Cancer. 2006;37:110–119. doi: 10.1007/s12029-007-0011-7. [DOI] [PubMed] [Google Scholar]
- 44.Yasumoto K, Yamada T, Kawashima A, Wang W, Li Q, Donev IS, Tacheuchi S, Mouri H, Yamashita K, Ohtsubo K, Yano S. The EGFR ligands amphiregulin and heparin-binding egf-like growth factor promote peritoneal carcinomatosis in CXCR4-expressing gastric cancer. Clin Cancer Res. 2011;17:3619–3630. doi: 10.1158/1078-0432.CCR-10-2475. [DOI] [PubMed] [Google Scholar]