Abstract
Milk contamination by phages, the susceptibility of the phages to pasteurization, and the high levels of resistance to phage infection of starter strains condition the evolution dynamics of phage populations in dairy environments. Approximately 10% (83 of 900) of raw milk samples contained phages of the quasi-species c2 (72%), 936 (24%), and P335 (4%). However, 936 phages were isolated from 20 of 24 (85%) whey samples, while c2 was detected in only one (4%) of these samples. This switch may have been due to the higher susceptibility of c2 to pasteurization (936-like phages were found to be approximately 35 times more resistant than c2 strains to treatment of contaminated milk in a plate heat exchanger at 72°C for 15 s). The restriction patterns of 936-like phages isolated from milk and whey were different, indicating that survival to pasteurization does not result in direct contamination of the dairy environment. The main alternative source of phages (commercial bacterial starters) does not appear to significantly contribute to phage contamination. Twenty-four strains isolated from nine starter formulations were generally resistant to phage infection, and very small progeny were generated upon induction of the lytic cycle of resident prophages. Thus, we postulate that a continuous supply of contaminated milk, followed by pasteurization, creates a factory environment rich in diverse 936 phage strains. This equilibrium would be broken if a particular starter strain turned out to be susceptible to infection by one of these 936-like phages, which, as a consequence, became prevalent.
Bacteriophage infection of the starters used in dairy fermentations is the main cause of disturbance in the manufacture of products such as cheese, yogurt, etc. (11, 23). The problem was initially recognized in 1935 (30) and led to the design and application of good manufacturing practices, such as direct inoculation of the starters in closed fermentation vats, use of antiphage media for starter propagation, and rotation of starter cultures (1). In addition, genes that encode natural resistance mechanisms were introduced into starter strains (2, 11, 14). As a consequence of all these measures, total loss of the final product is infrequent nowadays, although phages are still responsible for quality defects that affect the flavor, texture, and even safety of dairy foods (1, 11).
Of the variety of phage species that infect Lactococcus lactis, only three, c2, 936, and P335, are commonly found in dairy plants, and these phages are responsible for most milk fermentation failures (4, 7, 24). All three belong to the family Siphoviridae, although 936 and P335 have isometric capsids (morphotype B1), whereas c2 has a prolate head (morphotype B2) (17, 18). Only virulent representatives of the c2 and 936 groups are known, while P335 includes both temperate and lytic viruses. Of the three, phages belonging to the 936 quasi-species are most frequently isolated from dairy environments, followed by phages belonging to the c2 group.
The question of the origin of the phages that colonize dairy environments has not yet been fully answered. It appears that P335-like phages may originate from the starter strains themselves, which frequently harbor prophages that can become induced and produce progeny that are able to develop in members of the starter other than their lysogenic hosts (9). In fact, virulent P335 phages exhibit extensive homology with inducible or defective prophages (21; M. Trotter, personal communication). However, the persistence of c2- and 936-like phages might be considered surprising, given the control measures that are usually adopted by dairies. Two possibilities are that these phages are introduced with the milk to be processed (22) and that mixed starters harbor phages that coexist with the bacterial strains, resulting in continuous recontamination of the dairy environment (28).
In this paper, the results of a search for lactococcal phages in a significant number of milk samples obtained from a wide geographical area are reported. Also, nine starter cultures that are widely used in the area were investigated with respect to their bacterial complexity and the phage susceptibility or inducibility of their component lactococcal strains. Finally, whey samples from a number of artisan dairies were analyzed for their phage contents. The data obtained are discussed together with data resulting from the differential effect of pasteurization on phage viability. Based on the results, a model of how the prevalent phage populations become established in the dairy environment is proposed.
MATERIALS AND METHODS
Sampling, microorganisms, and culture conditions.
Nine hundred samples (100-μl aliquots) of raw milk, each derived from a different farm (from a 10,500-km2 region located in northern Spain), were added to 100-μl exponential cultures of L. lactis subsp. cremoris MG1614 and L. lactis subsp. lactis IL1403 growing in M17-glucose broth supplemented with 10 mM CaCl2 and 10 mM MgSO4 (MMC broth) and incubated for 4 h. Aliquots of these cultures (100 μl) were placed on plates containing solid MMC agar and overlaid with soft MMC agar (0.7% agar). Phages were reisolated twice from the resulting plaques, suspended in MMC broth, filtered, and stored as 50% glycerol suspensions at −80°C. A similar phage isolation procedure (with the enrichment step omitted) was used for 24 whey samples, each obtained from a different artisan cheese factory. P335, bIL170 (a 936-like virus), and c2 were used as representative phages of their groups and were used as positive propagation controls and for comparison with the newly isolated phages (Table 1). The nine mesophilic homolactic commercial starters used were provided as frozen suspensions. Serial dilutions of each of these suspensions were plated onto M17-lactose solid medium, and 20 randomly chosen colonies were subjected to plasmid analysis.
TABLE 1.
Bacterial strains and phages used in this work
| Strain or phage | Relevant characteristics | Reference or source |
|---|---|---|
| L. lactis strains | ||
| MG1614 | Plasmid free, host for phage c2 and c2-like phages | 15 |
| IL1403 | Plasmid free, host for bIL170 and other 936-like phages | 8 |
| F4-2 | Host for phage P335 and P335-like phages | S. Moineau |
| Bacteriophages | ||
| c2 | Prolate headed, c2 species, representative phage | 26 |
| 1101 | Prolate headed, c2 species, milk isolated | This study |
| 1370 | Prolate headed, c2 species, milk isolated | This study |
| 1390 | Prolate headed, c2 species, milk isolated | This study |
| 1133 | Prolate headed, c2 species, milk isolated | This study |
| bIL170 | Small isometric headed, 936 species, representative phage | 3 |
| 1240 | Small isometric headed, 936 species, milk isolated | This study |
| 154/2 | Small isometric headed, 936 species, milk isolated | This study |
| 174/5 | Small isometric headed, 936 species, milk isolated | This study |
| 224/14 | Small isometric headed, 936 species, milk isolated | This study |
| 244/15 | Small isometric headed, 936 species, milk isolated | This study |
| 9205 | Small isometric headed, 936 species, whey isolated | This study |
| P335 | Small isometric headed, P335 species, representative phage | 5 |
| 104/37 | Small isometric headed, P335 species, milk isolated | This study |
| 114/14 | Small isometric headed, P335 species, milk isolated | This study |
| 154/33 | Small isometric headed, P335 species, milk isolated | This study |
| 159/19 | Small isometric headed, P335 species, milk isolated | This study |
Representative cultures for each plasmid pattern were separately infected with a collection of 32 phages isolated from failed fermentations (donated by the starter manufacturing company Christian Hansen, Hoersholm, Denmark). These phages belonged to the 936 quasi-species (14 phages) or to the c2 group (10 phages) or were taxonomically undefined (8 phages) (T. Janzen, personal communication).
The lysogenic status of the strains isolated from the commercial starters was determined by measuring the induction of prophages from exponential cultures in MMC broth with 0.6 μg of mitomycin C per ml (20). Filter-sterilized culture supernatants were tested for the presence of phages by the double-layer method by using L. lactis MG1614, IL1403, and F4-2 as indicators.
DNA techniques.
Plasmid DNA isolation was performed by using 5 ml of an overnight culture in M17-lactose broth and the method reported by O'Sullivan and Klaenhammer (25). PCRs were performed with 5-μl aliquots of phage lysates (as DNA sources), the c2-, 936-, and P335-specific primers, and the conditions described by Labrie and Moineau (19). Bacteriophages were purified with isopycnic CsCl continuous gradients prior to DNA extraction (27). Restriction enzymes were obtained from Roche and were used in accordance with the supplier's instructions. Plasmid DNA, restriction fragments, and PCR products were separated by electrophoresis in 0.7 or 2% agarose gels, as required, by using Tris-borate-EDTA buffer as the electrolyte, stained with ethidium bromide, and visualized under UV light. Hybridization experiments were performed under high-stringency conditions (60°C in 2× SSC with washing in 0.1× SSC at the same temperature [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) by using the protocols described by Sambrook et al. (27). The target DNA was fixed to Hybond-N membranes (Amersham). Labeling and detection of the probes were performed with a nonradioactive digoxigenin DNA labeling and detection kit (Roche) used in accordance with the manufacturer's recommendations.
High-temperature resistance testing.
The heat susceptibility of phages was tested by two methods by using (i) a water bath and (ii) a plate heat exchanger. For the first method, 5-ml whole-fat ultrahigh-temperature milk aliquots were kept for 15 min in a bath at the appropriate temperature before they were contaminated with individual phage suspensions to obtain final concentrations of 105 to 106 PFU/ml. Samples, taken at different times, were immediately diluted in MMC broth at room temperature, and the survivors were scored by the double-layer method as described above. Whole-fat milk instead of medium was used to take into account any protective effect of raw milk on phage viability. For pasteurization in a heat exchanger, 70-liter whole-fat milk aliquots were inoculated with each of the phages to obtain a final concentration of approximately 105 PFU/ml. Treatment took place at 72°C for 15 s with a Stork-Interiberica apparatus (pilot plant model A). Samples (10 ml) were withdrawn before and after heating (five samples were taken from each treatment at regular intervals) and analyzed for phage survival within 2 h of withdrawal.
RESULTS
Phage isolation and classification.
Of the 900 samples of raw milk analyzed, 83 (9.2%) contained phage, as determined by plaque formation on lawns of L. lactis subsp. cremoris MG1614 and/or L. lactis subsp. lactis IL1403 (Table 2). Analysis of these phages by the specific multiplex PCR method described by Labrie and Moineau (19) indicated that 60 (72.3%) belonged to the c2 quasi-species and propagated exclusively on L. lactis subsp. cremoris MG1614. Of the remaining phages, 20 (24%) had amplicons characteristic of the 936 group. Sixteen of these phages developed only on L. lactis subsp. lactis IL1403, while three propagated on both hosts and one infected L. lactis subsp. cremoris MG1614 exclusively. Very few P335 phages were recovered; we obtained only three isolates (3.6%), all of which multiplied on both hosts. In addition, 12 of the 900 milk samples contained both 936- and c2-like phages. According to the PCR data, no phages that could not be classified in one of these three species were found.
TABLE 2.
Distribution of phages obtained from raw milk and whey samples as a function of the strain used for propagationa
| Substrate | Propagating host(s) | No. (%) of positive samples | No. (%) belonging to the following phage species:
|
||
|---|---|---|---|---|---|
| c2 | 936 | P335 | |||
| Milk | MG1614 | 61 | 60 | 1 | |
| IL1403 | 16 | 16 | |||
| Both | 6 | 3 | 3 | ||
| Totalb | 83 (9.21) | 60 (6.66) | 20 (2.22) | 3 (0.33) | |
| Whey | MG1614 | 1 | 1 | ||
| IL1403 | 19 | 19 | |||
| Both | 4 | 1 | 3 | ||
| Totalc | 24 (100) | 1 (4.17) | 20 (83.33) | 3 (12.5) | |
The phages, which were isolated by the double-layer propagation method, were classified by PCR by using species-specific primers (19).
The percentages are the percentages of 900 milk samples.
The percentages are the percentages of 24 whey samples.
In contrast, the 24 whey samples obtained from farmhouse dairies located in the same region contained phage (Table 2), although most of the phages were 936. In agreement with the host range data described above, the sole c2 isolate developed on L. lactis subsp. cremoris MG1614, while most of the 936 phages developed exclusively on L. lactis subsp. lactis IL1403. The three P335 phages found infected both hosts, as did one isolate that turned out to belong to the 936 quasi-species.
Analysis of starter culture strains with respect to phage susceptibility.
The shift of prevalent phages from c2 in milk to 936 in whey might have been due to greater susceptibility of the bacterial strains included in starter preparations to 936-like phages. To address this possibility, the L. lactis strains present in nine commercial starters that are widely used in our region were analyzed. The strains were differentiated on the basis of their plasmid contents (Table 3). Two of the starters (starters 1 and 2 [Table 3]) contained just one strain, which was the same strain in both cases, as judged from the identical plasmid patterns shown by the 20 colonies processed in each analysis. Two other preparations contained the same strain; in one case this strain was present with another strain at a ratio of 1:1 (starter 3 [Table 3]), while in the other case the two strains were accompanied by a third strain and the ratio was 2:1:1 (starter 4 [Table 3]). The other five starter formulations analyzed (starters 5 to 9 [Table 3]) showed greater variability, and the number of component strains varied between three and six. No isolate was common to any two of these five starters. Altogether, 24 strains were obtained, and 18 of these strains were resistant to infection by any of our 32 L. lactis-specific phages (Table 3). Two of the other six strains were susceptible to most phages and produced clear plaques. The other four strains seemed to be marginally susceptible to some of the phages, producing clearing on host lawns when they were inoculated at a high concentration but unable to produce single plaques on the plates. This general resistance of the starter strains to phage infection seems to rule out the possibility that the change in the prevalent phages from c2- to 936-like phages was due to differential susceptibility in favor of the latter group.
TABLE 3.
Strain composition of the starters and relationship of the susceptibility of the strains to phage infection and prophage inducibility with mitomycin C
| Starter | No. of strains | Relative abundance (%) | Sensitivitya | MitC induction (PFU/ml)b
|
|
|---|---|---|---|---|---|
| L. lactis IL1403 | L. lactis F4-2 | ||||
| 1 | 1 | 100c | R | ||
| 2 | 1 | 100c | R | ||
| 3 | 2 | 50c | R | ||
| 50d | R | ||||
| 4 | 3 | 50c | R | ||
| 25d | R | 5 × 101 | 8 × 101 | ||
| 25 | R | ||||
| 5 | 3 | 40 | R | ||
| 40 | PS | 1.3 × 102 | 3.2 × 102 | ||
| 20 | S | ||||
| 6 | 5 | 50 | S | 2 × 101 | 6 × 101 |
| 17 | R | ||||
| 17 | R | ||||
| 8 | R | 7 × 101 | 4 × 101 | ||
| 8 | R | ||||
| 7 | 4 | 18 | PS | ||
| 36 | R | 1.2 × 102 | 2.3 × 102 | ||
| 18 | R | 2 × 101 | 6 × 101 | ||
| 28 | R | ||||
| 8 | 3 | 33.5 | PS | 1 × 101 | 5 × 101 |
| 55.5 | PS | 6 × 101 | 1.2 × 102 | ||
| 11 | R | ||||
| 9 | 6 | 33 | R | ||
| 25 | R | ||||
| 17 | R | ||||
| 8.3 | R | 1.9 × 102 | 3.4 × 102 | ||
| 8.3 | R | ||||
| 8.3 | R | ||||
Susceptibility to a collection of 32 phages (see Materials and Methods). R, resistant; S, susceptible; PS, partial susceptibility (specific concentrated phage suspensions were able to produce clearings in lawns of the strain, but the same phages were unable to generate isolated plaques).
The values are means of data obtained in three independent induction experiments. L. lactis MG1614 was also used as host, but no plaques were obtained from any of the induced supernatants.
The same strain was present in starters 1, 2, 3, and 4.
The same strain was present in starters 3 and 4.
In addition, filtered supernatants of starter cultures grown overnight were separately plated on L. lactis strains MG1614, IL1403, and F4-2. No plaques were observed, which indicated the absence of pseudolysogeny (i.e., carryover of virulent phages in an infective equilibrium with one or more of the starter strains) in the commercial preparations.
Finally, the 24 strains obtained from the starters were incubated in the presence of mitomycin C to induce possible resident prophages, and their filtered supernatants were placed on lawns of the three indicator strains. While no plaques were observed on L. lactis MG1614 lawns, nine of the strains produced phage on IL1403 or F4-2 lawns. However, the phage titers were very low, between 101 and 3 × 103 PFU/ml (Table 3).
Effect of high temperature on phage survival.
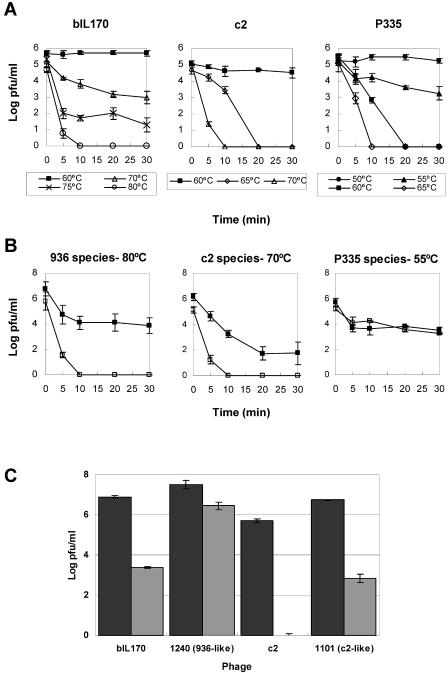
The striking difference between the frequencies of isolation of c2 and 936 phages from raw milk and whey might also have been due to different susceptibilities of these two kinds of phages to the temperature used in the pasteurization process, which is the only sanitizing treatment applied to the milk before inoculation of the starter cultures. This possibility was addressed by determination of the temperature susceptibilities of representative phages from the three groups. The phages were suspended in sterile whole-fat milk to mimic natural conditions (i.e., to take into account any protective effect of raw milk on phage viability). The results obtained (Fig. 1A) indicated that both the representative c2 and 936 phages (i.e., c2 and bIL170) resisted heating at 60°C for 30 min without any loss of viability, while 105-PFU/ml suspensions of P335 became completely inactivated after 20 min at that temperature. However, no viable c2 phages were observed after treatment at 70°C for 10 min; in contrast, similar 936 suspensions still contained phage after treatment for 30 min at 75°C.
FIG. 1.
Effect of high temperature on the viability of L. lactis bacteriophages suspended in whole-fat milk. (A) Treatment of representative phages belonging to the 936 (bIL170), c2, and P335 quasi-species in a water bath. (B) Comparison of the behavior of each representative phage (□) with the behavior of four milk isolates (▪) at the temperatures indicated above the graphs. (C) Pasteurization of phage-contaminated milk in a plate heat exchanger (72°C for 15 s). Dark gray bars, control suspensions; light gray bars, treated suspensions. Experiments were carried out in triplicate (A and B) or quintuplicate (C).
The different degrees of heat resistance shown by the representative phages could be a general feature of the groups or simply a peculiarity of the strains used. To test these two possibilities, several phages from each group, which were isolated from milk, were subjected to treatment at the critical temperatures (Fig. 1B). The data obtained indicate that the newly isolated c2 phages (1101, 1133, 1370, and 1390) and 936 phages (154/2, 174/5, 224/14, and 244/15) were more resistant than the representative phages (c2 and bIL170). However, the wild P335 phages tested (104/37, 114/14, 154/33, and 159/19) showed heat susceptibilities similar to that of the original strain. Furthermore, we concluded that 936-like phages are, on the whole, more resistant than the c2 phages isolated from the same environment.
Pasteurization of phage suspensions in a plate heat exchanger resulted in a generally drastic drop in viability (Fig. 1C), in spite of the relatively mild conditions used (72°C for 15 s) compared to the conditions used in the experiments described above. Under these conditions, no c2 (representative strain) survivors were obtained from a suspension that contained more than 105 PFU/ml. In contrast, a preparation of the wild c2 strain (1101) still contained viable phages after the treatment, although it was relatively susceptible to pasteurization (the titer was reduced by a factor close to 10−4). The titer of bIL170 (representative of 936 phages) was reduced by more than 3 log units upon milk pasteurization, while a newly isolated 936-like phage (1240) suffered only a modest reduction to 10−1 PFU/ml (i.e., it was about 100-fold more resistant than bIL170). Once again, the wild isolates appeared to be more resistant to high temperature than the representative strains. Furthermore, the data confirmed the greater intrinsic resistance of 936-like phages than of c2 and P335 to high temperatures and might explain the difference in prevalent phage populations between milk and whey upon pasteurization.
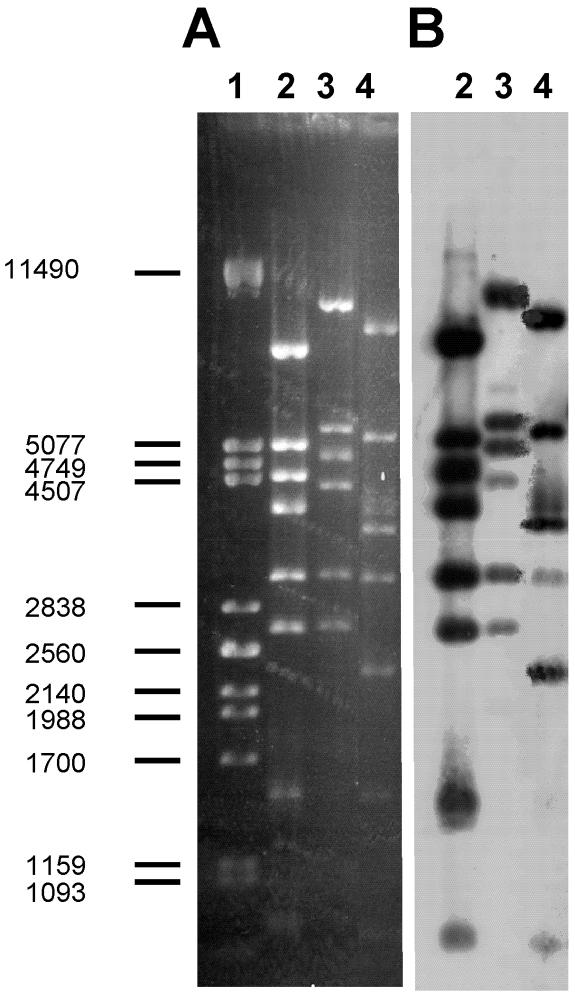
Genome comparison for 936-like phage isolates.
Wild 936 isolates were less susceptible to high temperatures than bIL170, the representative phage of the species. This phenotypic difference coincides with alterations in the corresponding restriction patterns of the genomes (Fig. 2A, compare lane 2 with lanes 3 and 4). On the other hand, 936-like phages isolated from milk and whey were quite dissimilar, as judged by this parameter (Fig. 2A, compare lanes 3 and 4), indicating that resistance to high temperatures might not be the only factor governing phage establishment and persistence in dairy factories. However, the degree of variation among the genomes of 936 phages isolated from milk and whey might not be as high as that suggested by the patterns shown in Fig. 2A, as all fragments obtained from the wild isolates hybridized with a probe made from the DNA of bIL170 (Fig. 2B).
FIG. 2.
(A) Restriction analysis (EcoRV) of genome DNA purified from 936-like phages. Lane 2, representative strain (bIL170); lane 3, milk isolate (1240); lane 4, whey isolate (9205). Lane 1 contained size markers in base pairs (lambda DNA restricted with PstI). (B) High-stringency hybridization with a probe made from bIL170 DNA.
DISCUSSION
To test the hypothesis that phage contamination of dairies is a consequence of the continuous supply of new viruses that might enter the factories either in the raw milk that is to be processed or in fermentation starters, a large number of milk samples obtained from farms located in a ca. 10,500-km2 area were analyzed along with nine commercial starters for phage contamination. An enrichment step was used to improve the recovery of phages from weakly contaminated milk samples. The strains used as hosts in the screening did not harbor any plasmids in order to avoid the presence of phage resistance determinants usually associated with extrachromosomal elements (2, 11, 14), and they belonged to different L. lactis subspecies in order to widen the host range. Under these conditions, almost 10% of the 900 samples processed yielded viable phages. All of these phages could be classified into one of the three groups most frequently found in industrial environments, and the most abundant phages were those belonging to the c2 quasi-species. This confirms that milk is an important source of phages, as reported previously for L. lactis (10, 13) and Streptococcus thermophilus (6). Presumably, raw milk becomes contaminated on the farm with wild L. lactis strains associated with the plant material used for livestock feed, on which phages could develop.
Based on the previous studies, the prevalence of c2-like phages might be due to the wider host range of prolate-headed phages (e.g., c2) than of isometric-headed phages (e.g., 936) (16). However, in our case there was a marked relationship between the phage group and the propagating host. For example, c2 isolates exclusively infected L. lactis subsp. cremoris MG1614, while most 936 phages propagated only in L. lactis subsp. lactis IL1403. It is striking that only six P335-related isolates were obtained and that all of these isolates developed on both hosts. The low frequency of isolation might reflect a low incidence or perhaps (given the temperate nature of P335-like phages as a whole) the resistance of the host strains to infection by viruses that might share their incompatibility group with the prophages present in their genomes (9, 12).
As pointed out above, c2-like phages were prevalent in milk. However, the most frequently isolated phages in dairies belonged to the 936 quasi-species, followed by c2-like and P335-like phages (Table 2), which is in line with previous reports (4, 7, 24). Three possible hypotheses were tested to explain this difference: (i) increased susceptibility of the strains included in the starters to 936-like phages; (ii) contamination of the starters with 936-related phages; and (iii) higher susceptibility of c2- and P335-like phages to milk pasteurization. The first hypothesis was discarded after we found a high level of resistance of the L. lactis strains isolated from nine starter formulations to infection by phages of any group; only 2 of 24 strains showed significant levels of susceptibility to members of a collection of 32 phages isolated from failed fermentations in Europe and the United States. The possibility of significant carryover of phages by the starter preparations was discarded in two steps. First, no evidence of pseudolysogeny was obtained. Second, the starter strains provided very poor phage yields upon induction of resident prophages (Table 3). To rule out the possibility that no plaques were observed due to a lack of appropriate indicator strains, the absorbance of induced cultures was continuously monitored, and we found no evidence of culture lysis (data not shown). However, the different degrees of heat resistance shown by the phages of the three quasi-species analyzed might, at least in part, promote the difference in the prevalent bacteriophage populations in milk and whey. In this respect, P335 phages were shown to be clearly susceptible to the conditions used for pasteurization (62°C for 30 min or 72°C for 15 s), but both the 936 and c2 representative phages were clearly resistant when they were heated in a water bath. However, c2 was marginally more sensitive than 936 (at 70°C, the decimal thermal reduction times were 1.3 min for c2 and 3.35 min for 936). This difference is probably not sufficient to conclude that thermal treatment is the main cause of the difference in prevalent phages between milk and whey. However, Chopin (10) reported that passage of milk through a plate heat exchanger, which is the most widely used method for pasteurization, was much more efficient for phage inactivation than heating in a water bath. Our data confirm this finding. Suspensions of c2 phages containing 105 PFU/ml were completely inactivated (representative strain) or exhibited a drastic reduction in viability (wild isolates), whereas 936 phages isolated from milk were only marginally susceptible, indicating that pasteurization may be a major factor in the switch of the predominant phage populations from c2 in milk to 936 in whey. However, pasteurization survival may not be sufficient for establishment of a particular 936 phage strain in the factory environment, as most of the bacterial strains included in the fermentation starters are resistant to infection (22 of 24 of these strains were refractory to phage-induced lysis). Under these conditions, the continuous supply of new viruses in the milk to be processed might induce a low (yet diverse) level of phage contamination in the dairy environment. The situation might change due to the presence in a starter of a bacterial strain susceptible to one phage that is endemic in that particular environment. This would result in massive multiplication of this phage and, as a consequence, in its prevalence in the factory environment. This might explain why the restriction patterns of 936 phages isolated from milk and whey are different.
It is noteworthy that all c2 and 936 phages tested during this work are more heat resistant than their representative counterparts, while all P335-related phages have similar susceptibilities. This finding might indicate that the former phages, which are virulent, are continuously subjected to natural selection, while the latter phages, which include temperate viruses, are protected from environmental stresses in the prophage form. This might also explain the occurrence of P335-like phages in whey, in spite of their susceptibility to high temperature. These phages might represent induced prophages that are residents in the starter strains (some degree of induction was observed upon treatment of some of the starter bacterial strains with mitomycin C), thus explaining the appearance of P335 phages in the pasteurized, fermenting milk.
It is difficult to compare our data on the relative abundance of phage in milk and/or whey with the data in previous reports, since most of the previous reports were published prior to assignment of the L. lactis bacteriophages to the specific groups recognized today (17). However, analysis of the data in previous reports allowed the identities of these phages to be predicted. For instance, 5 of the 17 industrial isolates analyzed by Chopin (10) were totally susceptible to heating at 72°C for 15 s (in a water bath), suggesting that they were P335-like phages. Three of the remaining isolates were partially resistant to treatment at 80°C for 15 s, while the other nine did not survive at all under these conditions. These data most probably place the phages in the 936 and c2 groups, respectively. Similarly, the four phages analyzed by Suarez and Reinheimer (29) may presumably be ascribed to the 936 (two phages), c2 (one phage), and P335 (one phage) quasi-species.
This report confirms that milk is probably the most important phage contamination source in dairy factories. However, the high levels of resistance of the starter strains to phage infection may hinder massive phage multiplication. Occasionally, the presence among the bacterial strains of a strain that is susceptible to a particular phage might result in the generation of a flawed product and/or in this phage spreading through the factory environment. This explains the success of starter rotation, which, upon introduction of new strains, should result in the inability of the currently predominant phage population to develop in subsequent batches of fermenting milk and in a return to the original situation.
Acknowledgments
The Laboratorio Interprofesional Lácteo de Asturias, T. Janzen (Christian Hansen, Hoersholm, Denmark), and A. Rodriguez (Instituto de Productos Lácteos de Asturias) are gratefully acknowledged for providing the dairy samples, the bacteriophage collection, and the starters, respectively, used in this work. S. Moineau (Université Laval, Quebec, Canada) and H. Neve (Federal Dairy Research Centre, Kiel, Germany) are warmly recognized for providing the representative phages. We also thank S. Boris for help during the processing of the milk samples, J. L. Caso for critical reading of the manuscript, and P. Barnes for proofreading the English.
This work was supported by grant PB-EXP01-23 from FICYT (Principado de Asturias) and by grant BIO2001-3621 from CICYT (Spanish Ministry of Education). C.M. is the recipient of a PRI fellowship from FICYT.
REFERENCES
- 1.Accolas, J. P., C. Peigney, G. K. Y. Limsowtin, P. J. Cluzel, and L. Séchaud. 1994. Lutte contre les bacteriophages dans l'industrie laitière, p. 473-492. In H. de Roissart and F. M. Luquet (ed.), Bacteries lactiques. Lorica, Uriage, France.
- 2.Allison, G. E., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226. [Google Scholar]
- 3.Bidnenko, E., S. D. Ehrlich, and M.-C. Chopin. 1995. Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J. Bacteriol. 177:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisonnette, F., S. Labrie, H. Deveau, M. Lamoureux, and S. Moineau. 2000. Characterization of mesophilic mixed starter cultures used for the manufacture of aged Cheddar cheese. J. Dairy Sci. 83:620-627. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., S. Hertwig, H. Neve, A. Geis, and M. Teuber. 1989. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. J. Gen. Microbiol. 135:2551-2560. [Google Scholar]
- 6.Bruttin, A., F. Desiere, N. d'Amico, J. P. Guerin, J. Sidoti, B. Huni, S. Lucchini, and H. Brussow. 1997. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 63:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey, C. N., E. Morgan, C. Daly, and G. F. Fitzgerald. 1993. Characterization and classification of virulent lactococcal bacteriophages isolated from a Cheddar cheese plant. J. Appl. Bacteriol. 74:268-275. [Google Scholar]
- 8.Chopin, A., M.-C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 9.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chopin, M. C. 1980. Resistance of 17 mesophilic lactic Streptococcus bacteriophages to pasteurization and to spray-drying. J. Dairy Res. 47:131-139. [DOI] [PubMed] [Google Scholar]
- 11.Coffey, A., and P. Ross. 2002. Bacteriophage resistance systems in dairy starter strains: molecular analysis to application. Antonie Leeuwenhoek 82:303-321. [PubMed] [Google Scholar]
- 12.Desiere, F., S. Lucchini, C. Canchaya, M. Ventura, and H. Brussow. 2002. Comparative genomics of phages and prophages in lactic acid bacteria. Antonie Leeuwenhoek 82:73-91. [PubMed] [Google Scholar]
- 13.Everson, T. C. 1991. Control of phages in the dairy plant. FIL-IDF Bull. 263:4-11. [Google Scholar]
- 14.Forde, A., and G. F. Fitzgerald. 1999. Bacteriophage defence in lactic acid bacteria. Antonie Leeuwenhoek 76:89-113. [PubMed] [Google Scholar]
- 15.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heap, H. A., and A. W. Jarvis. 1980. A comparison of prolate- and isometric-headed lactic streptococcal bacteriophages. N. Z. J. Dairy Sci. Technol. 15:75-81. [Google Scholar]
- 17.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 18.Josephsen, J., and H. Neve. 1998. Bacteriophages and lactic acid bacteria, p. 385-436. In S. Salminen and S. A. von Wright (ed.), Lactic acid bacteria. Microbiology and functional aspects. Marcel Dekker Inc., New York, N.Y.
- 19.Labrie, S., and S. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madera, C., P. Garcia, T. Janzen, A. Rodriguez, and J. E. Suarez. 2003. Characterisation of technologically proficient wild Lactococcus lactis strains resistant to phage infection. Int. J. Food Microbiol. 86:213-222. [DOI] [PubMed] [Google Scholar]
- 21.Madsen, S., D. Mills, G. Djordjevic, H. Israelsen, and T. Klaenhammer. 2001. Analysis of the genetic switch and replication region of a P335-type bacteriophage with an obligately lytic lifestyle on Lactococcus lactis. Appl. Environ. Microbiol. 67:1128-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntyre, K., H. A. Heap, G. P. Davey, and G. K. Y. Limsowtin. 1991. The distribution of lactococcal bacteriophage in the environment of a cheese manufacturing plant. Int. Dairy J. 1:183-197. [Google Scholar]
- 23.Moineau, S. 1999. Applications of phage resistance in lactic acid bacteria. Antonie Leeuwenhoek 76:377-382. [PubMed] [Google Scholar]
- 24.Moineau, S., M. Borkaev, B. J. Holler, S. A. Walker, J. K. Kondo, E. R. Vedamuthu, and P. A. Vanderbergh. 1996. Isolation and characterization of lactococcal bacteriophages from cultured buttermilk plants in the United States. J. Dairy Sci. 79:2104-2111. [Google Scholar]
- 25.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137:227-231. [DOI] [PubMed] [Google Scholar]
- 26.Pillidge, C. J., and A. W. Jarvis. 1988. DNA restriction maps and classification of the lactococcal bacteriophages c2 and sk1. N. Z. J. Dairy Sci. Technol. 23:411-416. [Google Scholar]
- 27.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Stahouders, J., and G. J. M. Leenders. 1984. Spontaneously developed mixed-strain cheese starters. Their behavior towards phages and their use in the Dutch cheese industry. Neth. Milk Dairy J. 38:157-181. [Google Scholar]
- 29.Suarez, V. B., and J. A. Reinheimer. 2002. Effectiveness of thermal treatments and biocides in the inactivation of Argentinian Lactococcus lactis phages. J. Food Prot. 65:1756-1759. [DOI] [PubMed] [Google Scholar]
- 30.Whitehead, H. R., and G. A. Cox. 1935. The occurrence of bacteriophage in lactic streptococci. N. Z. J. Dairy Sci. Technol. 16:319-320. [Google Scholar]