Abstract
It has been estimated that less than 1% of the microorganisms in nature can be cultivated by conventional techniques. Thus, the classical approach of isolating enzymes from pure cultures allows the analysis of only a subset of the total naturally occurring microbiota in environmental samples enriched in microorganisms. To isolate useful microbial enzymes from uncultured soil microorganisms, a metagenome was isolated from soil samples, and a metagenomic library was constructed by using the pUC19 vector. The library was screened for amylase activity, and one clone from among approximately 30,000 recombinant Escherichia coli clones showed amylase activity. Sequencing of the clone revealed a novel amylolytic enzyme expressed from a novel gene. The putative amylase gene (amyM) was overexpressed and purified for characterization. Optimal conditions for the enzyme activity of the AmyM protein were 42°C and pH 9.0; Ca2+ stabilized the activity. The amylase hydrolyzed soluble starch and cyclodextrins to produce high levels of maltose and hydrolyzed pullulan to panose. The enzyme showed a high transglycosylation activity, making α-(1, 4) linkages exclusively. The hydrolysis and transglycosylation properties of AmyM suggest that it has novel characteristics and can be regarded as an intermediate type of maltogenic amylase, α-amylase, and 4-α-glucanotransferase.
Soil microorganisms have been the source of many useful biomolecules. However, various molecular analyses such as 16S rRNA studies have confirmed that less than 1% of the microorganisms in soil can be cultured by conventional methods (3, 22, 26, 29). In order to search for new or improved bioactive products from unculturable biota, the sum total of soil microbial genomes, or metagenome, has been extracted to construct metagenomic libraries (10, 22, 26). Previous studies have shown that this strategy offers a large pool of novel genes ranging from small-sized genes conferring enzymatic activities, such as lipases (12), 4-hydroxybutyrate dehydrogenases (11), and amylases (24, 25, 28, 30), to complex gene clusters encoding the enzymes involved in antibiotic production (5, 9, 19, 25).
Because the average size of the structural genes for most enzymes is around 1 to 2 kb, the construction of metagenomic libraries by means of a high-copy-number plasmid vector has been used to search for novel enzymes, despite the relatively short lengths of DNA that can be obtained by this method (11). It is considered feasible to clone an entire pathway in one clone by using the bacterial artificial chromosome library system, especially to search for antibiotics or other complex gene clusters (19, 25). Nevertheless, large-sized DNA is not readily extracted from soil or well preserved in the process of metagenomic library construction with bacterial artificial chromosome vectors.
Amylases (EC 3.2.1.-) are enzymes that hydrolyze starch, and some of them perform transglycosylation or condensation as well as hydrolysis. Amylases and related enzymes have been among the most important enzymes in many industrial fields, especially in the food industry. The application of an amylase in industrial reactions depends on its unique characteristics, such as its action pattern, substrate specificity, major reaction products, optimal temperature, and optimal pH. Finding novel amylases is important with regard to economics as well as science.
In this study, metagenomic libraries were constructed with pUC19, which is a common cloning vector. DNA was extracted from the ground soil of Seoho stream, located in Suwon, South Korea. One novel amylase gene was found in the constructed libraries, and the enzyme was characterized.
MATERIALS AND METHODS
Bacterial strains and plasmids.
For the soil library construction, Escherichia coli DH5α was used as a host, and pUC19 was employed as a vector. pET29b (Novagen) was used as an overexpression vector to produce the target protein in E. coli BL21(DE3) from the target gene encoding amylase.
DNA manipulations and protein methods.
All DNA manipulations, including cloning, transformation into E. coli, PCRs, and DNA sequencing, were performed according to standard techniques (27) and manufacturers' instructions, unless indicated otherwise. Also, the fundamental protein preparation and analysis, including protein extraction from E. coli, protein quantification, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were performed in accordance with standard protocols (4).
Soil DNA preparation and construction of libraries.
Soil was sampled at the junction of the ground and the water of Seoho stream, located in Suwon, South Korea. The surface of the sampling site was covered with moss, and the soil sample was collected at a depth of 5 cm. The DNA isolation from soil samples was based on direct lysis methods (11, 34). The purified soil DNA was partially digested with Sau3AI and size fractionated by sucrose density centrifugation (10 to 40%). Fractions containing DNA fragments of 3 to 7 kb were ligated into BamHI-digested pUC19, and the products were transformed into E. coli DH5α. White colonies were collected to construct libraries in 96-well plates, and constructed libraries were stored in a deep freezer (−80°C) until screening.
Library screening.
Soil libraries in 96-well plates were replicated onto Luria-Bertani (LB) agar containing 50 μg of ampicillin per ml and 2% soluble starch by using a 48-replica pin (Sigma). Colonies were grown at 37°C for 1 day, and then 5 ml of top agar (0.6%) containing d-cycloserine (60 μg/ml) was overlaid to allow detection of the intracellular enzymes. The plates were incubated for one more day before phenotype determination. Amylase activity was detected by flooding the plates with Gram's iodine solution (0.203 g of I2 and 5.2 g of KI in 100 ml of aqueous solution). Active colonies were detected as bright clear haloes upon fluorescent light illumination (8). Plasmids were isolated from all positive clones of the initial screening and retransformed into cells to retest phenotypes. DNA sequencing was performed with a BigDye terminator cycle sequencing kit (Applied Biosystems) and the ABI Prism 3700 DNA analyzer (Perkin-Elmer) in the National Instrumentation Center for Environmental Management (Seoul, South Korea).
Enzyme overexpression and purification.
The putative amylase gene was amplified from the pS2A4 plasmid by using the primers 5′-TTTACATATGAAAAAATCCATCCT-3′ with an NdeI site at the 5′ end and 5′-CAGAAGTGTCGACTTAATCCTTC-3′ with a SalI site at the 3′ end. Amplified DNA was ligated into NdeI- and SalI-digested pET29b (Novagen), and the construct (p29AmyM) was transformed into E. coli BL21(DE3) cells. Originally, pET29 was designed to place an S tag at the N terminus and a six-histidine tag at the C terminus of the insert. In this study, the tagging regions were removed to avoid any structural modification of the original protein. The S tag was removed by digestion with NdeI; the six-histidine tag was removed by putting the termination codon before the tagging region. Transformed cells were grown in 10 ml of LB broth in a 100-ml flask at 37°C overnight; 7.5 ml of the seed culture was used to inoculate 750 ml of LB broth dispensed into three 1,000-ml flasks, which were incubated with agitation at 37°C until an optical density at 600 nm of 1.2 to 1.4 was reached. At this point, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 50 μM, and the flasks were incubated at 16°C for 7 h. Cells were harvested by centrifugation at 9,000 × g for 10 min at 4°C. Harvested cells were suspended with 40 ml of 50 mM glycine-NaOH buffer (pH 9.0) and lysed by ultrasonication (Sonifier 250; Branson). Cell debris was removed by centrifugation at 12,000 × g for 15 min at 4°C. The supernatant was mixed with 0.5 g of raw cornstarch to make a 1% (wt/vol) starch solution and agitated for 2 h at 4°C. After centrifugation at 12,000 × g for 15 min, the starch pellet containing the enzyme was washed three times in 50 ml of cold 50 mM glycine-NaOH buffer (pH 9.0), suspended with 20 ml of 20% (wt/vol) maltose solution, agitated for 2 h at 37°C, and centrifuged to remove starch. The eluate was loaded onto a Q-Sepharose column (anion exchanger; 1.6 by 40 cm) (Pharmacia) equilibrated with 50 mM Tris-HCl buffer (pH 7.5). The column was washed with the same buffer at a flow rate of 2 ml/min; bound protein was eluted with a linear gradient of 0 to 0.6 M NaCl in the same buffer. Active fractions from Q-Sepharose chromatography were pooled and concentrated to 10 ml by using Centriprep 50 (Amicon Co.). The concentrate was dialyzed against 15% glycerol and 0.2 M NaCl in 50 mM glycine-NaOH buffer (pH 9.0) for 24 h with buffer changes every 6 h. The preparation was stored at −20°C until use.
Amylase activity determination.
Determination of amylolytic activity was carried out by using the dinitrosalicylic acid method, which is a procedure for the determination of reducing sugar (21). The enzyme reaction mixture was composed of substrate and an appropriate quantity of enzyme in 20 mM glycine-NaOH buffer (pH 9.0); 500 μl of the enzyme mixture was incubated at the appropriate temperature for 10 min. The reaction was stopped by adding 500 μl of dinitrosalicylic acid solution (10.6 g of 3,5-dinitrosalicylic acid, 19.8 g of NaOH, 306 g of potassium sodium tartrate, 7.6 ml of phenol, 8.3 g of sodium metabisulfate, and 1,416 ml of distilled water). The reaction mixture was boiled for 5 min and cooled by placing the tubes on ice. Absorbance was measured at 575 nm in a 1-cm polystyrene cuvette by using a Hitachi U-1100 spectrophotometer (Tokyo, Japan). One unit of hydrolyzing activity was defined as the amount of enzyme required to produce 1 μmol of maltose-equivalent reducing sugar in 1 min.
For the determination of optimal pH, the following buffers were used for the different pH ranges: pH 4.5 to 6.0, 50 mM sodium acetate; pH 5.5 to 8.0, 50 mM sodium phosphate; pH 7.5 to 9.0, 50 mM Tris-HCl; pH 8.5 to 10, 50 mM CHES [(2-(cyclohexylamino)ethanesulfonic acid]; pH 9.0 to 11.0, 50 mM glycine-NaOH; and pH 10.5 to 12.5, 50 mM Na2HPO4-NaOH.
Thin layer chromatography (TLC).
An appropriate amount of enzyme was incubated with substrate at 42°C. A silica gel K6F thin-layer chromatography plate (Whatman) was activated by incubation at 110°C for 30 min. Prepared samples were spotted on the silica gel plate with a pipette; the plate was placed in a TLC chamber containing a solvent mixture of isopropanol-ethyl acetate-water (3:1:1, vol/vol/vol) and developed at room temperature. The plate was dried completely and dipped rapidly into a methanol solution containing 3 g of N-(1-naphthyl)-ethylenediamine and 50 ml of concentrated H2SO4 solution per liter. The plate was dried and placed at 110°C for 10 min for visualization.
HPIC.
Reaction mixtures were heated in boiling water for 5 min and filtered through a 4.5-μm-pore-size syringe filter. A CarboPac (PA1) column (0.4 by 25 cm; Dionex) and an electrochemical detector (ED40; Dionex) were used for high-performance ion chromatography (HPIC) analysis. Buffer A (150 mM aqueous NaOH solution) and buffer B (600 mM sodium acetate solution in buffer A) were used to make a linear gradient for the elution of saccharides.
Computer programs and services used.
Sequence similarity searches were conducted with BLAST provided by the National Center for Biotechnology Information (2). Sequence manipulation, open reading frame (ORF) searches, and multiple alignments among similar enzymes were conducted with DNASTAR software. Signal peptides were analyzed with SignalP version 1.1 (Center for Biological Sequence Analysis, Technical University of Denmark [http://www.cbs.dtu.dk]).
Nucleotide sequence accession number.
The nucleotide sequence of the insert of pS2A4 has been deposited in the GenBank database under the accession number AY383543.
RESULTS
Construction of metagenomic libraries.
DNA samples were prepared from soil by direct DNA extraction and by using a Wizard DNA Clean-up kit. Approximately 17 μg of DNA was extracted from 1 g of soil. This yield is in the same range as that reported previously for the isolation of DNA from other soils (11). Approximately 600 clones were obtained from 1 μg of stream ground soil DNA. To test the quality of the library, 70 clones were randomly selected, and the recombinant plasmids were prepared. The average insert size was 3.5 kb, and sizes ranged from 2 to 7 kb; the percentage of plasmids containing inserts was approximately 85%.
Screening for amylase activity.
Amylase screening resulted in one amylolytic clone (pS2A4) from among 30,000 clones on an LB agar plate containing soluble starch. The amylolytic activity was also detected without d-cycloserine treatment, which we used to permeabilize the cells, so it can be inferred that this enzyme has a signal sequence that is functional in E. coli.
Molecular analysis of the insert DNA cloned in pS2A4.
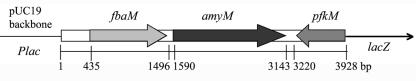
The clone harboring pS2A4 was further characterized. The insert DNA was sequenced and compared with the sequences in the GenBank database by using BLASTX (2). The length of the insert DNA was 3,928 bp, and there were two complete ORFs and one incomplete ORF (Fig. 1). The first complete ORF (fbaM; 1,062 bp) showed the highest similarity with the fructose bisphosphate aldolase class II gene from Yersinia pestis (67% identity and 78% positives). The second complete ORF (amyM; 1,551 bp) was adjacent to fbaM, and it showed the highest similarity with the neopullulanase gene from Paenibacillus polymyxa (47% identity and 64% positives). This ORF was thought to encode a putative amylase and was subjected to further analyses in this study. The incomplete ORF (pfkM) was missing an N-terminal fragment and showed the highest similarity with the phosphofructokinase I gene from Thermus thermophilus (49% identity and 62% positives). The putative functions of all three genes were related to sugar utilization.
FIG. 1.
Schematic diagram of the DNA insert cloned in pS2A4. fbaM is a putative fructose bisphosphate aldolase gene, amyM is a gene encoding the amylase (AmyM) characterized in this work, and pfkM is a fraction of a gene encoding a putative phosphofructokinase.
Molecular analysis of the putative amylase.
The putative amylase gene (amyM) was 1,551 bp long; the molecular mass of the translated protein was estimated to be 59,405 Da. The first 14 amino acid residues were predicted by SignalP analysis to be a signal peptide cleaved in gram-negative bacteria at the ALVAF site. The BLASTP search results for the amino acid sequence of AmyM showed that AmyM had high similarity with the neopullulanase from P. polymyxa (32, 33), α-amylase from Bacillus megaterium (6, 7), α-amylase A from Halothermothrix orenii (20), AmyC from Dictyoglomus thermophilum (14), and periplasmic α-amylase from Xanthomonas campestris K-11151 (1). These enzymes are characterized as α-amylase-like enzymes that have transferring activity, and in some cases they can also hydrolyze cyclodextrin and pullulan, although their similarity to the pullulanase or neopullulanase family is low. Other genes with similarities higher than 30% included those encoding several other α-amylases, oligo-1,6-glucosidases, trehalose synthases, and 4-α-glucanotransferases from thermophilic bacteria.
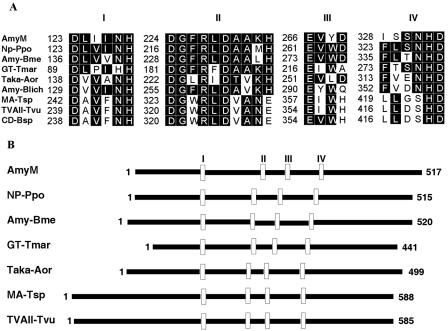
AmyM had four conserved regions that are common in the α-amylase family. Figure 2 shows the comparisons of the four conserved regions with other enzymes belonging to the α-amylase family.
FIG. 2.
Comparisons of amino acid sequences of AmyM with those of related amylases. (A) Amino acid sequences in conserved regions. Highly conserved residues are boxed in black. (B) Schematic alignment of the primary structures. Four conserved regions are marked with open boxes. NP-Ppo, neopullulanase of P. polymyxa (accession no. AAD05199; 47% identity); Amy-Bme, α-amylase of B. megaterium (P20845; 45% identity); GT-Tmar, 4-α-glucanotransferase of Thermotoga maritima (S60618; 33% identity); Taka-Aor, Taka-amylase A of Aspergillus oryzae (P10529; 25% identity); Amy-Blich, α-amylase of Bacillus lichenifomis (P06278; 24% identity); MA-Tsp, maltogenic amylase of Thermus sp. strain IM6501 (AAC15072; 27% identity); TVAII-Tvu, neopullulanase of Thermoactinomyces vulgaris (Q08751; 28% identity); and CD-Bsp, cyclomaltodextrinase of a Bacillus sp. (AAA92925; 27% identity).
Purification of the putative amylase.
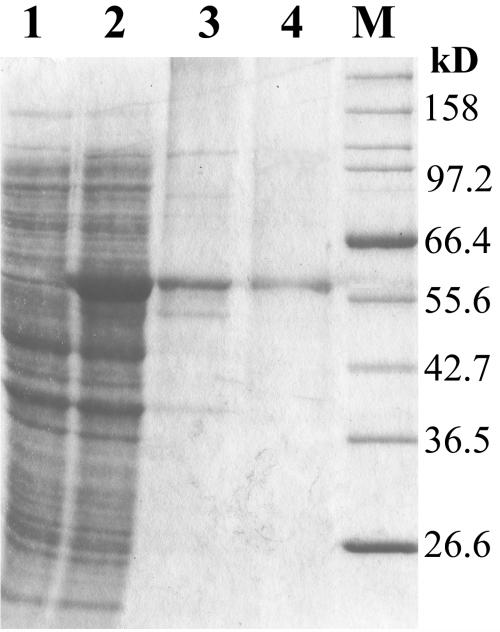
AmyM was overexpressed by using pET29 and purified by adsorption to raw starch and Q-Sepharose column chromatography. The raw cornstarch was confirmed to adsorb approximately 90% of the amylase; however, the yield after 20% maltose elution was relatively low (30 to 40%). The final yield obtained from the cell extract to the Q-Sepharose column chromatography was 20.5%, and the purification factor was 14.2-fold. The size of AmyM, as determined by SDS-PAGE, was about 58 kDa, which is close to the molecular mass calculated from the primary structure (Fig. 3).
FIG. 3.
SDS-PAGE analysis of AmyM at each purification step. Lane 1, cell extract of E. coli harboring p29AmyM before IPTG induction; lane 2, cell extract after IPTG induction; lane 3, the eluted protein after starch adsorption; lane 4, the purified protein after Q-Sepharose column chromatography; lane M, size marker proteins.
Physicochemical properties of AmyM.
The initial reducing sugar assay showed that AmyM was active toward soluble starch, β-cyclodextrin, and pullulan. AmyM showed the highest activity at pH 9.0 toward soluble starch and β-cyclodextrin (Fig. 4A). The optimal reaction temperature was approximately 42°C with starch and 35°C with β-cyclodextrin and pullulan. The optimal reaction temperature was increased by 5°C in the presence of calcium ion (Fig. 4B). The calcium ion was critical to the thermal stability of AmyM, increasing the decimal reduction time at 40°C from 10 (0 mM) to 83 min (1 mM). In the presence of 1 mM Ca2+, the decimal reduction times at 40, 45, 50, and 55°C were 83, 27, 4, and 2.5 min, respectively. AmyM was completely inactivated by a 5-min incubation at 65°C. These results indicate that AmyM is a typical mesophilic enzyme.
FIG. 4.
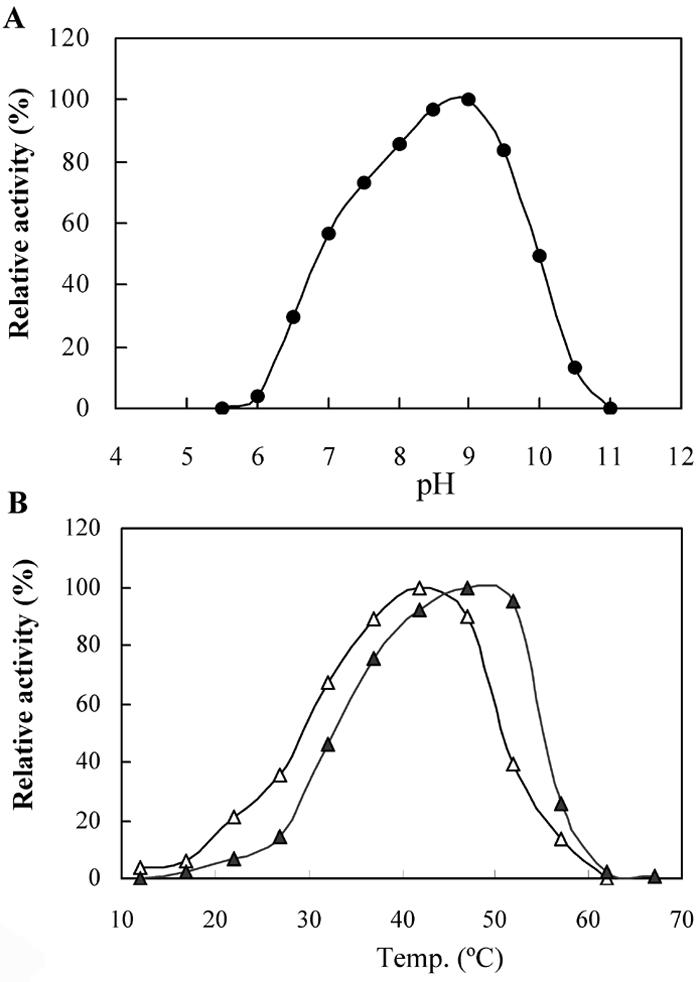
Effect of pH and temperature on activity of AmyM. (A) pH-dependent activity of AmyM. Enzyme activity was measured at 42°C in the corresponding buffer described in the Materials and Methods. (B) Temperature-dependent activity of AmyM toward soluble starch with (▴) and without (▵) 1 mM calcium ion. Enzyme activity was measured at each temperature in 20 mM glycine-NaOH buffer (pH 9.0). The relative activities are the enzyme activities at each temperature divided by the maximal activity under each calcium ion condition.
To determine the effect of metal ions on AmyM activity, enzyme activity was measured at 32°C in the presence of various metal ions. By adding 1 mM calcium ion, enzyme activity was increased dramatically both on soluble starch (197%) and on β-cyclodextrin (168%). Alkali metal ions, such as Li+, Na+, K+, and Cs+, showed either negligible effects or a slight enhancement of enzyme activity toward both soluble starch and β-cyclodextrin. Interestingly, divalent ions differentially affected AmyM activity toward soluble starch and β-cyclodextrin. At a concentration of 1 mM, divalent ions including Mg2+, Ba2+, Fe2+, Co2+, Ni2+, and Cu2+ scarcely influenced the enzyme activity toward soluble starch but inhibited the activity toward β-cyclodextrin. However, at 5 mM, all transition metal ions completely inhibited enzyme activity toward both soluble starch and β-cyclodextrin. In particular, Hg2+ and Fe3+ were highly inhibitory and inactivated the enzyme completely when used in a 1 mM concentration. The addition of 1 mM chelating agents, such as EDTA and EGTA, totally inhibited enzyme activity.
Hydrolysis properties of AmyM.
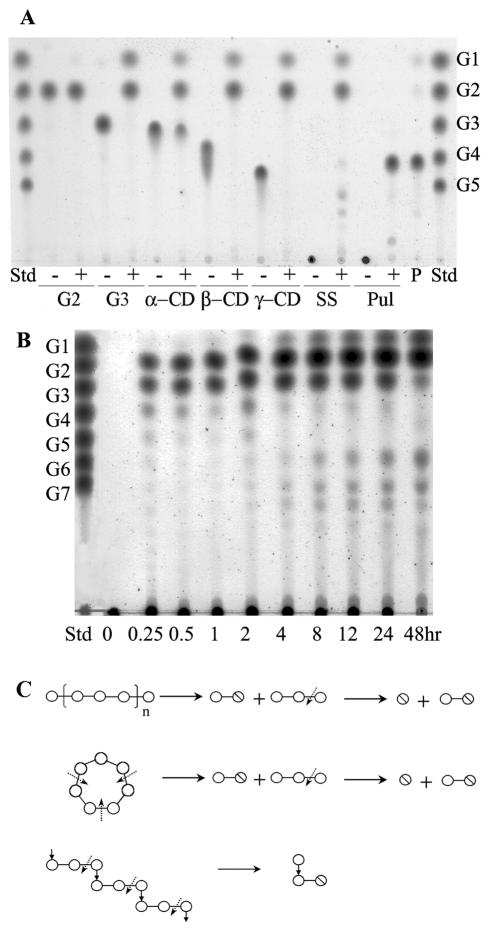
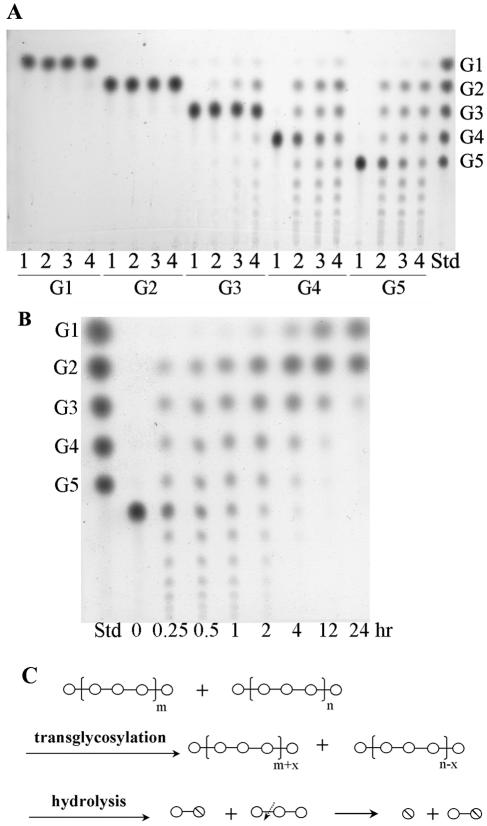
At optimal temperatures, enzyme hydrolysis activity was highest toward soluble starch rather than toward cyclodextrins or pullulan (Table 1). Among the cyclodextrins, γ-cyclodextrin is preferred over β-cyclodextrin or α-cyclodextrin as a substrate. TLC analyses showed the AmyM hydrolysis patterns of various substrates (Fig. 5A). AmyM hydrolyzed soluble starch, cyclodextrins, and maltotriose to produce glucose and maltose. Additionally, the enzyme hydrolyzed pullulan to panose units, which were confirmed by HPIC analysis (data not shown). However, this enzyme barely hydrolyzed maltose.
TABLE 1.
Hydrolysis activities of AmyM on various substrates
| Substrate | Activity (U/mg) | Relative activity (%) |
|---|---|---|
| Soluble starch | 16.5 | 100.0 |
| α-Cyclodextrin | 6.6 × 10−2 | 0.4 |
| β-Cyclodextrin | 1.82 | 11.0 |
| γ-Cyclodextrin | 5.53 | 33.5 |
| Pullulan | 1.07 | 6.5 |
FIG. 5.
TLC analyses of the hydrolyzed products of AmyM action. (A) Hydrolysis activity of AmyM. Each substrate (0.5%, wt/vol) was incubated without (− lanes; substrate controls) or with (+ lanes) the enzyme (2 U/ml) at 42°C for 12 h. G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose; α-CD, α-cyclodextrin; β-CD, β-cyclodextrin; γ-CD, γ-cyclodextrin; SS, soluble starch; Pul, pullulan. Lane Std, maltooligosaccharide standards from glucose (G1) to maltopentaose (G5); lane P, panose standard. (B) Time course analysis of hydrolyzed products of soluble starch. Soluble starch (1%, wt/vol) was incubated with the enzyme (0.2 U/ml) at 42°C. The spots larger than maltotriose generated at later stages were confirmed by HPIC as branched oligosaccharides. (C) Simplified diagram describing the hydrolysis of starch, β-cyclodextrin, and pullulan by AmyM. Circle, glucose unit; slashed circle, reducing end of the molecule; dotted arrows, cutting sites by AmyM; line, α-(1,4) glycosidic linkage; ↓, α-(1,6) glycosidic linkage. n, arbitrary natural number.
A time course analysis with soluble starch showed that this enzyme hydrolyzed starch to maltose and maltotriose units at the early stage of hydrolysis and subsequently hydrolyzed maltotriose to accumulate glucose and maltose as final products. (Fig. 5B).
Transglycosylation properties of AmyM.
To determine whether AmyM has transglycosylation activity, a high concentration (5%, wt/vol) of maltooligosaccharides was incubated with different amounts of enzyme, and the products were analyzed by TLC (Fig. 6A). Maltooligosaccharides larger than maltotriose were easily converted to yield a series of glucose polymers from maltose, even at the lowest enzyme concentration (0.5 U/ml). These products are typical ones made by the glycosylation to the C-4 position of the acceptor, i.e., disproportionation (17). However, only a small amount of transglycosylation product was obtained from maltotriose, even at the highest enzyme concentration (2.5 U/ml). Although glucose was not released by the disproportionation reaction, a small amount of glucose was detected in overreacted samples, due to the hydrolysis activity of AmyM (Fig. 6A, lanes 4 of G4 and G5), which is obvious in the time course analysis of maltohexaose (Fig. 6B). Maltohexaose was disproportionated successfully by AmyM during the first few hours. However, at a later stage of the reaction, large maltooligosaccharides were hydrolyzed to glucose and maltose, which became the major reaction products at 24 h.
FIG. 6.
TLC analyses of transferred products of AmyM action. (A) Transglycosylation activity of AmyM. Each substrate (5%, wt/vol) was incubated with different amounts of enzyme at 42°C for 2 h. Lane 1, 0 U; lane 2, 0.5 U; lane 3, 1 U; lane 4, 2.5 U of AmyM added per ml of substrate solution; lane Std, maltooligosaccharide standards from glucose (G1) to maltopentaose (G5). G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose. (B) Dual action of AmyM for transglycosylation and hydrolysis of maltohexaose. Maltohexaose (5%, wt/vol) was incubated with the enzyme (2.5 U/ml) at 42°C. Lane Std, maltooligosaccharide standards from glucose (G1) to maltopentaose (G5). (C) Simplified diagram showing the dual action of AmyM on maltooligosaccharides. Circle, glucose unit; slashed circle, reducing end of the molecule; dotted arrows, cutting sites by AmyM; line, α-(1,4) glycosidic linkage. n, m, and x are arbitrary natural numbers.
To check the availability of other sugars as acceptors, a 5% (wt/vol) solution of each sugar was incubated with AmyM in the presence of maltotriose as a donor (Fig. 7). AmyM transferred glycosyl units to α-methyl glucoside, cellobiose, lactose, and maltitol to produce one or two kinds of transfer products (Fig. 7, arrows). Mannose, xylose, galactose, sucrose, trehalose, raffinose, and melibiose could not be used as acceptor molecules (data not shown).
FIG. 7.
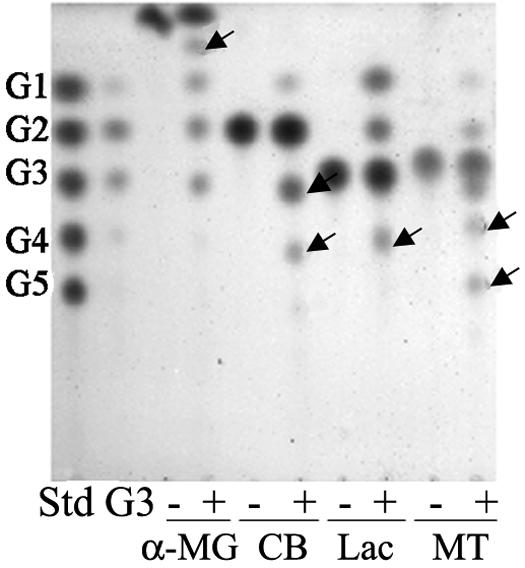
Transglycosylation reaction of AmyM in the presence of different acceptors. Substrate solution containing 1% (wt/vol) maltotriose as donor and 5% (wt/vol) (each) acceptor was incubated with the enzyme (0.2 U/ml) at 42°C for 12 h. Lane Std, maltooligosaccharide standards from glucose (G1) to maltopentaose (G5); lane G3, the reaction control without acceptor; lanes −, the reaction controls without donor; lanes +, the reaction products with both donor and corresponding acceptor. α-MG, α-methyl glucoside; CB, cellobiose; Lac, lactose; MT, maltitol.
DISCUSSION
In this work, metagenomic libraries were constructed by using the vector pUC19, and one novel amylase gene was isolated. The one positive clone, pS2A4, had two complete genes (fbaM, encoding a putative fructose bisphosphate aldolase, and amyM, encoding a putative amylase) and one incomplete gene (pfkM, encoding a putative phosphofructokinase); all three genes were different from any other genes deposited in the GenBank database. Two of the genes were located adjacently in the same direction, and only one possible promoter sequence was found upstream. However, Shine-Dalgano sequences were not found in either gene, probably because this DNA is from a microorganism that is phylogenetically distant from E. coli, and the ribosome-binding site may be quite different.
AmyM was characterized as an amylase with interesting properties. AmyM exhibits multifunctional and mixed properties of several different amylases belonging to the glycoside hydrolase family 13, such as α-amylase, maltogenic amylase (or neopullulanase), and 4-α-glucanotransferase (13). AmyM hydrolyzes α-(1,4) glycosidic bonds in starch, as does α-amylase (Fig. 5A). AmyM also hydrolyzes cyclodextrins to glucose and maltose and pullulan to panose, which are unique properties of maltogenic amylase (15). However, AmyM has a higher hydrolysis activity toward starch than toward cyclodextrins and pullulan, unlike maltogenic amylase, which is most active toward cyclodextrin (Table 1). In addition to the hydrolysis activities mentioned above, AmyM shows a disproportionation activity toward high concentrations of maltooligosaccharides, producing a series of maltohomologues (Fig. 6A). In fact, the TLC pattern of AmyM products is identical to the patterns of well-known disproportionating enzymes, such as 4-α-glucanotransferases (17). AmyM disproportionates maltotetraose and larger maltooligosaccharides more easily than small molecules like maltotriose (Fig. 6A), indicating that maltotetraose and larger maltooligosaccharides are good donor molecules for transglycosylation by AmyM. The series of maltooligosaccharide products are distributed equally, except that glucose is not produced at the early reaction stage. These behaviors are very similar to those of the 4-α-glucanotransferases from Thermotoga maritima and a Bacillus sp. reported previously (18, 23). However, the major difference between them is that AmyM also exhibits hydrolysis activity.
Due to the dual activity of AmyM, the final products of the disproportionation reaction are glucose and maltose, because the larger molecules produced by the action of transglycosylation are hydrolyzed by the same enzyme (Fig. 6B). There are some amylases known to have dual transglycosylation and hydrolysis activities. Cyclodextrin glucanotransferase not only hydrolyzes starch to oligosaccharides but also transfers the glucanosyl segment to an acceptor molecule to form transfer products or cyclodextrins (17). However, cyclodextrin glucanotransferase cannot hydrolyze pullulan, which is a substrate for AmyM. Maltogenic amylase, or neopullulanase, is another enzyme with dual activity, but transfer products are formed by the enzyme mainly via α-linkages (1, 6, 15). AmyM, by contrast, transfers the glucanosyl segment of the donor to an acceptor sugar only via α-linkages (1, 4). AmyM can use other acceptor molecules, such as α-methyl glucoside, cellobiose, lactose, and maltitol (Fig. 7); one or two kinds of transfer products are produced from these acceptors. Interestingly, the action pattern with lactose was somewhat different from the patterns with other acceptors in that more glucose was produced (Fig. 7, lane Lac +). This is probably because of the subsequent hydrolysis of the β-linkage (1, 4) of the lactose transfer product. We are investigating this in more detail, as there is currently no report of an amylolytic enzyme able to hydrolyze both α- and β-linkages (1, 4).
Similar action patterns have been reported with a neopullulanase of P. polymyxa (32) and an α-amylase of B. megaterium (6, 7); these amylases have high amino acid similarity with AmyM, according to a BLASTP search. However, only the pullulan-hydrolyzing activity of the P. polymyxa neopullulanase was tested and not the transglycosylation activity. The disproportionation activity of the α-amylase from B. megaterium was not reported, and its optimal pH (pH 7.0) is quite different from that of AmyM (pH 9.0). Actually, the optimal pH of AmyM (pH 9.0) is much higher than all pH values previously reported (pH 4 to 7).
The physicochemical properties of AmyM toward soluble starch and β-cyclodextrin differ somewhat. The optimal temperature for the hydrolysis of soluble starch is 7°C higher than that for the hydrolysis of β-cyclodextrin, and the activity on β-cyclodextrin is more sensitive to metal ions than is that of soluble starch. In many maltogenic amylases, the unique substrate-dependent properties exhibited by an enzyme are reportedly due to the oligomeric state of the enzyme, which is influenced by the extra domain at the N terminus (16). AmyM does not have this extra domain, however.
The unique characteristics mentioned above are presumably related to the amino acid sequences in the conserved regions of the enzymes (Fig. 2). Comparisons of these regions indicate that AmyM shares more conserved sequences with the α-amylases and 4-α-glucanotransferases than with the maltogenic amylases, neopullulanases, or cyclodextrinases. The isoleucine-serine-serine residues in the conserved region IV of AmyM are unique. In most amylases, isoleucine (I-328) is replaced with phenylalanine or leucine, and serine (S-329) is replaced with hydrophobic amino acids. The hydrophobic or hydrophilic environment at an active site determines whether the reaction mode is hydrolysis or transglycosylation (31).
In conclusion, by using a metagenomic approach, a novel amylolytic gene was isolated. AmyM has mixed characteristics of the α-amylase, 4-α-glucanotransferase, and neopullulanase families. AmyM can hydrolyze starch, cyclodextrins, and pullulan. Also, the enzyme has a transglycosylation activity that is able to disproportionate maltooligosaccharides. These unique properties of AmyM might be due to hydrophobic differences at its catalytic site, compared to the primary structures of other related amylases. Further studies of directed evolution, three-dimensional structure, and kinetics are expected to explain the uniqueness of AmyM and ultimately to deepen our understanding of amylases and related enzymes.
Acknowledgments
This work was supported by the Korean Ministry of Science and Technology (21st Century Frontier Microbial Genomics and Applications Program). J. Yun, S. Kang, S. Park, and H. Yoon have been the recipients of a graduate fellowship provided by the Ministry of Education through the Brain Korea 21 Project.
REFERENCES
- 1.Abe, J., Y. Shibata, M. Fujisue, and S. Hizukuri. 1996. Expression of periplasmic α-amylase of Xanthomonas campestris K-11151 in Escherichia coli and its action on maltose. Microbiology 142:1505-1512. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollag, D. M., and S. J. Edelstein. 1991. Protein methods. Wiley-Liss Inc., New York, N.Y.
- 5.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 6.Brumm, P. J., R. E. Hebeda, and W. M. Teague. 1991. Purification and characterization of the commercialized, cloned Bacillus megaterium α-amylase. Part I: purification and hydrolytic properties. Starch Staerke 43:315-319. [Google Scholar]
- 7.Brumm, P. J., R. E. Hebeda, and W. M. Teague. 1991. Purification and characterization of the commercialized, cloned Bacillus megaterium α-amylase. Part II: transferase properties. Starch Staerke 43:319-323. [Google Scholar]
- 8.Cho, H. Y., Y. W. Kim, T. J. Kim, H. S. Lee, D. Y. Kim, J. W. Kim, Y. W. Lee, S. B. Lee, and K. H. Park. 2000. Molecular characterization of a dimeric intracellular maltogenic amylase of Bacillus subtilis SUH4-2. Biochim. Biophys. Acta 1478:333-340. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie, D. E., S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R. Rondon, J. Clardy, R. M. Goodman, and J. Handelsman. 2002. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 11.Henne, A., R. Daniel, R. A. Shmitz, and G. Gottschalk. 1999. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl. Environ. Microbiol. 65:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henne, A., R. A. Schmitz, M. Bomeke, G. Gottschalk, and R. Daniel. 2000. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horinouchi, S., S. Fukusumi, T. Ohshima, and T. Beppu. 1988. Cloning and expression in Escherichia coli of two additional amylase genes of a strictly anaerobic thermophile, Dictyoglomus thermophilum, and their nucleotide sequences with extremely low guanine-plus-cytosine contents. Eur. J. Biochem. 176:243-253. [DOI] [PubMed] [Google Scholar]
- 15.Kim, I. C., J. H. Cha, J. R. Kim, S. Y. Jang, B. C. Seo, T. K. Cheong, D. S. Lee, Y. D. Choi, and K. H. Park. 1992. Catalytic properties of the cloned amylase from Bacillus licheniformis. J. Biol. Chem. 267:22108-22114. [PubMed] [Google Scholar]
- 16.Kim, T. J., V. D. Nguyen, H. S. Lee, M. J. Kim, H. Y. Cho, Y. W. Kim, T. W. Moon, C. S. Park, J. W. Kim, B. H. Oh, S. B. Lee, B. Svensson, and K. H. Park. 2001. Modulation of the multisubstrate specificity of Thermus maltogenic amylase by truncation of the N-terminal domain and by a salt-induced shift of the monomer/dimer equilibrium. Biochemistry 40:14182-14190. [DOI] [PubMed] [Google Scholar]
- 17.Kitahata, S., and I. Maeda. 1988. Cyclomaltodextrin glucanotransferase and disproportionating enzymes, p. 154-172. In The Amylase Research Society of Japan (ed.), Handbook of amylases and related enzymes: their sources, isolation methods, properties and applications. Pergamon Press, New York, N.Y.
- 18.Liebl, W., R. Feil, J. S. Gabelsberger, J. Kellermann, and K. H. Schleifer. 1992. Purification and characterization of a novel thermostable 4-α-glucanotransferase of Thermotoga maritima cloned in Escherichia coli. Eur. J. Biochem. 207:81-88. [DOI] [PubMed] [Google Scholar]
- 19.MacNeil, I. A., C. L. Tiong, P. R. August, T. H. Grossman, K. A. Loiacono, B. A. Lynch, T. Phillips, S. Narula, R. Sundaramoorthi, A. Tyler, T. Aldredge, H. Long, M. Gilman, D. Holt, and M. S. Osburne. 2001. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J. Mol. Microbiol. Biotech. 3:301-308. [PubMed] [Google Scholar]
- 20.Mijts, B. N., and B. K. C. Patel. 2002. Cloning, sequencing and expression of an α-amylase gene, amyA, from the thermophilic halophile Halothermothrix orenii and purification and biochemical characterization of the recombinant enzyme. Microbiology 148:2343-2349. [DOI] [PubMed] [Google Scholar]
- 21.Miller, G. L. 1959. Use of dinitrosalycylic acid for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 22.Osburne, M. S., T. H. Grossman, P. R. August, and I. A. MacNeil. 2000. Tapping into microbial diversity for natural products drug discovery. ASM News 66:411-417. [Google Scholar]
- 23.Pazur, J. H., and S. Okada. 1968. The isolation and mode of action of a bacterial glucanosyltransferase. J. Biol. Chem. 243:4732-4738. [PubMed] [Google Scholar]
- 24.Richardson, T. H., X. Tan, G. Frey, W. Callen, M. Cabell, D. Lam, J. Macomber, J. M. Short, D. E. Robertson, and C. Miller. 2002. A novel, high performance enzyme for starch liquefaction. J. Biol. Chem. 277:26501-26507. [DOI] [PubMed] [Google Scholar]
- 25.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing in the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rondon, M. R., R. M. Goodman, and J. Handelsman. 1999. The Earth's bounty: assessing and accessing soil microbial diversity. Trends Biotechnol. 17:403-409. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schmeisser, C., C. Stoeckigt, C. Raasch, J. Wingender, K. N. Timmis, D. F. Wenderoth, H.-C. Flemming, H. Liesegang, R. A. Schmitz, K.-E. Jaeger, and W. R. Streit. 2003. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 69:7298-7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torsvik, V., R. Sorheim, and J. Gokoyr. 1996. Total bacterial diversity in soil and sediment communities-a review. J. Ind. Microbiol. 17:170-178. [Google Scholar]
- 30.Voget, S., C. Leggewie, A. Uesbeck, C. Raasch, K.-E. Jaeger, and W. R. Streit. 2003. Prospecting for novel biocatalysts in a soil metagenome. Appl. Environ. Microbiol. 69:6235-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto, T. 1994. Enzyme chemistry and molecular biology of amylases and related enzymes. CRC Press, Boca Raton, Fla.
- 32.Yebra, M. J., J. Arroyo, P. Sanz, and J. A. Prieto. 1997. Characterization of novel neopullulanase from Bacillus polymyxa. Appl. Biochem. Biotechnol. 68:113-120. [Google Scholar]
- 33.Yebra, M. J., A. Blasco, and P. Sanz. 1999. Expression and secretion of Bacillus polymyxa neopullulanase in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 170:41-49. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]