Abstract
Sodium lactate additions to a trichloroethene (TCE) residual source area in deep, fractured basalt at a U.S. Department of Energy site have resulted in the enrichment of the indigenous microbial community, the complete dechlorination of nearly all aqueous-phase TCE to ethene, and the continued depletion of the residual source since 1999. The bacterial and archaeal consortia in groundwater obtained from the residual source were assessed by using PCR-amplified 16S rRNA genes. A clone library of bacterial amplicons was predominated by those from members of the class Clostridia (57 of 93 clones), of which a phylotype most similar to that of the homoacetogen Acetobacterium sp. strain HAAP-1 was most abundant (32 of 93 clones). The remaining Bacteria consisted of phylotypes affiliated with Sphingobacteria, Bacteroides, Spirochaetes, Mollicutes, and Proteobacteria and candidate divisions OP11 and OP3. The two proteobacterial phylotypes were most similar to those of the known dechlorinators Trichlorobacter thiogenes and Sulfurospirillum multivorans. Although not represented by the bacterial clones generated with broad-specificity bacterial primers, a Dehalococcoides-like phylotype was identified with genus-specific primers. Only four distinct phylotypes were detected in the groundwater archaeal library, including predominantly a clone affiliated with the strictly acetoclastic methanogen Methanosaeta concilii (24 of 43 clones). A mixed culture that completely dechlorinates TCE to ethene was enriched from this groundwater, and both communities were characterized by terminal restriction fragment length polymorphism (T-RFLP). According to T-RFLP, the laboratory enrichment community was less diverse overall than the groundwater community, with 22 unique phylotypes as opposed to 43 and a higher percentage of Clostridia, including the Acetobacterium population. Bioreactor archaeal structure was very similar to that of the groundwater community, suggesting that methane is generated primarily via the acetoclastic pathway, using acetate generated by lactate fermentation and acetogenesis in both systems.
Inadequate disposal practices of chlorinated solvents have resulted in the widespread contamination of groundwater with trichloroethene (TCE), a known toxin and suspected carcinogen. In situ biostimulation via anaerobic microbial reductive dechlorination is an attractive remediation strategy for TCE due to low operation and maintenance costs and minimal secondary-waste production relative to traditional extraction methods. Direct addition of nutrients such as electron donors to the contaminated subsurface can stimulate the activity of indigenous microbial communities. After oxygen is consumed, anaerobic populations reduce alternative electron acceptors such as nitrate, ferric iron, sulfate, and carbon dioxide (10). Chloroethenes can also be used as terminal electron acceptors (37, 41, 48, 49), a process known as dehalorespiration, whereby hydrogen atoms sequentially replace chlorine atoms, leading to detoxification of the chloroethene molecule.
Numerous pure strains that either respire or cometabolize chloroethenes have been isolated. Cometabolism of chlorinated compounds, although poorly understood, appears to be common among some anaerobic Bacteria such as the Clostridia and the homoacetogens (7, 19, 78, 80). The enzymatic reactions responsible for these fortuitous degradations, however, may be limited to the highly chlorinated compounds tetrachloroethene (PCE) and TCE. Conversely, dehalorespiring organisms couple chloroethene reduction to energy generation, which increases degradation rates and often results in reduction of the less chlorinated compounds cis-dichloroethene (DCE) and vinyl chloride (VC) (37, 39, 77). Many gram-negative Bacteria can couple the oxidation of organic acids or hydrogen to the reductive dechlorination of PCE or TCE to cis-DCE, including Desulfuromonas chloroethenica (45), Desulfuromonas michiganensis (76), Sulfurospirillum multivorans (previously classified as Dehalospirillum multivorans) (51, 67), and the facultative anaerobe strain MS-1 of the family Enterobacteraceae (69). In addition to the gram-negative Proteobacteria, several species of gram-positive bacteria, including Dehalobacter restrictus (38) and members of the genus Desulfitobacterium (24, 28, 47), dehalorespire using PCE or TCE as the terminal electron acceptor.
Efficient, complete reductive dechlorination of PCE and TCE to ethene has been documented for numerous mixed cultures (5, 25, 64, 77). The first isolate found to be capable of energetic dechlorination of PCE to ethene was Dehalococcoides ethenogenes strain 195, although VC reduction was cometabolic (55, 56). In contrast, the isolate Dehalococcoides sp. strain BAV1 demonstrated energetic dechlorination of cis-DCE and VC, but dechlorinated TCE only cometabolically (32, 33). To date, all isolated Dehalococcoides spp. have been demonstrated to use only hydrogen as an electron donor and halogenated organic compounds as electron acceptors (1, 33, 56).
While efforts to understand the dechlorinating physiology of isolated microorganisms are of unequivocal value, it is likely that interactions among multiple populations in complex communities are essential for dechlorination in the field. The nature of these complex communities and interactions pertinent to the dechlorination process remain largely unknown. Detailed characterization of an actively dechlorinating community from a field site may provide insight into interspecies relationships, thereby guiding bioremediation optimization efforts.
A plume of TCE-contaminated groundwater lies beneath Test Area North (TAN) at the U.S. Department of Energy's Idaho National Engineering and Environmental Laboratory. The plume emanates from a residual source that resulted from the disposal of mixed wastes composed of TCE, raw sewage, and radionuclides through an injection well (58). The aquifer resides deep within fractured basalt, making contaminant characterization and ex situ remediation difficult and expensive. TCE concentrations of at least 32,000 μg liter−1 have been measured within the plume that spans an area of approximately 1 by 3 km and extends from 64 m to greater than 155 m in depth (74).
In 1999, a field evaluation of in situ biostimulation of anaerobic reductive dechlorination was begun at TAN. Lactate was injected into the TCE source area on a weekly basis from January until early September. The lactate was rapidly fermented to acetate and propionate, with subsequent production of small quantities of butyrate in the areas of highest electron donor concentration (54, 72). At the end of 8 months of regular lactate additions, molar concentrations of ethene generally exceeded the initial TCE concentrations throughout the treatment zone (73). Bioremediation was selected to replace the pump-and-treat method for remediation of the source area, and lactate has been injected every 6 to 8 weeks from February 2000 to the present. The purpose of the study presented herein was to describe the complex microbial community at TAN that has been enriched from regular lactate amendments to the subsurface and that is responsible for the complete dechlorination of TCE to ethene in situ and to compare and contrast a TCE-dechlorinating enrichment community to the field community from which it was derived in order to determine its representativeness.
MATERIALS AND METHODS
Collection of groundwater from TAN.
Well TAN-25 is located within the residual source area of the TCE plume, approximately 15 m downgradient of the lactate injection well. From June to December 2001, 1-liter samples were collected monthly by using aseptic precautions from TAN-25 at a depth of 67 m beneath the ground surface; the water table resides at approximately 64 m. Groundwater was collected with a Redi-Flo 2 submersible pump (Grundfos) by a low-flow technique. Three in-line volumes were purged from the well before the samples were collected in sterile screw-top bottles, filled completely to minimize exposure to air.
Establishment of a TCE-dechlorinating consortium in the laboratory.
An anaerobic TCE-dechlorinating culture was established in the laboratory in a bioreactor designed entirely of glass and Teflon to ensure anaerobic conditions and minimize adsorption of chlorinated organic compounds to the system. To fabricate the bioreactor, the top of a glass vacuum flask was sealed shut and a 1-cm (inside diameter [i.d.]) glass sampling port with a glass stopcock and Teflon septum was attached to the bottom for removing liquid samples from the bioreactor. A second glass port was attached to the flask near the top and sealed with a Teflon-lined screw cap for headspace sampling.
TAN groundwater (1 liter) collected in June 2001 was directly transferred into the bioreactor in an anaerobic glove box containing 90% N2, 5% CO2, and 5% H2 by a sterile technique. The bioreactor culture was amended with lactate (10 mM, nominal concentration) and TCE (nominally 7.6 μM) and incubated statically at 12°C to mimic the average in situ temperature of the aquifer. Liquid samples (10 ml) were collected weekly and analyzed for lactate, acetate, propionate, butyrate, TCE, cis- and trans-DCE, VC, ethene, methane, and pH. One quarter of the total volume was removed weekly via fill and draw and replaced with fresh TAN groundwater collected monthly between June 2001 and November 2001 and amended with TCE (7.6 μM) and lactate (10 mM). This was performed to ensure recruitment of the populations present in the field to the laboratory culture. After 4 months of incubation, reductive daughter products (cis-DCE, VC, and ethene) were not observed in the bioreactor. It was suspected that short hydraulic retention times (approximately 4 weeks) were insufficient to allow growth of dechlorinators at 12°C. Therefore, in November 2001, one-half of the volume was replaced with fresh groundwater and the headspace was flushed for 1 h with sterile N2 (100%) to remove hydrogen introduced in the glove box. The bioreactor was amended with lactate (10 mM), TCE was increased to 76 μM, and the consortium was incubated at 12°C as before. Samples were removed regularly for chemical analyses (10 ml) and DNA extraction (30 ml); however, the hydraulic retention time was increased to approximately 100 weeks. The volume removed was replaced with fresh groundwater collected in December 2001 and January 2002.
Chemical analyses.
TCE, cis- and trans-DCE, and VC were analyzed by the solid-phase microextraction (SPME) technique (4). Briefly, liquid samples (5 ml) were placed in glass serum bottles (25 ml), which were evacuated and sealed with gray butyl Teflon-lined septa, and equilibrated for 3 h at room temperature. A 0.75-μm-pore-size Carboxen polydimethylsiloxane-coated fiber (Supelco) was used to sorb headspace gas for 15 min and was then desorbed into the injector of a Hewlett-Packard model 5890 series II gas chromatograph (GC) equipped with a 30-m, 0.32-mm-i.d. (1.8-μm film thickness) RTx-624 chromatography column (Restek). The injector was fitted with a 1-mm SPME liner and maintained at 250°C. Helium was used as the carrier gas at a flow rate of 2 ml min−1. The column temperature was initially maintained at 60°C for 6.5 min and then increased to 180°C at a rate of 70°C min−1. Analytes were detected with a flame ionization detector (FID) maintained at 280°C. The minimum detection limit for TCE, DCE, and VC was 5 ppb. Aqueous-phase concentrations of the analytes were calculated with Henry's constants (29).
Analysis of ethene, ethane, and methane was performed by injecting a headspace sample (100 μl) from a 5-ml aqueous sample prepared for the chloroethene analysis above into the Hewlett-Packard model 5890 series II GC equipped with an FID and a 0.53-mm (i.d.) Rt alumina column (Restek). Helium was used as the carrier gas at a flow rate of 6.5 ml min−1. Throughout the assay, the temperature of the column was maintained at 80°C, the injector was maintained at 250°C, and the detector was maintained at 275°C. The minimum detection limit for ethene, ethane, and methane was 1 ppb by volume.
Samples for acetate, propionate, and butyrate measurements were prepared by filtering (0.2-μm-pore-size filter) samples and adjusting the pH to 2.0 with concentrated phosphoric acid. Typically, 100 μl of acid was added per 0.5 ml of filtrate, and 1.0 μl of the acidified solution was injected directly into the model 5890 GC equipped with an FID and a 30-m, 0.53-mm-i.d. (0.5-μm film thickness) Nukol column (Supelco). Helium, which was used as the carrier gas, was delivered at a flow rate of 8.2 ml min−1. The column temperature was maintained at 125°C, and the injector and detector temperatures were maintained at 225 and 250°C, respectively. The minimum detection limit for acetate, propionate, and butyrate was 5 ppm.
Lactate concentration was measured in groundwater filtrate with a Dionex 4500i ion chromatograph equipped with an IonPac ICE-AS6 column and a conductivity detector. A 0.4 mM nitric acid solution was used as the eluant at a flow rate of 1.5 ml min−1, and 5.0 mM tetrabutylammonium hydroxide was used as the anion suppression regenerant. The minimum detection limit for lactate was 0.2 ppm.
Solutions of TCE (98.6%; Supelco), cis-DCE (99.0%; Supelco), trans-DCE (99.0%; Supelco), and VC (2,000 ppm; AccuStandard) were used for calibration standards. Certified ethene, ethane, and methane standards (Scotts Specialty Gas) were used to calibrate the gas analyses. Certified standard mixes of acetic, propionic, and butyric acids in methanol were obtained from the Environmental Resource Association and AccuStandard.
Extraction of nucleic acids.
Triplicate 35-ml aliquots of TAN groundwater and 10-ml aliquots of laboratory culture were centrifuged (20,000 × g) for 20 min to obtain cell pellets. The groundwater and culture samples contained approximately 107 cells ml−1 (determined by direct counts with acridine orange). The supernatant was decanted, and the cell pellets were resuspended in 500 μl of DNase-free water (Promega). Community DNA was extracted with the FastDNA SPIN kit for soil (Bio 101) according to the manufacturer's instructions and eluted in nuclease-free water (50 μl). The triplicate extractions were pooled and stored at −20°C.
16S ribosomal DNA (rDNA)-targeted PCR.
PCR was used to amplify nearly full-length 16S rRNA genes from Bacteria and Archaea. Each 50-μl PCR mixture included 0.4 mg of molecular-grade bovine serum albumin (Sigma Chemicals) ml−1, 1× PCR buffer (Promega), 1.5 mM MgCl2, 0.5 μM (each) forward and reverse primer (Invitrogen), 1 U of Taq DNA polymerase (Promega), 0.2 mM (each) deoxynucleoside triphosphate (Invitrogen), 1 μl of template DNA, and molecular-grade water (Promega). Universal primer 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) was used in conjunction with either eubacterial primer 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′) or archaeal primer 4F (5′-TCC GGT TGA TCC TGC CRG-3′) (12), targeting the 16S rRNA genes of Bacteria and Archaea, respectively. Amplification was performed on a Perkin-Elmer model 9600 thermocycler by the following regimen: 95°C (5 min), followed by 25 cycles of 95°C (1 min), 53.5°C (1 min), and 72°C (1 min). The reaction was finished with an additional 7 min at 72°C. PCR products were examined in a 1.2% agarose gel prior to clone library construction to confirm specificity of the amplification reactions. Dehalococcoides spp. were targeted with primers Fp DHC 1 (5′-GAT GAA CGC TAG CGG CG-3′) and Rp DHC 692 (5′-TCA GTG ACA ACC TAG AAA AC-3′), corresponding to base positions 1 to 17 and 692 to 673, respectively (35).
PCR for terminal restriction fragment length polymorphism (T-RFLP) employed primers 8F, modified with phosphoramidite fluorochrome 5-carboxyfluorescein (FAM; Invitrogen), and 907R (5′-CCG TCA ATT CMT TTR AGT TT-3′) for the Bacteria and 4F, modified with FAM, and 958R (5′-YCC GGC GTT GAM TCC AAT T-3′) (12) for the Archaea. Each PCR (50-μl mixture) was performed in triplicate by the PCR and thermocycler protocols described above.
Construction of bacterial and archaeal 16S rDNA clone libraries.
Nearly full-length 16S rDNA PCR products amplified from the TAN groundwater community and nearly 700-bp fragments from Dehalococcoides-specific PCR were purified (QIAquick PCR purification kits; QIAGEN) and cloned into the p-GemT Easy vector (Promega) according to the manufacturer's instructions. Transformants were selected with standard blue-white screening on plates of S-Gal (3,4-cyclohexenoesculetin-β-d-galactopyranoside; Sigma) with ampicillin. Plasmids were purified from 139 transformants from the bacterial library, 45 from the archaeal library, and 5 from the Dehalococcoides library. Plasmid DNA was extracted and purified from cultures of each clone grown in 1 ml of TPYNG medium (66) containing ampicillin with the QIAprep Spin Miniprep kit (QIAGEN). The purified plasmids were subjected to PCR with primers M13F (5′-GTA AAA CGA CGG CCA G-3′) and M13R (5′-CAG GAA ACA GCT ATG AC-3′), flanking the insertion site on the vector, to reamplify the insert; PCR reagents and conditions were as described above. Fifteen microliters of reamplified inserts was digested in NEB2 buffer (New England Biolabs) with the restriction endonucleases MspI (1 U) and HinpII (1 U) (37°C, 6 h) for restriction fragment length polymorphism (RFLP) analysis, and resolved in a 3% agarose gel (NuSieve).
Sequencing and phylogenetic analysis.
One representative of each unique RFLP type (phylotype) from the bacterial and archaeal libraries and two from the single Dehalococcoides RFLP type were sequenced with primers M13F, M13R, 515F (5′-GTG CCA GCM GCC GCG GTA A-3′) (63), 519R (5′-ATT ACC GCG GCT GCT GG-3′), 906F (5′-GAA ACT TAA AKG AAT TG-3′) (63), and 907R (5′-CCG TCA ATT CCT TTR AGT TT-3′) (46) to obtain greater-than-twofold average coverage of the entire insert. Sequencing reactions employed the ABI PRISM BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems) and model 3700 automated DNA sequencer (Applied Biosystems). The sequences were assembled and aligned with EditSeq and SeqMan software (DNASTAR). Sequences were initially aligned against known sequences (GenBank database) with the BLAST tool (3) provided by the National Center for Biotechnology Information prior to phylogenetic analyses. The Ribosomal Database Project (9) Chimera Check program and secondary structure determination were used to check the partial 16S rRNA gene sequences for potential chimeric artifacts. Potentially chimeric sequences were not given further consideration. Sequences with ≥97% sequence similarity to one another were considered indistinguishable and were viewed as belonging to a single phylotype (75).
The determined sequences of all nonchimeric cloned 16S rRNA genes from the TAN groundwater community were aligned with sequences included in the ARB database or obtained from the EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk) with the tools implemented in the ARB package (http://www.arb-home.de). The resulting alignment was visually inspected, and potential errors were subsequently corrected manually. Evolutionary distances were calculated based on the algorithms of Jukes and Cantor (43) or Felsenstein (21) using neighbor-joining and parsimony methods of tree reconstruction, as implemented in the ARB program package.
T-RFLP analysis of TAN groundwater and bioreactor communities.
The triplicate PCR products generated from each DNA sample (combined triplicate extractions) were combined and purified with the QIAquick PCR purification kits (QIAGEN). PCR product concentration was estimated by measurement of absorbed UV light at a λ of 260 nm. Approximately 200 ng of PCR product was digested with MspI (1 U) or HhaI (1 U) (New England BioLabs; 37°C, 3 h) for Bacteria and Archaea, respectively. The digested fragments were purified by ethanol precipitation as described elsewhere (66), and the DNA pellet was resuspended in 10 μl of DNase-free water. Samples were denatured by heating to 95°C for 3 min followed by submersion in an ice bath. The denatured DNA (2 μl), along with the internal standard Rox 1000 (Applied Biosystems), was loaded onto a model 377 DNA sequencer (Applied Biosystems). The resulting data were analyzed with Genescan, version 2.1 (Applied Biosystems).
Each PCR product was digested once, and each digest was run in triplicate on the DNA sequencer to generate replicate T-RFLP profiles of the TAN groundwater and bioreactor samples. These replicates were aligned manually, and composite profiles were generated as described previously (16, 17). Methods described by Dunbar et al. (16, 17) were used to ensure that the fragments compared between the groundwater and laboratory cultures were real and not artifacts of the method. Therefore, only those fragments exhibiting high reproducibility were evaluated. Briefly, terminal restriction fragments (T-RFs) in different replicates that differed by less than 0.5 bp were considered identical and assigned the average size and peak heights of the fragments making up the composite T-RF. Any T-RF not present in all of the replicate profiles was discarded. Likewise, any T-RF within a composite profile with peak height averaging ≤25 fluorescence units was also discarded. The sum of all T-RF peak heights ≥25 (Bacteria) or ≥50 (Archaea) florescence units was used as an indicator of the total DNA quantity represented, and peak heights were normalized to the replicate with the lowest total fluorescence.
Many of the T-RF peaks represented in the TAN groundwater and bioreactor were indirectly identified by digesting individual clones from 16S rDNA libraries and identifying their corresponding T-RFs. The PCR and T-RFLP protocols were the same as those described above except that 25 ng of plasmid was used as the PCR template. Although each clone generated a single T-RF, multiple clones frequently generated the same size T-RF (18). Theoretical T-RF sizes were also determined in silico by locating restriction sites within the clones.
Diversity indices.
Once the community profiles were normalized, statistical parameters to measure diversity, relative abundance, and similarity were calculated for the T-RFLP profiles and for the clone libraries so that the communities could be compared as described previously (17) by using the Jaccard coefficient to estimate the similarity of the communities and the Shannon and Simpson indices to estimate diversity.
Nucleotide sequence accession numbers.
All clone sequences have been deposited in the GenBank, DDBJ, and EMBL databases under accession numbers AY667249 to AY667274, as indicated in Tables 1 and 2.
TABLE 1.
Bacterial 16S rDNA clones from TAN groundwater and identification of T-RFs
| Clone | GenBank accession no. | T-RF length (bp) | Frequencya | Putative class/order or candidate division | Closest GenBank matchb |
|---|---|---|---|---|---|
| TANB55 | AY667261 | 218, 224, 300 | 32 | Clostridia/Clostridiales | Acetobacterium sp. strain HAAP-1c (99%) |
| TANB77 | AY667263 | 315 | 6 | Clostridia/unclassified | |
| TANB7 | AY667252 | 288 | 5 | Clostridia/Clostridiales | Clone WCHB1-82d (98%) |
| TANB5 | AY667250 | 230 | 4 | Clostridia/Clostridiales | Clone DCE25c (98%) |
| TANB101 | AY667266 | 520 | 3 | Clostridia/Clostridiales | Clostridium haemolyticum (96%) |
| TANB44 | AY667258 | 464 | 3 | Clostridia/unclassified | |
| TANB115 | AY667268 | 520 | 2 | Clostridia/Clostridiales | Clostridium puniceum (98%) |
| TANB107 | AY667265 | 453 | 1 | Clostridia/Clostridiales | Clone P3IB-23 (97%) |
| TANB127 | AY667269 | 295 | 1 | Clostridia/Clostridiales | Clone vadinHB04 (95%) |
| TANB3 | AY667249 | 90 | 9 | Sphingobacteria/Sphingobacteriales | Clone BD1-16 (95%) |
| TANB53 | AY667260 | 90 | 2 | Sphingobacteria/Sphingobacteriales | |
| TANB59 | AY667262 | 92 | 2 | Bacteroides/Bacteroidales | |
| TANB6 | AY667251 | 469 | 2 | ɛ-Proteobacteria/Campylobacterales | Sulfurospirillum multivoransf (98%) |
| TANB142 | AY667270 | 509 | 1 | δ-Proteobacteria/Desulfuromonadales | Trichlorobacter thiogenesg (99%) |
| TANB18 | AY667253 | 123 | 2 | Spirochaetes/Spirochaetales | Clone DCE33h (99%) |
| TANB52a | AY667259 | 544 | 2 | Spirochaetes/Spirochaetales | |
| TANB22 | AY667254 | 262 | 7 | OP11 | Clone d153i (99%) |
| TANB35 | AY667256 | 262 | 3 | OP11 | |
| TANB37 | AY667257 | 262 | 1 | OP11 | Clone d153i (97%) |
| TANB108 | AY667267 | 262 | 1 | OP11 | |
| TANB84 | AY667264 | 1489 | 1 | OP3 | |
| TANB25 | AY667255 | 279 | 3 | Mollicutes/Acholeplasmatales | |
| TANDhc2j | 514 | NA | Unclassified | Dehalococcoides sp. strain FL2k (100%) |
Frequency out of 93 nonchimeric bacterial clones from TAN groundwater. NA, not applicable.
Sequences in the database with ≥95% similarity to TAN clones are listed, along with the sequence similarity percentages.
An RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine)-degrading homoacetogen isolated from a methanogenic consortium (2).
From a chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation (14).
From a cis-DCE-dechlorinating consortium (31).
Previously classified as Dehalospirillum multivorans (51).
A trichloroacetate-dechlorinating isolate (12).
From TCE- and cis-DCE-dechlorinating consortia (31).
From a TCE-contaminated site undergoing intrinsic dechlorination (50).
Clone TANDhc2 was not a part of the general bacterial library; rather it was amplified separately with Dehalococcoides-specific primers Fp DHC 1 and Rp DHC 692 (35).
Strain FL2 is the dehalogenating population in a defined coculture (48).
TABLE 2.
Archaeal 16S rDNA clones from TAN groundwater and identification of T-RFs
| Clone | GenBank accession no. | T-RF length (bp) | Frequencya | Putative order/family | Closest GenBank matchb |
|---|---|---|---|---|---|
| TANA2 | AY667272 | 196 | 24 | Methanosarcinales/Methanosaetaceae | Methanosaeta concilii (98%) |
| TANA5 | AY667273 | 335 | 10 | Methanomicrobiales/Methanospirillaceae | Methanospirillum hungatei (96%) |
| TANA1 | AY667271 | 329 | 5 | Methanosarcinales/Methanosarcinaceae | Methanosarcina lacustris (97%) |
| TANA6 | AY667274 | 335 | 4 | Methanomicrobiales/unclassified | Clone 57-1 (97%) |
Frequency out of 43 nonchimeric bacterial clones from TAN groundwater.
Sequences in the database with ≥95% similarity to TAN clones are listed here along with the sequence similarity percentages.
RESULTS
TAN groundwater chemistry.
Groundwater chemical characteristics in well TAN-25 are described in detail elsewhere (54, 72). Briefly, lactate injection resulted in dramatic increases in chemical oxygen demand and organic acids, a subsequent decrease in oxidation-reduction potential, complete removal of sulfate, an increase in dissolved iron, generation of methane, and a reduction of all aqueous-phase TCE to ethene, which has been maintained since 1999. The sample from 8 November 2001 was collected 1 week following a lactate injection.
Bacterial community structure.
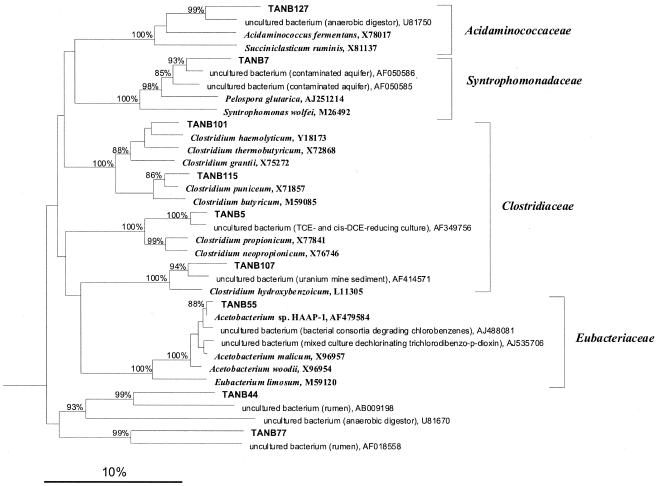
DNA was extracted from groundwater at well TAN-25 and used to construct a clone library of nearly full-length bacterial 16S rRNA genes. One hundred thirty-nine clones were screened by RFLP analysis and grouped according to RFLP type. Highly similar sequences (≥97% sequence similarity) were considered indistinguishable (75), resulting in the identification of 53 unique phylotypes within the bacterial library. Approximately 82% of the clones were observed multiple times, suggesting that reasonably sufficient coverage of the library had been achieved (53). Of the 53 bacterial phylotypes, 31 were potentially chimeric (discussed further below) and were thus removed from diversity analyses. The 22 nonchimeric phylotypes, representing 93 of the 139 clones, were aligned against the GenBank database and are described in Table 1. The bacterial library was predominated by members of the Clostridia, which constituted 57 of the 93 clones, or 61% of the library. Of these, the most abundant phylotype was represented by clone TANB55, most closely related to the homoacetogen Acetobacterium sp. strain HAAP-1 (2) (99.5% sequence similarity, 32 of 93 clones); Acetobacterium malicum is the most closely related organism identified to the species level (98.7% sequence similarity). Clone TANB7 had 98% sequence similarity with another clone, WCHB1-82, recovered from a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing natural attenuation (14). Similarly, clones TANB5 and TANB18 had ≥98% similarity with clones from TCE- and cis-DCE-dechlorinating consortia (31).
Eleven clones representing two phylotypes were most closely affiliated with a class of common soil and water Bacteria, the Sphingobacteria. Another 13 clones appeared to be affiliated with the proposed phyla OP11 and OP3, of which, to our knowledge, there are currently no isolated representatives (61). Clones TANB22 and TANB37 had high sequence similarity with clone d153, recovered from a TCE-contaminated site undergoing natural attenuation (50). The remaining phylotypes were most similar to species of Bacteroides, Spirochaetes, Mollicutes, and Proteobacteria. Only two proteobacterial clones were observed. Clone TANB142 was nearly identical to the δ-Proteobacterium Trichlorobacter thiogenes, a trichloroacetic acid-degrading microorganism in the proposed family Geobacteraceae (13, 71). The other Proteobacterium, TANB6, was most similar to the PCE and TCE dechlorinator Sulfurospirillum multivorans (previously classified as Dehalospirillum multivorans and recently transferred to the genus Sulfurospirillum) (51, 67).
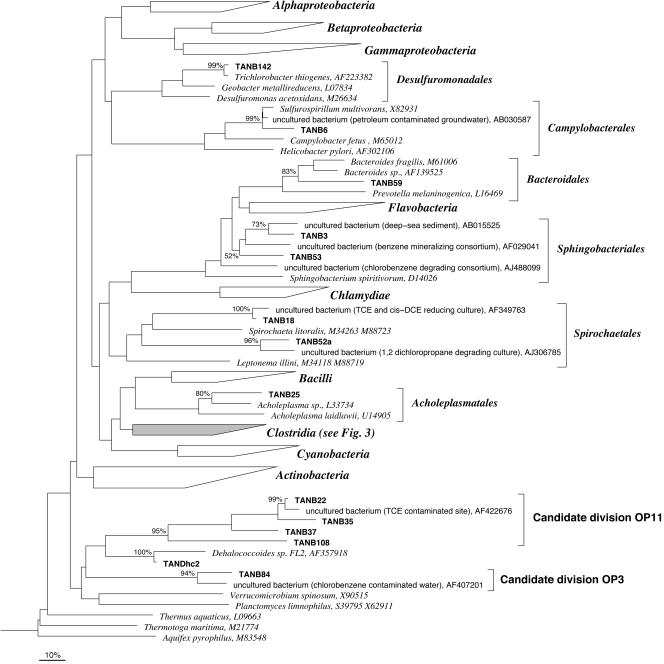
The phylogenetic trees in Fig. 1 and 2 depict the relationships among the 23 bacterial phylotypes (including Dehalococcoides) from TAN groundwater and include several reference strains from the Bacteria, including some organisms known to carry out reductive dechlorination (e.g., Sulfurospirillum multivorans, Trichlorobacter thiogenes, and Dehalococcoides ethenogenes). There is a notable lack of representatives from the Proteobacteria in TAN groundwater (Fig. 1). Several phylotypes are deeply branching, with only distant relationships to known organisms. For example, clones TANB3 and TANB53 in the Sphingobacteriales form a distinct branch with clones from deep-sea sediments and a benzene-mineralizing consortium. Likewise, the clones affiliated with candidate phyla OP11 and OP3 have no pure-culture representatives. Clones TANB18 and TANB52a are only distantly related to Spirochaeta litoralis. This tendency toward phylogenetically distinct clones is most clear in the Clostridia (Fig. 2). Indeed, only three of the nine representatives of this division bear any significant relatedness (≥95% sequence similarity) to cultured organisms (Table 1).
FIG. 1.
Global phylogenetic tree based on 16S rRNA gene sequences showing the affiliation of TAN clone sequences to the main lines of descent of Bacteria. The positions of TAN clone sequences affiliated to the class Clostridia are shown in Fig. 2. The tree is based on a selection of 207 species representing the various depicted classes and orders of Bacteria, including several species of Archaea used to define the root (not shown). The reconstructed tree is a consensus tree based on maximum-parsimony and neighbor-joining calculations. Bootstrap values (1,000 resamplings) are shown only at the branching points of TAN clone sequences and refer to the neighbor-joining treeing method (21) using the algorithm described by Jukes and Cantor (43) to calculate phylogenetic distances. Scale bar, 10% estimated sequence divergence.
FIG. 2.
Phylogenetic tree based on 16S rRNA gene sequences showing the positions of TAN clone sequences among members of the class Clostridia. Brackets, established families of the order Clostridiales. The sequence of Escherichia coli (GenBank accession no. L10328) was used to define the root (not shown). The neighbor-joining method of Saitou and Nei (65) was used to reconstruct the matrix, and the algorithm described by Jukes and Cantor (43) was used to calculate phylogenetic-distance values. Only bootstrap values above 80% (1,000 resamplings for each node) are shown at the respective branching points. Scale bar, 10% estimated sequence divergence.
PCR amplification of similar DNA sequences can result in the formation of chimeras (75). One reason that a large proportion of our bacterial library was potentially chimeric (46 of 139 clones) may be the prevalence of very similar sequences, which has been shown to increase the incidence of chimeras (75). Although it is difficult to make sense of chimeric sequences, fragments of these clones were similar to those of some identifiable organisms, most of which were represented by nonchimeric clones in this study. The largest fragments aligned best with 16S rRNA genes from Acetobacterium spp., Clostridium spp., and other Clostridia, Bacteroides, δ- and ɛ-Proteobacteria, candidate phyla OP3 and OP11, Mollicutes, and Verrucomicrobia (data not shown). Of these, the clone containing the Verrucomicrobia fragment was a member of the only taxon not represented in the portion of the library used for analysis.
Detection of Dehalococcoides spp.
Approximately 700-bp PCR amplicons were generated from TAN groundwater with primers Fp DHC 1 and Rp DHC 692, specific for the genus Dehalococcoides (35). These PCR products were then cloned, and five 16S rRNA gene clones were screened via RFLP. All of the clones had the same RFLP pattern, and two of the clones were sequenced to confirm their affiliation with the genus Dehalococcoides. The clones had 99% sequence similarity with one another, and one of the two was 100% identical to the 16S rRNA gene from the cocultured organism Dehalococcoides sp. strain FL2 (48) (Table 1 and Fig. 1).
Archaeal community structure.
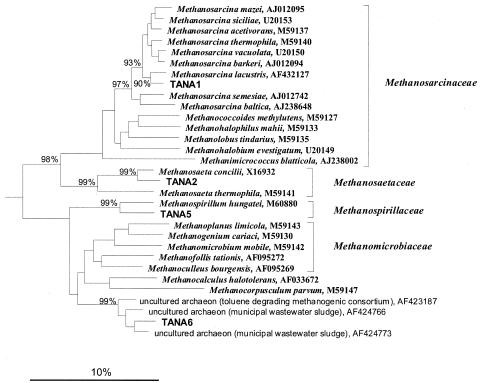
Forty-five archaeal 16S rRNA gene clones were screened by RFLP and grouped into RFLP-defined phylotypes. Fully 100% of the archaeal clones were observed multiple times. By applying a minimum threshold of 3% sequence dissimilarity, five unique phylotypes were identified, one of which was potentially chimeric and thus disregarded. All four nonchimeric sequences had ≥96% similarity with 16S rRNA genes from methanogens in the GenBank database (Table 2), displaying far less diversity than pristine regions of the aquifer (60). The library was predominated (more than 50% of the archaeal clones) by a phylotype represented by clone TANA2, which has 98% sequence similarity with the acetoclast Methanosaeta concilii. The remaining clones were most similar to Methanospirillum hungatei, Methanosarcina lacustris, and an environmental clone affiliated with the Methanomicrobiales. The largest fragment of the chimeric clone was most similar to Methanospirillum hungatei (data not shown).
The tree in Fig. 3 depicts relationships among the four archaeal 16S rDNA phylotypes from TAN groundwater and reference strains from the Archaea. Clone TANA6 is part of a deep branch within the Methanomicrobiales that has no cultured representatives. All of the archaeal clones appear to be affiliated with methanogenic microorganisms.
FIG. 3.
Phylogenetic tree based on 16S rRNA gene sequences showing the positions of TAN clone sequences among members of the orders Methanosarcinales and Methanomicrobiales of the Euryarchaeota. Brackets, established families of methanogens. Sequences of members of the Euryarchaeota were used to define the root (not shown). The neighbor-joining method of Saitou and Nei (65) was used to reconstruct the matrix, and the algorithm described by Jukes and Cantor (43) was used to calculate phylogenetic-distance values. Only bootstrap values above 80% (1,000 resamplings for each node) are shown at the respective branching points. Scale bar, 10% estimated sequence divergence.
Development of TCE-dechlorinating activity in the laboratory.
Groundwater from well TAN-25 was used to inoculate a bioreactor in the laboratory that was monitored for chloroethenes, organic acids, and methane (data not shown). Once the hydraulic retention time was increased in November 2001 (see Materials and Methods), the development of TCE-dechlorinating activity within the laboratory culture occurred in two distinct stages: (i) nearly stoichiometric dechlorination of TCE to cis-DCE within 25 days of inoculation and (ii) dechlorination of cis-DCE to VC and ethene after 48 days.
The fate of the electron donor within the bioreactor was also tracked throughout the 67-day sampling period. Similar to what was observed at the TAN field site (54, 72), lactate was fermented in the bioreactor to propionate and acetate in a ratio slightly greater than 1:1 and maintained during the entire study period.
Methane was also observed in the bioreactor. Methane concentrations within the culture peaked 2 weeks after the change in hydraulic retention time (0.26 mM) but then declined, achieving stable concentrations that were subsequently maintained throughout the remainder of the experimental period (0.07 to 0.13 mM).
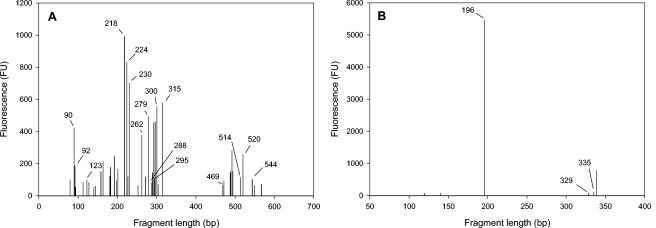
T-RFLP analysis of the TAN groundwater community.
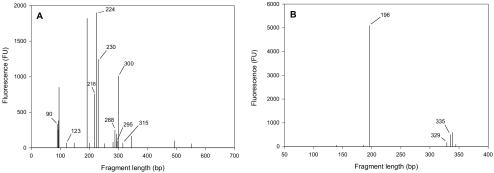
T-RFLP analysis was performed with TAN groundwater DNA in triplicate. The composite bacterial profile contained 43 repeatable T-RFs (Fig. 4A). The values for peak heights generated in the replicate profiles for each T-RF were within 10% of each other. The T-RFs were compared to those generated from the TAN groundwater clone library to provide tentative identification. Fifteen of the 43 community T-RFs corresponded to at least one clone in the library, accounting for 59% of the total fluorescence. Twelve of these could be confidently assigned to a single clone, while the other three corresponded to multiple clones (T-RFs 90, 262, and 520; Table 1). The 28 unidentified T-RFs accounted for the remaining 41% of the total fluorescence.
FIG. 4.
T-RFLP analysis of Bacteria and Archaea in TAN groundwater. (A) Bacterial T-RFLP profile of TAN groundwater generated with primers 8F-FAM and 907R and the restriction endonuclease MspI. (B) Archaeal T-RFLP profile of TAN groundwater generated with primers 4F-FAM and 958R and the restriction endonuclease HhaI. Labeled peaks correspond to clones reported in Tables 1 and 2. Fluorescence units (FU) are arbitrary.
Three of the largest peaks within the TAN groundwater bacterial profile, T-RFs 218, 224, and 300, all corresponded to the phylotype represented by clone TANB55, which was most similar to Acetobacterium sp. strain HAAP-1. Three separate clones generated these three T-RFs, comprising 24% of the total fluorescence, but the sequences were greater than 97% similar to one another and were thus pooled into one phylotype. T-RFs corresponding to clones from the class Clostridia (including Acetobacterium spp.) accounted for 42% of the total fluorescence within the groundwater T-RFLP profile. The 514-bp T-RF predicted for the Dehalococcoides clone (TANDhc2) based on sequence analysis was present in the groundwater bacterial T-RFLP profile and was not produced by any of the other clones in the library (Fig. 4A; Table 1). The only other known dehalorespirer detected was Sulfurospirillum multivorans (T-RF 469). Other T-RFs that were correlated with the clone library represent five additional orders and are shown in Table 1.
The groundwater archaeal T-RFLP profile contained six reproducible T-RFs, three of which could be assigned to one or more of the clones in the library (Fig. 4B; Table 2). The predominant T-RF in the archaeal profile, responsible for 83% of the total fluorescence, was T-RF 196, which corresponded to clone TANA2 (most similar to Methanosaeta concilii). T-RF 329 corresponded to archaeal clone TANA1 (Methanosarcina lacustris) and comprised 1% of the total fluorescence. T-RF 335 was generated by two clones, TANA5 and TANA6 (Methanomicrobiales spp.) and comprised 1.6% of the total fluorescence. Three T-RFs from the groundwater community were not identified based on the clone library and contributed 14% of the total fluorescence.
T-RFLP analysis of the dechlorinating laboratory consortium.
T-RFLP analysis was performed in triplicate on the dechlorinating consortium that was enriched in the bioreactor 67 days after the increase in hydraulic retention time (Fig. 5). T-RFLP analysis of the bioreactor consortium enriched with a 4-week hydraulic retention showed far less diversity than that shown in Fig. 5 (data not shown). The composite bacterial profile from the bioreactor contained 22 reproducible T-RFs (Fig. 5A). Nine of the bacterial T-RFs could be correlated to T-RFs identified in the groundwater clone library (Fig. 5A; Table 1). Fluorescence from bioreactor T-RFs corresponding to clone TANB55 (218, 224, and 300 bp), most closely affiliated with Acetobacterium sp. strain HAAP-1, comprised 34% of the total fluorescence from all bacterial T-RFs in the consortium. T-RFs corresponding to groundwater clones affiliated with the class Clostridia (218, 224, 300, 315, 288, 230, 520, and 295 bp) accounted for 54% of the total fluorescence. Other bacterial fragments corresponding to groundwater clones included T-RF 90 (clones TANB3 and TANB53, affiliated with the order Sphingobacteriales) and T-RF 123 (clone TANB18, affiliated with the order Spirochaetales) (Table 1).
FIG. 5.
T-RFLP analysis of Bacteria and Archaea in the dechlorinating bioreactor consortium. (A) Bacterial T-RFLP profile of the bioreactor consortium generated with primers 8F-FAM and 907R and the restriction endonuclease MspI. (B) Archaeal T-RFLP profile of the bioreactor consortium generated with primers 4F-FAM and 958R and the restriction endonuclease HhaI. Labeled peaks correspond to clones reported in Tables 1 and 2. Fluorescence units (FU) are arbitrary.
The T-RF corresponding to Dehalococcoides clone TANDhc2 (514 bp) was not present in the laboratory consortium T-RFLP profile (Fig. 5A). Note, however, that amplification of groundwater and bioreactor DNA with the Dehalococcoides-specific primers generated an amplicon of the expected size (data not shown), suggesting that Dehalococcoides cells were present in the consortium but were below the T-RFLP method detection limit.
The archaeal T-RFLP profile in the bioreactor consortium contained seven repeatable T-RFs, three of which could be assigned to clones from the TAN archaeal clone library (Fig. 5B; Table 2). T-RF 196, associated with clone TANA2 (affiliated with Methanosaeta concilii), contributed 78% of the total fluorescence in the bioreactor T-RFLP profile. T-RF 335, corresponding to clones TANA5 and TANA6 (Methanomicrobiales spp.), accounted for 8% of the total fluorescence. The T-RF corresponding to clone TANA1, most similar to Methanosarcina lacustris, contributed only 2.4% of the total fluorescence. The remaining four T-RFs contributed 11.6% of the total fluorescence.
Comparison of microbial diversity in the laboratory bioreactor consortium to that in the TAN groundwater community.
For the diversity calculations, it was assumed that every T-RF represented a different species within the microbial communities and that the proportion of each T-RF's fluorescence intensity (peak height) to the total fluorescence of the community profile was reflective of that species' relative abundance within the community. In the present study, three T-RFs (90, 520, and 262 bp) were correlated to multiple clone phylotypes, but all of the phylotypes were related (Sphingobacteriales, Bacteroidales, and candidate division OP11), and therefore the Jaccard coefficient measured the presence or absence of these groups. Relative abundance is used here not to estimate the actual abundance of a given phylotype in the community but rather to track changes in predominance from the groundwater community to the bioreactor consortium. The relative proportion of a given RFLP pattern from the TAN groundwater clone library was used as another indicator of relative abundance for comparison to the fluorescence approach.
Richness of Bacteria in TAN groundwater estimated from the T-RFLP profile was double that estimated from the clone library data (43 versus 22 unique phylotypes; Table 3). This is partially the result of having pooled clones with ≥97% similarity to one another (e.g., TANB55 is represented by three T-RFs). Another explanation of the lower richness in the clone library is the poorer sensitivity of clone screening compared to that of T-RFLP; for a phylotype to be recognized in a library of 100 randomly sampled clones it must make up at least 1% of the library. The richness of Bacteria in the bioreactor consortium was estimated at 22 according to the T-RFLP profile, about one-half that of the TAN groundwater Bacteria T-RFLP.
TABLE 3.
Bacterial and archaeal 16S rDNA-based indices of diversity for TAN groundwater and the derivative anaerobic enrichment culture
| Sample | Richness | Sa | Hb | Ec | Dd |
|---|---|---|---|---|---|
| TAN bacterial clone library | 22 | NA | 3.59 | 0.80 | 0.85 |
| TAN bacterial T-RFLP | 43 | NA | 4.90 | 0.87 | 0.96 |
| Bioreactor bacterial T-RFLP | 22 | 0.33 | 3.57 | 0.64 | 0.89 |
| TAN archaeal clone library | 4 | NA | 1.69 | 0.85 | 0.64 |
| TAN archaeal T-RFLP | 6 | NA | 0.89 | 0.30 | 0.30 |
| Bioreactor archaeal T-RFLP | 7 | 0.83 | 1.22 | 0.41 | 0.38 |
Jaccard similarity coefficient comparing the T-RFLP profiles of Archaea and Bacteria in the bioreactor with those in the field at TAN. NA, not applicable.
Shannon index.
Shannon function.
Simpson index.
The Jaccard similarity coefficient, which considers only the presence or absence of a population and not relative abundance, indicated that the two bacterial communities shared only 33% of their total species, suggesting considerable differences in community composition. The Shannon (H) and Simpson (D) indices, which consider both richness and evenness, were greatest for the Bacteria in TAN groundwater based on T-RFLP data (Table 3). The Shannon function, which weighs evenness more heavily than the Shannon or Simpson indices (1.0 indicates perfect evenness), was greater for the field community (0.87) than for the laboratory consortium (0.64) (Table 3).
Diversity of Archaea in TAN groundwater and in the bioreactor consortium was considerably lower than that of Bacteria (Table 3). The Jaccard coefficient of 0.83 suggests that the archaeal T-RFLP from the bioreactor was very similar to that of TAN groundwater. Shannon and Simpson diversity indices for the archaeal communities were highest for the clone library from TAN groundwater, probably due to the poor evenness of the T-RFLP profiles of groundwater and bioreactor Archaea. Only the TAN groundwater clone library displayed substantial evenness (Shannon function; E) among the Archaea.
DISCUSSION
In this study, molecular analyses allowed elucidation of the structure of a microbial community that reductively dechlorinates TCE to ethene in deep, fractured basalt due to biostimulation with sodium lactate and also of the laboratory culture derived from this groundwater community. Comparison of the laboratory consortium with the field community illustrates that, although the laboratory consortium was much less diverse, the major populations involved in electron donor utilization appeared to be preserved. The indices demonstrate that some bacterial diversity, including both richness and evenness, was lost during transfer to laboratory conditions. Accounting for this loss of diversity may be important when interpreting data from laboratory enrichments used to simulate field processes. While the desired function (dechlorination) was retained in this case, a function being evaluated could be lost during laboratory enrichment, particularly if the microbial community has not already been enriched in the field. Furthermore, if a new set of conditions is applied to a laboratory enrichment community whose diversity is much less than that of the field community, the response of the enrichment community may not be representative of the field.
The TAN groundwater community had many similarities with those of other dechlorinating sites. Although not as dominant as they were at TAN, Clostridia were also present in an aquifer where TCE was being intrinsically degraded predominantly to cis-DCE at the Lawrence Livermore National Laboratory (50), in an aquifer undergoing intrinsic natural attenuation of hydrocarbons and chlorinated alkanes at Wurtsmith Air Force Base (14), and in a predominantly Fe(III)-reducing TCE-contaminated aquifer (36, 85). All three of these other sites also had members of candidate division OP11, and the two of the three sites where complete declorination was observed had green nonsulfur Bacteria (the group to which Dehalococcoides is most similar). A significant difference between the biostimulated TAN community and the others was the predominance of Proteobacteria in the three previous studies without biostimulation, as opposed to the predominance of Clostridia at TAN.
Of particular interest is the Wurtsmith Air Force Base study (14) because it is the only study for which Archaea were characterized. Interestingly, as observed at TAN, the archaeal communities were predominated by one clone that was most closely related to the acetoclastic Methanosaeta concilii. As one of the abundant bacterial clones was most similar to species of the acetogenic genus Syntrophus, the authors suggested a syntrophic relationship between these two populations, although the tie to attenuation of organic contaminants was not discussed.
To our knowledge, there have been no published characterizations of field communities undergoing biostimulation; therefore, direct comparison of the TAN field community to other biostimulated field communities cannot be made. However, the TAN laboratory enrichment can be directly compared to other laboratory enrichments undergoing dehalogenation to provide insight into the similarities and differences of functional groups active in these systems. Similar to what was found for TAN, Richardson et al. reported that, in an enriched TCE-dechlorinating community from Alameda Naval Air Station in California, low-G+C gram-positive species were a significant proportion of the total community, as were species associated with the Cytophaga/Flavobacterium/Bacteroides cluster (64). One distinct difference between this culture and that from TAN, however, was that Desulfovibrio spp. were implicated in lactate fermentation, providing hydrogen and acetate to Dehalococcoides. In the TAN culture, however, it appears that Acetobacterium spp. largely play this role.
In a PCB-dechlorinating mixed community, low-G+C gram-positive species and Dehalococcoides spp. were also present (62). As in the Richardson et al. culture (64), this culture contained fermentative sulfate reducers (including Desulfovibrio and Desulfuromonas spp.), as opposed to the homacetogens present in the TAN community. It is interesting that the Richardson et al. (64) and Pulliam-Holoman et al. (62) enrichments were incubated at 25 to 30°C, in contrast to the incubation of the TAN enrichment at 12°C. Archaea in the Pulliam-Holoman culture were also characterized and were dominated by methanogens, including the strictly acetoclastic Methanosaeta concilii, which comprised 6 of 24 RFLP types.
Similar to what has been seen at other sites, lactate additions at TAN led to dechlorination of TCE to ethene in two distinct stages (72). In the first stage, TCE was reduced to cis-DCE. Microorganisms with enzymes from the acetyl-coenzyme A pathway, including acetogens and acetoclastic methanogens, may play a role in this first stage of dechlorination due to an abundance of transition metal corrinoid cofactors (15, 20, 42). Corrinoid-dependent dechlorination has been demonstrated in methanogenic and acetogenic consortia and isolates (6, 19, 24, 26, 40, 41, 79-82, 84), as well as dehalorespiratory isolates (1, 27, 41, 44, 52, 59, 67, 70). Our knowledge of microorganisms that can participate in the second stage of dechlorination, namely, the reduction of cis-DCE and VC, is currently limited to the genus Dehalococcoides (11, 33, 56). This taxon was detected at TAN and is likely supported by homoacetogenic Bacteria, which provide a source of fixed carbon and hydrogen. Corrinoids may also provide a source of vitamin B12, an essential nutrient for Dehalococcoides (55).
Homoacetogenic Bacteria in the TAN groundwater and laboratory culture appear to maintain a syntrophic or commensal relationship with acetoclastic and acetate-assimilating methanogens and/or dechlorinators. Homoacetogens produce acetate as the primary end product from energy-yielding metabolism of a variety of substrates, including H2 and CO2 and/or lactate (15). The fermentation of lactate by homoacetogens or other fermenters may proceed according to the reaction (79)
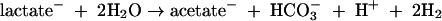 |
(1) |
Acetate can also be produced from H2-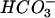 by homoacetogenic Bacteria (15, 79) according to the reaction
by homoacetogenic Bacteria (15, 79) according to the reaction
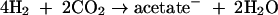 |
(2) |
Propionate is present at TAN in a nearly 1:1 ratio with acetate (54), suggesting that lactate is also being fermented by nonhomoacetogens according to the reaction (30, 68)
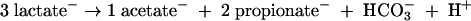 |
(3) |
The propionate generated can also serve as a source of acetate by fermentation in the reaction (23)
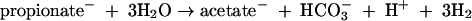 |
(4) |
Acetate produced by reactions 1 through 4 is available for acetoclastic methanogenesis, which proceeds according to the reaction (83)
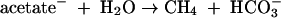 |
Molecular H2, however, is generally thought to be responsible for most dehalorespiratory activity, not acetate. Hydrogen can be generated from reactions 1 and 4, as well as through acetate oxidation via the reaction (34, 86)
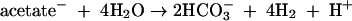 |
and almost certainly stimulates dehalorespiring microorganisms at TAN, such as Sulfurospirillum multivorans (67) and Dehalococcoides ethenogenes (55, 56). The methanogens at TAN were all either acetoclastic (clones TANA1 and TANA2) or acetate assimilating (clones TANA5 and TANA6) (83), including a clone closely affiliated with the obligate acetoclast Methanosaeta concilii, making up more than one-half of the library. Methanospirillum hungatei has even been shown to exhibit chemotaxis toward acetate (57). Obligately hydrogenotrophic methanogenic taxa were not detected. Characterization of other methanogenic communities has shown that the acetoclastic pathway for methanogenesis is favored over the hydrogenotrophic pathway at low temperatures (15°C) in contrast to high temperatures (30°C) (8, 23). Similarly, at lower temperatures hydrogenotrophic acetogenesis is less favored than acetogenesis from fermentation of complex sugars (23). The year-round low temperatures in the TAN aquifer (ca. 12°C), mimicked in the laboratory culture, along with the accumulation of acetate from several of the reactions described above, may thus suppress hydrogenotrophic acetogenesis and methanogenesis, minimizing competition with hydrogenotrophic dechlorinators. This suggests that hydrogenotrophic methanogenesis, which is often portrayed as being detrimental to dechlorination because of competition with hydrogenotrophic dechlorinators (22), is less important at TAN than acetoclastic methanogenesis. Likewise, efficient dechlorination of the TCE residual source at TAN over the last 5 years in conjunction with active methane production suggests that the methanogenesis occurring at TAN does not negatively impact dechlorination over the long term. As additional insight on relationships between dechlorinators and other populations in complex, dechlorinating field communities is gained, the ability to optimize contaminant degradation during biostimulation or bioaugmentation will be significantly improved.
Acknowledgments
We extend our gratitude to Cathy Rae for performing chloroethene and organic acid analyses and to Erin O'Leary-Jepsen of the Molecular Research Core Facility at Idaho State University for performing the T-RFLP analyses. The manuscript was improved significantly by the thoughtful criticism of Mike Lehman and Rick Colwell.
This work was funded by a grant from the U.S. Department of Energy through the INEEL LDRD program under DOE Idaho Operations Office contract DE-AC07-99ID13727.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Gorisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, N. R., and C. M. Arnett. 2004. Anaerobic biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Acetobacterium malicum strain HAAP-1 isolated from a methanogenic mixed culture. Curr. Microbiol. 48:332-340. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Arthur, C. L., D. W. Potter, K. D. Buchholz, S. Motlagh, and J. Pawliszyn. 1992. Solid-phase microextraction for the direct analysis of water: theory and practice. LC GC 10:656-661.
- 5.Ballapragada, B. S., H. D. Stensel, J. A. Puhakka, and J. F. Ferguson. 1997. Effect of hydrogen on reductive dechlorination of chlorinated ethenes. Environ. Sci. Technol. 31:1728-1734. [Google Scholar]
- 6.Bouwer, E. J., and P. L. McCarty. 1983. Transformations of 1- and 2-carbon halogenated aliphatic organic compounds under methanogenic conditions. Appl. Environ. Microbiol. 45:1286-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. C., M. Hatsu, K. Jung, Y. S. Yoo, and K. Takamizawa. 2000. Isolation and characterization of a tetrachloroethylene dechlorinating bacterium, Clostridium bifermentans DPH-1. J. Biosci. Bioeng. 89:489-491. [DOI] [PubMed] [Google Scholar]
- 8.Chin, K.-J., T. Lukow, S. Stubner, and R. Conrad. 1999. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15 and 30oC). FEMS Microbiol. Ecol. 30:313-326. [DOI] [PubMed] [Google Scholar]
- 9.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cord-Ruwisch, R., H.-J. Seitz, and R. Conrad. 1988. The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch. Microbiol. 149:350-357. [Google Scholar]
- 11.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl. Environ. Microbiol. 69:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Wever, H., J. R. Cole, M. R. Fettig, D. A. Hogan, and J. M. Tiedje. 2000. Reductive dehalogenation of trichloroacetic acid by Trichlorobacter thiogenes gen. nov., sp. nov. Appl. Environ. Microbiol. 66:2297-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drake, H. L. 1994. Acetogenesis, acetogenic bacteria, and the acetyl-CoA “Wood/Ljungdahl” pathway: past and current perspectives. In H. L. Drake (ed.), Acetogenesis. Chapman and Hall, New York, N.Y.
- 16.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egli, C., T. Tschan, R. Scholtz, A. M. Cook, and T. Leisinger. 1988. Transformation of tetrachloromethane to dichloromethane and carbon dioxide by Acetobacterium woodii. Appl. Environ. Microbiol. 54:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelmann, T., F. Kaufmann, and G. Diekert. 2001. Isolation and characterization of a veratrol:corrinoid protein methyl transferase from Acetobacterium dehalogenans. Arch. Microbiol. 175:376-383. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein, J. 1982. Numerical methods for inferring phylogenetic trees. Q. Rev. Biol. 57:379-404. [Google Scholar]
- 22.Fennell, D. E., J. M. Gossett, and S. H. Zinder. 1997. Comparison of butyric acid, ethanol, lactic acid, and propionic acid as hydrogen donors for the reductive dechlorination of tetrachloroethene. Environ. Sci. Technol. 31:918-926. [Google Scholar]
- 23.Fey, A., and R. Conrad. 2000. Effect of temperature on carbon and electron flow and on the archaeal community in methanogenic rice field soil. Appl. Environ. Microbiol. 66:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finneran, K. T., H. M. Forbush, C. G. Van Praagh, and D. R. Lovley. 2002. Desulfitobacterium metallireducens sp. nov., an anaerobic bacterium that couples growth to the reduction of metals and humic acids as well as chlorinated compounds. Int. J. Syst. Evol. Microbiol. 52:1929-1935. [DOI] [PubMed] [Google Scholar]
- 25.Freedman, D. L., and J. M. Gossett. 1989. Biological reductive dechlorination of tetrachloroethylene and trichloroethylene to ethylene under methanogenic conditions. Appl. Environ. Microbiol. 55:2144-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gander, J. W., G. F. Parkin, and M. M. Scherer. 2002. Kinetics of 1,1,1-trichloroethane transformation by iron sulfide and a methanogenic consortium. Environ. Sci. Technol. 36:4540-4546. [DOI] [PubMed] [Google Scholar]
- 27.Gantzer, C. J., and L. P. Wackett. 1991. Reductive dechlorination catalyzed by bacterial transition-metal coenzymes. Environ. Sci. Technol. 25:715-722. [Google Scholar]
- 28.Gerritse, J., V. Renard, T. M. Pedro Gomes, P. A. Lawson, M. D. Collins, and J. C. Gotschal. 1996. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165:132-140. [DOI] [PubMed] [Google Scholar]
- 29.Gossett, J. M. 1987. Measurement of Henry's Law constants for C1 and C2 chlorinated hydrocarbons. Environ. Sci. Technol. 21:202-208. [Google Scholar]
- 30.Gottschalk, G. 1986. Bacterial metabolism, 2nd ed. Springer-Verlag, New York, N.Y.
- 31.Gu, A. Z., B. P. Hedlund, J. T. Staley, S. E. Strand, and H. D. Stensel. 2004. Analysis and comparison of the microbial community structures of two enrichment cultures capable of reductively dechlorinating TCE and cis-DCE. Environ. Microbiol. 6:45-54. [DOI] [PubMed] [Google Scholar]
- 32.He, J., K. M. Ritalahti, M. R. Aiello, and F. E. Löffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He, J., K. M. Ritalahti, K. L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 34.He, J., Y. Sung, M. E. Dollhopf, B. Z. Fathepure, J. M. Tiedje, and F. E. Löffler. 2002. Acetate versus hydrogen as direct electron donors to stimulate the microbial reductive dechlorination process at chloroethene-contaminated sites. Environ. Sci. Technol. 36:3945-3952. [DOI] [PubMed] [Google Scholar]
- 35.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hohnstock-Ashe, A. M., S. M. Plummer, R. M. Yager, P. Baveye, and E. L. Madsen. 2001. Further biogeochemical characterization of a trichloroethene-contaminated fractured dolomite aquifer: electron source and microbial communities involved in reductive dechlorination. Environ. Sci. Technol. 35:4449-4456. [DOI] [PubMed] [Google Scholar]
- 37.Holliger, C. 1995. The anaerobic microbiology and biotreatment of chlorinated ethenes. Curr. Opin. Biotechnol. 6:347-351. [Google Scholar]
- 38.Holliger, C., D. Hahn, H. Harmsen, W. Ludwig, W. Schumacher, B. Tindall, F. Vazquez, N. Weiss, and A. J. B. Zehnder. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 39.Holliger, C., and G. Schraa. 1994. Physiological meaning and potential for application of reductive dechlorination by anaerobic bacteria. FEMS Microbiol. Rev. 15:297-305. [DOI] [PubMed] [Google Scholar]
- 40.Holliger, C., G. Schraa, E. Stupperich, A. Stams, and A. Zehnder. 1992. Evidence for the involvement of corrinoids and factor F430 in the reductive dechlorination of 1,2-dichloroethane by Methanosarcina barkeri. J. Bacteriol. 174:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holliger, C., G. Wohlfarth, and G. Diekert. 1999. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 42.Jablonsky, P. E., and J. G. Ferry. 1992. Reductive dechlorination of trichloroethylene by CO-reduced CO dehydrogenase enzyme complex from Methanosarcina thermophila. FEMS Microbiol. Lett. 96:55-60. [DOI] [PubMed] [Google Scholar]
- 43.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules. In H. N. Murano (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 44.Krasotkina, J., T. Walters, K. A. Maruya, and S. W. Ragsdale. 2001. Characterization of the B-12- and iron-sulfur-containing reductive dehalogenase from Desulfitobacterium chlororespirans. J. Biol. Chem. 276:40991-40997. [DOI] [PubMed] [Google Scholar]
- 45.Krumholz, L., R. Sharp, and S. Fishbain. 1996. A freshwater anaerobe coupling acetate oxidation to tetrachloroethylene dehalogenation. Appl. Environ. Microbiol. 62:4108-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid-determination of 16S ribosomal-RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA. 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, T., T. Tokunaga, A. Suyama, and K. Furukawa. 2001. Efficient dechlorination of tetrachloroethylene in soil slurry by combined use of an anaerobic Desulfitobacterium sp. strain Y-51 and zero-valent iron. J. Biosci. Bioeng. 92:453-458. [DOI] [PubMed] [Google Scholar]
- 48.Löffler, F. E., J. M. Tiedje, and R. A. Sanford. 1999. Fraction of electrons consumed in electron acceptor reduction and hydrogen thresholds as indicators of halorespiratory physiology. Appl. Environ. Microbiol. 65:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Louie, T. M., and W. W. Mohn. 1999. Evidence for a chemiosmotic model of dehalorespiration in Desulfomonile tiedjei DCB-1. J. Bacteriol. 181:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowe, M., E. L. Madsen, K. Schindler, C. Smith, S. Emrich, F. Robb, and R. U. Halden. 2002. Geochemistry and microbial diversity of a trichloroethene-contaminated Superfund site undergoing intrinsic in situ reductive dechlorination. FEMS Microbiol. Ecol. 40:123-134. [DOI] [PubMed] [Google Scholar]
- 51.Luijten, M. L. G. C., J. deWeert, H. Smidt, H. T. S. Boschker, W. M. deVos, G. Schraa, and A. J. M. Stams. 2003. Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int. J. Syst. Evol. Microbiol. 53:787-793. [DOI] [PubMed] [Google Scholar]
- 52.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 54.Martin, J. P., K. S. Sorenson, and L. N. Peterson. 2001. Favoring efficient in situ TCE dechlorination through amendment injection strategy, p. 265-272. In V. Magar, D. Fennell, J. L. Morse, B. Alleman, and A. Leeson (ed.), Anaerobic degradation of chlorinated solvents. Battelle Press, Columbus, Ohio.
- 55.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maymó-Gatell, X., Y.-T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 57.Migas, J., K. L. Anderson, D. L. Cruden, and A. J. Markovetz. 1989. Chemotaxis in Methanospirillum hungatei. Appl. Environ. Microbiol. 55:264-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.National Research Council. 2000. Research needs in subsurface science. National Academy Press, Washington, D.C.
- 59.Neumann, A., A. Seibert, T. Trescher, S. Reinhardt, G. Wohlfarth, and G. Diekert. 2002. Tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans: substrate specificity of the native enzyme and its corrinoid cofactor Arch. Microbiol. 177:420-426. [DOI] [PubMed] [Google Scholar]
- 60.O'Connell, S. P., R. M. Lehman, O. L. Snoeyenbos-West, V. D. Winston, D. E. Cummings, M. E. Watwood, and F. S. Colwell. 2003. Detection of Euryarchaeota and Crenarchaeota in an oxic basalt aquifer. FEMS Microbiol. Ecol. 44:165-173. [DOI] [PubMed] [Google Scholar]
- 61.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 62.Pulliam-Holoman, T. R., M. A. Elberson, L. A. Cutter, H. D. May, and K. R. Sowers. 1998. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl. Environ. Microbiol. 64:3359-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reysenbach, A.-L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson, R. E., V. K. Bhupathiraju, D. L. Song, T. A. Goulet, and L. Alvarez-Cohen. 2002. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ. Sci. Technol. 36:2652-2662. [DOI] [PubMed] [Google Scholar]
- 65.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 66.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 67.Scholz-Muramatsu, H., A. Neumann, M. Meßmer, E. Moore, and G. Diekert. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163:48-56. [Google Scholar]
- 68.Seeliger, S., P. H. Janssen, and B. Schink. 2002. Energetics and kinetics of lactate fermentation to acetate and propionate via methylmalonyl-CoA or acrylyl-CoA. FEMS Microbiol. Lett. 211:65-70. [DOI] [PubMed] [Google Scholar]
- 69.Sharma, P., and P. McCarty. 1996. Isolation and characterization of a facultatively aerobic bacterium that reductively dehalogenates tetrachloroethene to cis-1,2-dichloroethene. Appl. Environ. Microbiol. 62:761-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siebert, A., A. Neumann, T. Schubert, and G. Diekert. 2002. A non-dechlorinating strain of Dehalospirillum multivorans: evidence for a key role of the corrinoid cofactor in the synthesis of an active tetrachloroethene dehalogenase. Arch. Microbiol. 178:443-449. [DOI] [PubMed] [Google Scholar]
- 71.Snoeyenbos-West, O. L., C. G. Van Praagh, D. R. Lovley, H. De Wever, J. R. Cole, M. R. Fettig, D. A. Hogan, and J. M. Tiedje. 2001. Trichlorobacter thiogenes should be renamed as a Geobacter species. Appl. Environ. Microbiol. 67:1020-1021. (Letter to the editor.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song, D. L., M. E. Conrad, K. S. Sorenson, and L. Alvarez-Cohen. 2002. Stable carbon isotope fractionation during enhanced in situ bioremediation of trichloroethene. Environ. Sci. Technol. 36:2262-2268. [DOI] [PubMed] [Google Scholar]
- 73.Sorenson, K. S. 2002. Enhanced bioremediation for treatment of chlorinated solvent residual source areas. ACS Symp. Ser. 837:119-131. [Google Scholar]
- 74.Sorenson, K. S., L. N. Peterson, R. E. Hinchee, and R. L. Ely. 2000. An evaluation of aerobic trichloroethene attenuation using first-order rate estimation. Bioremed. J. 4:337-357. [Google Scholar]
- 75.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sung, Y., K. M. Ritalahti, R. A. Sanford, J. W. Urbance, S. J. Flynn, J. M. Tiedje, and F. E. Löffler. 2003. Characterization of two tetrachloroethene-reducing, acetate-oxidizing anaerobic bacteria and their description as Desulfuromonas michiganensis sp. nov. Appl. Environ. Microbiol. 69:2964-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tandoi, V., T. D. DiStefano, P. A. Bowser, J. M. Gossett, and S. H. Zinder. 1994. Reductive dehalogenation of chlorinated ethenes and halogenated ethanes by a high-rate anaerobic enrichment culture. Environ. Sci. Technol. 23:973-979. [DOI] [PubMed] [Google Scholar]
- 78.Terzenbach, D. P., and M. Blaut. 1994. Transformation of tetrachloroethylene to trichloroethylene by homoacetogenic bacteria. FEMS Microbiol. Lett. 123:213-218. [DOI] [PubMed] [Google Scholar]
- 79.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Traunecker, J., A. Preuß, and G. Diekert. 1991. Isolation and characterization of a methyl chloride utilizing, strictly anaerobic bacterium. Arch. Microbiol. 156:416-421. [Google Scholar]
- 81.Vogel, T. M., and P. L. McCarty. 1985. Biotransformation of tetrachloroethylene to trichloroethylene, dichloroethylene, vinyl chloride, and carbon dioxide under methanogenic conditions. Appl. Environ. Microbiol. 49:1080-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weathers, L. J., G. F. Parkin, and P. J. Alvarez. 1997. Utilization of cathodic hydrogen as electron donor for chloroform cometabolism by a mixed, methanogenic culture. Environ. Sci. Technol. 31:880-885. [Google Scholar]
- 83.Whitman, W. B., T. L. Bowen, and D. R. Boone. May 1999, revision date. The methanogenic bacteria. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, release 3.0. [Online.] Springer-Verlag, New York, N.Y.
- 84.Wild, A. P., W. Winkelbauer, and T. Leisinger. 1995. Anaerobic dechlorination of trichloroethene, tetrachloroethene and 1,2-dichloroethene by an acetogenic mixed culture in a fixed-bed reactor. Biodegradation 6:309-318. [DOI] [PubMed] [Google Scholar]
- 85.Yager, R. M., S. E. Bilotta, C. L. Mann, and E. L. Madsen. 1997. Metabolic adaptation and in situ attenuation of chlorinated ethenes by naturally occurring microorganisms in a fractured dolomite aquifer near Niagara Falls, New York. Environ. Sci. Technol. 31:3138-3147. [Google Scholar]
- 86.Zinder, S. H. 1994. Syntrophic acetate oxidation and “reversible acetogenesis,” p. 387-415. In H. L. Drake (ed.), Acetogenesis. Chapman and Hall, New York, N.Y.