Abstract
A new real-time PCR method is presented that detects and quantifies three tetracycline resistance (Tcr) genes [tet(O), tet(W), and tet(Q)] in mixed microbial communities resident in feedlot lagoon wastewater. Tcr gene real-time TaqMan primer-probe sets were developed and optimized to quantify the Tcr genes present in seven different cattle feedlot lagoons, to validate the method, and to assess whether resistance gene concentrations correlate with free-tetracycline levels in lagoon waters. The method proved to be sensitive across a wide range of gene concentrations and provided consistent and reproducible results from complex lagoon water samples. The log10 of the sum of the three resistance gene concentrations was correlated with free-tetracycline levels (r2 = 0.50, P < 0.001; n = 18), with the geometric means of individual resistance concentrations ranging from 4- to 8.3-fold greater in lagoon samples with above-median tetracycline levels (>1.95 μg/liter by enzyme-linked immunosorbent assay techniques) than in below-median lagoon samples. Of the three Tcr genes tested, tet(W) and tet(Q) were more commonly found in lagoon water samples. Successful development of this real-time PCR assay will permit other studies quantifying Tcr gene numbers in environmental and other samples.
The tetracycline class of antibiotics is used frequently for the treatment and/or prevention of bacterial disease and for growth promotion in the cattle and swine industries (24, 25). For example, since the mid-1990s, more than 106 kg of tetracycline has been used per year in the U.S. livestock industry alone (9). There is growing evidence that such use of antibiotics for therapy, prophylaxis, and animal growth promotion may be resulting in the selection of resistant animal pathogens and commensals in the environment (reviewed by Chopra and Roberts [9] and Wegener [26]) because of the selection of tetracycline-resistant (Tcr) strains in the rumina and intestines of exposed animals. A recent study on integrated livestock-fish farming indicated a significant increase in bacterial resistance to six antimicrobials in water-sediment samples from fish ponds exposed to manure from antibiotic-fed chickens (18). However, little quantitative information is available on the numbers of resistance genes in the environment.
Phenotypic resistance testing of tetracycline and other antibiotics has been performed for years, although these methods tend to detect only resistance in culturable bacterial species and do not directly detect specific genes that confer the resistance. Previous studies have used nonquantitative PCR with Tcr gene-specific primers for the detection of tetracycline efflux genes (2, 5); however, such systems are not optimal for sensitive detection or for precise quantification. Therefore, a quantitative method for tracking resistance genes in environmental samples is needed to help determine whether antibacterial resistance in the environment is truly increasing because of anthropogenic practices.
In this study, a new quantitative real-time PCR assay was developed for the detection and quantification of three specific Tcr genes in cattle feedlot wastewater. This is a potential source of resistance genes because antibiotics are frequently in feedlot operations and cattle feces, urine, and discarded feed readily washed off into wastewater lagoons by natural drainage. Hence, bacteria (and any resistance genes) might be concentrated in such lagoons and thus lagoons represent a composite sample of resistance genes from the larger feedlot environment. Real-time DNA PCR was chosen as the method for quantification because it is sensitive and allows quantification through the use of unambiguous standard curves developed from defined quantities of cloned Tcr plasmids. Further, this method does not require bacterial cultivation, thus reducing the bias due to the detection of Tcr genes only present in culturable species.
Our general research strategy was to target three specific genes for the development of a real-time PCR method for estimating Tcr gene concentrations in feedlot wastewater treatment lagoons (also in the excreta of treated animals) (3, 8, 10). The genes were chosen from the ribosomal protection protein supergroup, which includes tet(M), tet(S), tet(O), tet(W), tet(Q), tet(T), tetB(P), and otr(A) (13, 22). The specific genes selected for this developmental study were tet(O), tet(W), and tet(Q) because they appear to be particularly common in cattle, often being found in the rumina or intestinal tracts of cattle (3, 20). Furthermore, all three genes have been found to be promiscuous in environmental organisms via different transfer mechanisms (7, 9, 12, 14), which makes them suitable candidates for broader monitoring of resistance gene numbers in the environment. To our knowledge, this is the first demonstration of sensitive detection and quantification of Tcr genes by real-time PCR for community DNA derived from cattle feedlot lagoons.
MATERIALS AND METHODS
Design of real-time PCR primer-probe sets.
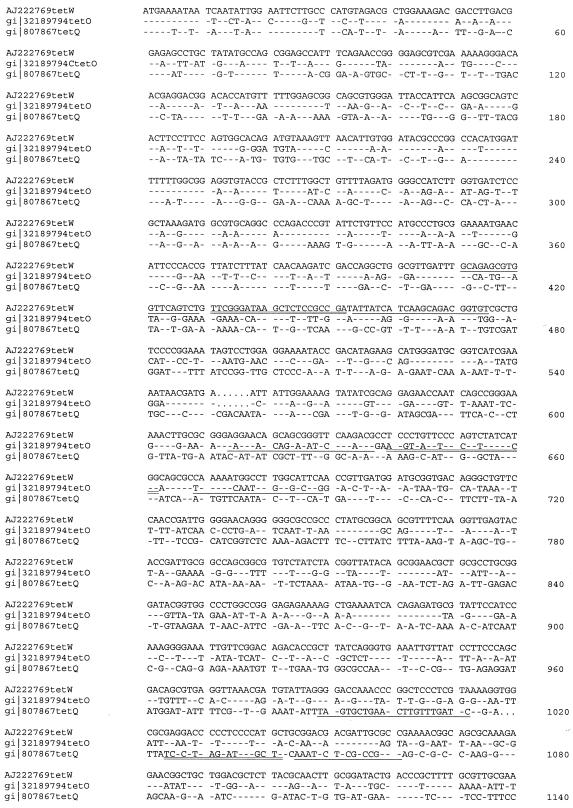
The nucleotide and amino acid coding sequences of tet(O) (GenBank accession no. M188960) (15), tet(Q) (GenBank accession no. X58717) (17), and tet(W) (GenBank accession no. AJ222769) (6) were aligned by the use of ClustalW (23). Regions corresponding to high homology in the N terminus (amino acids 1 to 131) and near the C terminus (amino acids 435 to 520) were excluded from the nucleotide sequences (Fig. 1) for the purpose of designing specific fluorogenic 5′ nuclease (TaqMan) primer-probe sets in regions of lower sequence homology between Tcr gene types. Primers and probe sets were then designed by the use of Primer Express 2.0 software (ABI, Foster City, Calif.). Following verification by BLASTN, which demonstrated that they were within a highly conserved region of each Tcr gene type, primers and probes were synthesized by Invitrogen (Carlsbad, Calif.) and ABI, respectively, with a 5′ FAM label and a 3′ TAMRA quencher for probes.
FIG. 1.
Alignment of the nucleotide sequences of the three Tcr genes examined in this study, tet(W) (GenBank accession no. AJ222769), tet(O) (GenBank accession no. M18896), and tet(Q) (GenBank accession no. X58717). ClustalW was used for the alignment, and nucleotides conserved relative to those of tet(W) are indicated by hyphens while gaps in the alignment are indicated by dots. Primers and TaqMan probes designed for real-time PCR assays are indicated, with the primers singly underlined and the probes doubly underlined. The nucleotide alignment is abbreviated at nucleotide 1140.
Standards.
Plasmids containing tet(O), tet(W), and tet(Q) were transformed into and grown in Escherichia coli (Stratagene XL10-Gold) in Terrific Broth (19) and purified with the High Pure plasmid kit (Roche, Indianapolis, Ind.) to serve as positive controls. Copy numbers were calculated after DNA quantification by UV spectrophotometry, and standards for copy number determinations were prepared by serially diluting purified plasmid DNAs in 30 μg of nuclease-free yeast tRNA (Roche) per ml for copy numbers ranging from 6 × 106 to 6 copies per reaction.
Collection and processing of lagoon water samples.
Water column samples were obtained by permission from seven lagoons located at five different feedlots in the midwestern United States. The study lagoons were selected on the basis of the shape, depth, consistency in water level, and presumed upstream antibiotic use patterns at each feedlot. Specifically, it was desired to have lagoons of roughly the same size and volume while also providing similar site access to allow consistent collection of samples. It was also desired that different feedlot tetracycline use strategies be represented in the study, ranging from semicontinuous subtherapeutic use to therapeutic use only or to no use at all; therefore, the lagoons were ultimately chosen to balance desired physical lagoon characteristics and differing antibiotic use tendencies for the study.
All field samples were collected with 1.2-m-long, 25-mm-diameter polyvinyl chloride tubes equipped with a check valve at the bottom end (11). Typically, samples were obtained by wading to a predefined location in each lagoon, slowly inserting the sampler through the water column, and then carefully withdrawing the samples, avoiding stirring up sediments during sample collection. Duplicate 500-ml samples were collected per lagoon per sample day. Samples were stored in presterilized amber bottles that were then stored in the dark on ice until return to the laboratory. Upon return, 2 ml of collected lagoon water was pelleted immediately at 20,000 × g for 10 min and subsequently stored at −20°C prior to DNA extraction. Total DNA was extracted from the pellets (<40 mg of pellets for 2-ml samples) with the QIAGEN DNA Stool Mini-Kit (QIAGEN, Inc., Valencia, Calif.), which resulted in a 200-μl DNA solution.
The remainder of the collected lagoon water samples were segregated for other analyses and stored at 4°C prior to the quantification of free tetracycline by enzyme-linked immunosorbent assay (ELISA) techniques, total suspended solids and volatile suspended solids (VSS) to quantify lagoon solids in each sample, and anaerobic plate counts to provide parallel phenotypic data on the specific samples analyzed by real-time PCR. Anaerobic plate counts were used for this comparison because all of the lagoons were functionally anaerobic and it was felt that anaerobic plating conditions would best enumerate both anaerobes and facultative anaerobes in the water samples. Total suspended solids and VSS were analyzed in accordance with standard methods (1), whereas ELISAs and plate count assays were performed as described below.
Real-time PCR assays for tet gene copy numbers.
Duplicate real-time PCRs were run with the ABI 7700 sequence detection system and ABI TaqMan Universal PCR Master Mix under reaction conditions of 50°C for 2 min for the uracil-DNA glycosylase step, 95° for 10 min, and 44 two-step cycles consisting of 95°C for 15 s and 60°C for 30 s. Primer concentrations were optimized for each tet gene assay with constant probe concentrations (21). The primer concentrations in the reaction mixtures were 900 nM (for each forward and reverse primer) for tet(O) and tet(W) and 300 nM for tet(Q). Duplicate 25-μl reaction mixtures including 5 μl of extracted DNA (corresponding to 50 μl of original lagoon water) were run in parallel with reaction mixtures of six 10-fold dilutions of the specific tet plasmid standards as described previously.
PCR samples were analyzed in a blinded fashion, without knowledge of the sample source, measured tetracycline level, or phenotypic culture results. Absolute gene copy numbers were initially obtained on the basis of the total DNA that could be extracted from the 2-ml volume of original feedlot water. This 2-ml volume for extraction was based upon preliminary screening of both 2- and 10-ml lagoon sample volumes. However, it was found that extracts from 10-ml sample volumes resulted in suppressed amplification relative to the 2-ml samples (probably due to overloading of the DNA purification columns) and the 2-ml volume was adopted for all samples. To verify that the 2-ml samples did not have background levels of compounds inhibitory of PCR, two types of control experiments were performed: (i) dilution series of selected samples analyzed for quantities of tet(W) DNA and (ii) mixing experiments involving extracted lagoon DNAs with fixed amounts of plasmid DNA containing the Tcr gene. Both tests indicated that no inhibition was prevalent at the 2-ml volume.
Normalization of real-time PCR assays for tet gene copy number.
To compare gene copy numbers between lagoons and with ambient tetracycline levels, the absolute number of resistance genes from each PCR were normalized on the basis of two metrics: per fixed volume of original lagoon water (i.e., gene copy number per 50 μl) and per milligram of volatile solids in each sample (i.e., gene copy number per milligram of VSS). These two measurements provided both actual gene copy concentrations in the lagoon samples and the number of copies per unit of volatile-solid mass (to assess the effect of the level of solids in the lagoon on the observed copy numbers). The statistical significance of differences between observed Tcr gene copy numbers (normalized to the lagoon volume or the mass of VSS) from the below-median and above-median tetracycline lagoons was tested with the nonparametric Wilcoxon rank sum test (4). Nonparametric tests were applied because analyses of the log-transformed data revealed a few of the sample sets (e.g., above- or below-median groups) were not normally distributed.
Phenotypic resistance assays.
Phenotypic resistance assays were performed to assess whether the real-time PCR method provided results similar to those of traditional methods. Typically, 1 ml of lagoon sample was serially diluted (typically to 10−3 to 10−4) and plated in triplicate (50 μl of each) onto Brucella medium amended with 0 to 16 μg of tetracycline per ml in accordance with NCCLS recommendations (16). Plates were incubated under anaerobic conditions (90% N2, 5% CO2, 5% H2; Coy Laboratory Products, Inc., Grass Lake, Mich.) for 7 days prior to CFU enumeration.
ELISAs for free tetracycline.
Free-tetracycline levels were determined with the Ridascreen ELISA tetracycline detection kit (R-Biopharm, Darmstadt, Germany) in accordance with the manufacturer's instructions. Typically, original samples were diluted 1:50 prior to analysis, although samples that had higher organic content (typically related to solids levels) were diluted up to 1:100 to minimize interference. Lagoon samples with known low baseline tetracycline levels were spiked intermittently with tetracycline standards to verify the efficacy of the method.
RESULTS AND DISCUSSION
Probe design: preliminary testing.
Table 1 presents the sequences of the primers and TaqMan probes designed for the three chosen Tcr genes. To verify the specificity of the primer-probe sets, all three target genes (on plasmids) were tested at a high copy number (6 ×106) with the other two primer-probe sets. With an annealing temperature of 60°C, all three sets of primers and probes were found to be highly specific and did not cross-amplify the other two tet gene-containing plasmids (data not shown). Standard curves for each target gene with defined quantities of cloned Tcr plasmids over 6 orders of magnitude were obtained for each experiment, consistently providing significant correlations (r2, always >0.99; data not shown).
TABLE 1.
Sequences of primers and TaqMan probes used for detection of three Tcr genes
| Gene (GenBank accession no.) | Forward primer | Fluorogenic probe | Reverse primer | Gene location (nucleotides of CDSa) |
|---|---|---|---|---|
| tet(O) M18896 | 5′AAGAAAACAGGAGATTCCAAAACG | 5′FAM-ACGTTATTTCCCGTTTATCACGG-Tamra | 5′CGAGTCCCCAGATTGTTTTTAGC | 607-682 |
| tet(Q) X58717 | 5′AGGTGCTGAACCTTGTTTGATTC | 5′FAM-TCGCATCAGCATCCCGCTC-Tamra | 5′GGCCGGACGGAGGATTT | 989-1057 |
| tet(W) AJ222769 | 5′GCAGAGCGTGGTTCAGTCT | 5′FAM-TTCGGGATAAGCTCTCCGCCGA-Tamra | 5′GACACCGTCTGCTTGATGATAAT | 411-476 |
CDS, coding sequence.
Application of real-time PCR method in feedlot lagoons.
To verify the utility of the new primer-probe sets for quantification of Tcr genes in complex lagoon waters, samples were collected from seven cattle feedlot lagoons over a 2-month period. The lagoons reflected different antibiotic use strategies, thus providing a variety of possible resistance gene responses and tetracycline levels. For the sake of performing simple statistical tests, the quantitative resistance gene data were first sorted by water column free-tetracycline levels. Specifically, the median tetracycline level from all samples was determined (i.e., 1.95 μg/liter) and the samples were then recategorized as either above or below the median tetracycline level for statistical comparison of the resistance gene copy levels of the above- versus below-median tetracycline lagoons.
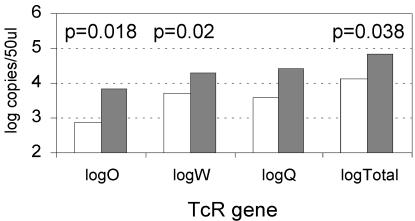
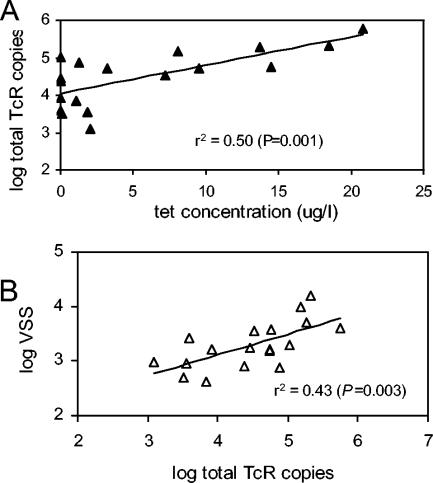
Figure 2 summarizes the observed concentrations (per 50 μl of lagoon water) of the tet(O), tet(Q), and tet(W) genes and the sum of the three genes in all of the samples collected (n = 18; 2 or 3 samples per lagoon). Total resistance gene levels (sum of the three genes) were significantly greater in the above-median tetracycline lagoons versus the below-median tetracycline lagoons (P = 0.038; Wilcoxon rank sum test performed on log-transformed data). Likewise, the levels of two individual resistance genes were also significantly higher in the above-median lagoons than those in the below-median lagoons (Wilcoxon rank sum test on log-transformed data), where P = 0.020 for tet(W), no significant difference for tet(Q), and P = 0.018 for tet(O). These results are consistent with the hypothesis that resistance genes would be positively selected after greater apparent exposure to tetracycline, predicted by the presence of intact tetracycline in the lagoon waters. Figure 3A further shows that total resistance gene concentrations were significantly correlated with free-tetracycline levels (r2 = 0.50, P < 0.001) for all pooled samples, although the data scatter was fairly high.
FIG. 2.
Geometric means of Tcr gene copy numbers were determined by real-time PCR assay of DNAs extracted from feedlot lagoon samples. Levels are shown for lagoons with above- and below-median soluble-tetracycline levels (median free-tetracycline level = 1.95 μg/liter). The mean and median tetracycline levels in the above-median lagoons were 10.8 and 9.5 μg/liter, and those in the below-median lagoons were 0.49 and 0.03 μg/liter, respectively. The genes represented include tet(O), tet(Q), and tet(W), and the total of these three genes is also indicated. Comparisons of above- and below-median numbers of gene copies were performed with the nonparametric Wilcoxon rank sum test.
FIG. 3.
Relationships between the total Tcr gene copy numbers per 50 μl of lagoon water and free-tetracycline levels, as well as the VSS, present in the respective lagoons were tested by linear regression. (A) Relationship between the log of the total Tcr gene copy number and the tetracycline concentration with the linear regression. Points represent two or three time points each for seven lagoons. (B) Relationship between the log of the total Tcr gene copy number and the log of the VSS.
When resistance gene numbers were normalized to the ambient VSS level, a measurement of the total particulate organic materials present in the lagoon waters, the relationship to free-tetracycline levels was considerably weaker (r2 = 0.15; data not shown). The normalization effort was an attempt to account for differing cattle numbers and distances from distinct lagoons; however, the lack of a strong relationship appears to be consistent with the hypothesis that tetracycline-resistant bacteria make up only a small fraction of the total particulate matter in the lagoon waters. However, the total numbers of Tcr copies were higher at higher VSS values: the regression of the log of the total number of Tcr copies against the log of the VSS value had an r2 value of 0.43 and a P value of 0.003 (Fig. 3B).
Of the three individual Tcr genes quantified here, tet(W) was more commonly found in the above-median lagoons (on the basis of the number of gene copies per unit of volume) at a 95% confidence level, as indicated by pairwise statistical testing with the Wilcoxon rank sum test [tet(O) versus tet(W), P = 0.029].
Comparisons of real-time PCR assessments and phenotypic plate assays.
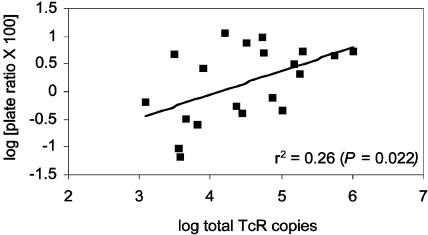
The primary goal of this project was to develop a real-time PCR method for monitoring Tcr genes in the environment; however, it was also important to determine whether the new method provided resistance data similar to those obtained by conventional detection methods. We examined the relationship between log-transformed total resistance gene copy numbers for each lagoon time point and the log of the percentage of colonies on anaerobic plates exposed to 2 μg of tetracycline per ml compared to 0 μg of tetracycline per ml. The two indicators of resistance were significantly correlated (r2 = 0.22, P = 0.022), with a degree of scatter (Fig. 4). Possible reasons for the degree of scatter include (i) the large observed variability in the individual plate count estimates (i.e., the sample size is small relative to the observed variance); (ii) the measurement by the plate count assay of only organisms that could be cultured under anaerobic conditions, producing results that may not reflect all of the organisms present that might bear Tcr genes; and (iii) the quantification of only three Tcr genes in this preliminary study; thus, other, undetected, resistance genes might have been present that were not accounted for.
FIG. 4.
Relationship between the log of the total Tcr gene copy number and the log of the percent reduction in plate colony counts at a tetracycline exposure level of 2 μg/ml relative to those at 0 μg/ml. Points represent 18 independent time points for seven lagoons.
General observations.
The results suggest that the new primer-probe sets for tet(O), tet(Q), and tet(W) effectively quantify these three Tcr genes in lagoon water samples by real-time PCR. The method appears to provide a useful parallel measurement of Tcr to phenotypic plate assays, and it has the advantage of detecting resistance genes in all organisms rather than just resistance in culturable species and is reliable over 6 orders of magnitude. The new method is limited in that it cannot detect genes in specific pathogens without their first being cultured, although more testing should improve correlations between detectable resistance genes and genes borne by specific pathogens.
This new method confirmed a broad relationship between the free-tetracycline level (reflecting upstream tetracycline use) and resistance gene copy concentrations in these cattle feedlot lagoons. Many factors are clearly uncontrollable in studying lagoons, such as varying weather conditions, the scale of the feedlot operation, routine operating practices at each site, the mode of operation of any given lagoon, and ambient levels of solids in lagoons. However, statistically significant differences between resistance gene concentrations in above- and below-median tetracycline lagoons are apparent and clearly warrant further investigation. On the basis of the early results reported here, the sampling program has recently been expanded to increase the database on relationships between downstream Tcr gene levels and feedlot operations. Furthermore, methods for additional Tcr genes are under development to expand the utility of these real-time PCR methods to other environmental and clinical settings.
Acknowledgments
We acknowledge student support from Kansas Biomedical Research Infrastructure Network grant 5P20RR016475-02 and grant P20RR16443 from the COBRE program, both of the National Center for Research Resources; from the NIH (R.K.Y.); and from the Environmental Protection Agency (grant CP-98722801-0 to D.W.G. and J.C.G.). Plasmids tet(O) and tet(W) were kindly provided by Karen Scott (Aberdeen, Scotland), and the tet(Q) plasmid was provided by Nadja Shoemaker (University of Illinois, Urbana). We also thank Diana Aga (State University of New York at Buffalo) for advice on the use of ELISA for tetracycline detection, Val H. Smith (University of Kansas) for helpful discussions, and Dan Nagengast at the Kansas Rural Center for coordination and leadership on this project.
REFERENCES
- 1.American Public Health Association. 1998. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, D.C.
- 2.Aminov, R. I., J. C. Chee-Sanford, N. Garrigues, B. Teferedegne, I. J. Krapac, B. A. White, and R. I. Mackie. 2002. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68:1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Analytical Software. 2003. Statistix 8. User's manual. Analytical Software, Tallahassee, Fla.
- 5.Andersen, S. R., and R. A. Sandaa. 1994. Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl. Environ. Microbiol. 60:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 7.Billington, S. J., J. G. Songer, and B. H. Jost. 2002. Widespread distribution of a Tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 46:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmund, G. K., S. M. Morrison, D. W. Grant, and S. M. Nevins. 1971. Role of excreted chlortetracycline in modifying the decomposition process in feedlot waste. Bull. Environ. Contam. Toxicol. 6:129-132. [DOI] [PubMed] [Google Scholar]
- 11.Graham, W. H., D. W. Grant, F. deNoyelles, Jr., V. H. Smith, C. K. Larive, and E. M. Thurman. 1999. Metolachlor and alachlor breakdown product formation patterns in aquatic field mesocosms. Environ. Sci. Technol. 33:4471-4476. [Google Scholar]
- 12.Krapac, I. G., W. S. Dey, C. A. Smyth, and W. R. Roy. 1998. Impacts of bacteria, metals, and nutrients on groundwater at two hog confinement facilities, p. 29-50. In Proceedings of the National Ground Water Association Animal Feeding Operations and Groundwater: Issues, Impacts, and Solutions—a Conference for the Future. National Groundwater Association, St. Louis, Mo.
- 13.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipsitch, M., R. S. Singer, and B. R. Levin. 2002. Antibiotics in agriculture: when is it time to close the barn door? Proc. Natl. Acad. Sci. USA 99:5752-5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manavathu, E. K., K. Hiratsuka, and D. E. Taylor. 1988. Nucleotide sequence analysis and expression of a tetracycline-resistance gene from Campylobacter jejuni. Gene 62:17-26. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 2004. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard, sixth edition. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 17.Nikolich, M. P., N. B. Shoemaker, and A. A. Salyers. 1992. A Bacteroides tetracycline resistance gene represents a new class of ribosome protection tetracycline resistance. Antimicrob. Agents Chemother. 36:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen, A., J. S. Andersen, T. Kaewmak, T. Somsiri, and A. Dalsgaard. 2002. Impact of integrated fish farming on antimicrobial resistance in a pond environment. Appl. Environ. Microbiol. 68:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, M. S., Y. Niu, Z. Li, I. Adany, D. M. Pinson, Z. Q. Liu, T. Berry, D. Sheffer, F. Jia, and O. Narayan. 2002. Systemic infection and limited replication of SHIV vaccine virus in brains of macaques inoculated intracerebrally with infectious viral DNA. Virology 301:130-135. [DOI] [PubMed] [Google Scholar]
- 22.Taylor, D. E., and A. Chau. 1996. Tetracycline resistance mediated by ribosomal protection. Antimicrob. Agents Chemother. 40:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.U.S. Department of Agriculture. 1996. Swine ’95. II. Reference of 1995 U.S. grower/finisher health and management practices. USDA APHIS VS CEAH. National Animal Health Monitoring System, Fort Collins, Colo.
- 25.U.S. Department of Agriculture. 2000. III. Health management and biosecurity in U.S. feedlots 1999. No. N336.1200. USDA APHIS VS CEAH. National Animal Health Monitoring System, Fort Collins, Colo.
- 26.Wegener, H. C. 2003. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 6:439-445. [DOI] [PubMed] [Google Scholar]