Abstract
Phenotypic characterization of aggregation phenotypes of Lactobacillus coryniformis revealed that strain DSM 20001T coaggregated with Escherichia coli K88, Campylobacter coli, and Campylobacter jejuni but not with other human pathogens. In addition, cells of these pathogens aggregated in the presence of the spent culture supernatant (SCS) of strain DSM 20001T. Cells of E. coli K88 remained viable in the coaggregates and aggregates for up to 24 h. Both coaggregation and aggregation (co/aggregation) occurred at pH 3.5 to 7.5 and was sensitive to heat (85°C for 15 min) and proteinase K. The co/aggregation-promoting factor (Cpf) was purified, and the gene was identified by PCR with degenerate primers derived from internal amino acid sequences. The cpf gene encoded a 19.9-kDa preprotein with a sec-dependent leader and an isoelectric point of 4.4. The amino acid sequence had no significant similarity to proteins with known functions. Northern analysis revealed not only major transcription from the promoter of cpf but also major transcription from the promoter of the preceding insertion element, ISLco1 belonging to the IS3 family. Recombinant Cpf produced in E. coli mediated aggregation of pathogens comparable to the aggregation obtained with purified Cpf or SCS of strain DSM 20001T. Cpf could be removed from cells of strain DSM 20001T by treatment with 5 M LiCl and could be subsequently reattached to the cell surface by using SCS or recombinant Cpf, which resulted in restoration of the co/aggregation property. These results together with those of the amino acid sequence analysis suggest that Cpf is a novel surface protein of L. coryniformis that mediates co/aggregation of some pathogens.
Lactic acid bacteria (LAB) grow in a variety of habitats, such as the mucosa and intestines of humans and animals, as well as fermenting foods and feed (13). Their ability to form multicellular aggregates has been shown to play an important role in colonization of the oral cavity (23) and the urogenital tract (27, 36), as well as in genetic exchange via conjugation (e.g., in Enterococcus faecalis [2] and Lactococcus lactis [11]). The aggregation ability comprises autoaggregation, characterized by clumping of cells of the same strain, and coaggregation, in which genetically distinct cells are involved (22, 38). Both types of aggregation have been described previously for lactobacilli, including Lactobacillus crispatus, Lactobacillus gasseri, and Lactobacillus reuteri (4, 8, 20, 37, 47).
Studies on the mechanism of autoaggregation in lactobacilli showed that proteins present in the culture supernatant (20, 37, 38) and proteins or lipoproteins located on the cell surface (5, 24) are involved in the cell aggregation. Furthermore, it was observed that spent culture supernatants (SCS) of autoaggregating lactobacilli mediate not only the aggregation of cells of the producer strain but also aggregation of other LAB (4, 37) and even Escherichia coli cells (20, 21). Thus, cell aggregation mediated by the SCS of a genetically distinct organism can be considered a third aggregation type (simply referred to as aggregation in this paper). In L. gasseri and Lactobacillus johnsonii the genes which encode an extracellular aggregation-promoting factor (Apf) involved in autoaggregation, as well as aggregation of other LAB (37), have been characterized. The Apf proteins have recently been described as novel surface proteins (48). In L. gasseri the Apf is involved in the maintenance of the cell shape (19). On the other hand, little data are available about the coaggregation of lactobacilli and its mechanism. Intestinal and vaginal lactobacilli have been found to coaggregate with each other (47) or with E. coli (20, 21, 35) and to aggregate E. coli cells in the presence of their SCS (6, 21).
Regarding the importance of the various aggregation types in lactobacilli, it was shown for L. crispatus that its aggregating phenotype enhances the gastrointestinal persistence of the organism in vivo, as well as its adhesion to epithelial cells in vitro (8). Furthermore, the ability of lactic acid bacteria to coaggregate with intestinal pathogens and uropathogenic bacteria in vitro (5, 35) and in vivo (36) is of special interest, as this feature may constitute a protective mechanism against infection (18). Remarkably, the previous descriptions of coaggregation phenomena have been limited to lactobacilli of human or animal origin (5, 37), and so far there are no data about coaggregation of food- or feed-associated bacteria with pathogens.
In this study we investigated the aggregation capability of Lactobacillus coryniformis. Strains of this species are commonly found in agricultural habitats, such as silage, cow dung, dairy barn air (1), and various food products, including cheese (15), salami (40), and Turkish boza (14). Two strains coaggregated with E. coli K88 and Campylobacter spp. Additionally, the SCS of strain DSM 20001T mediated aggregation of these pathogens. We characterized the coaggregation- and aggregation (co/aggregation)-promoting factor (Cpf) of strain DSM 20001T at the genotypic and phenotypic level and observed that Cpf is a novel surface protein of lactobacilli.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains used in this study are listed in Table 1. E. coli and salmonella were grown in Luria-Bertani (LB) medium (39) at 37°C, lactobacilli were grown in MRS medium (Difco Laboratories) at 30°C, listeria was grown in brain heart infusion (Merck) at 37°C, Campylobacter spp. were grown in Preston medium (Oxoid) under microaerophilic conditions (Anaerocult C; Merck) at 42°C, and clostridia were grown in reinforced clostridial medium (Merck) under anaerobic conditions (Anaerocult A; Merck) at 37°C. For protein isolation, L. coryniformis DSM 20001T was grown in MRS medium without Tween 80. When required, ampicillin and kanamycin were added to the culture media at concentrations of 100 and 30 μg/ml, respectively.
TABLE 1.
Bacterial strains used in this study
| Strain | Characteristicsa | Source or referenceb |
|---|---|---|
| Lactobacillus bifermentans DSM 20003T | Isolated from spoiled cheese | DSMZ |
| Lactobacillus coryniformis subsp. coryniformis DSM 20001T | Cpf-positive strain, isolated from silage | DSMZ |
| Lactobacillus coryniformis LTH 1750 | Isolated from sauerkraut | This study |
| Lactobacillus coryniformis subsp. torquens DSM 20004T | Isolated from air, of cow shed | DSMZ |
| Lactobacillus curvatus LTH 684 | Isolated from Hungarian salami | This study |
| Lactobacillus gasseri 4B2 | Apf positive | 37 |
| Lactobacillus johnsonii La1 | Apf positive | 48 |
| Lactobacillus plantarum LTH 1389 | Isolated from fermented sausage | This study |
| Escherichia coli LTH 1577 | ETEC, O149:K88, canine isolate | H. Karch |
| E. coli LTH 4781 | ETEC, O147:H19:K88, porcine isolate | H. Karch |
| E. coli 4140-86 | EAEC, O44:H18, human isolate | RKI |
| E. coli 10281/98 | EIEC, O144; human isolate | RKI |
| E. coli 10282/98 | EIEC, O152:H, human isolate | RKI |
| E. coli 789/01 | EPEC, O125:H6, human isolate | RKI |
| E. coli 9899/00 | EPEC, O157:H45, human isolate | RKI |
| E. coli 117/86 | ETEC, O6:H16, human isolate | RKI |
| E. coli 147/1 | ETEC, O28:H, human isolate | RKI |
| E. coli 97-14606 | EHEC, O30:H, human isolate | RKI |
| E. coli 97-10085 | EHEC, O157:H7, human isolate | RKI |
| E. coli 97-16152 | EHEC, O103:H2, human isolate | RKI |
| E. coli BL21(DE3) | Expression host, T7 DNA polymerase | Novagen |
| Clostridium perfringens LTH 934 | Human isolate | This study |
| C. perfringens LTH 5226 | Spoiled preserve | This study |
| C. perfringens LTH 5228 | Spoiled preserve | This study |
| Listeria monocytogenes LTH 4592 | Serotype 1, minced meat | Matforsk |
| L. monocytogenes LTH 4593 | Serotype 4, minced meat | Matforsk |
| L. monocytogenes LTH 4594 | Serotype 4b, minced meat | Matforsk |
| Salmonella enterica serovar Typhimurium LTH 790 | Food isolate | This study |
| S. enterica serovar Typhimurium LTH 1578 | Food isolate | This study |
| S. enterica serovar Typhimurium LTH 2750 | Food isolate | This study |
| Campylobacter coli DSM 4689T | Penner serovar 4; pig isolate | DSMZ |
| Campylobacter jejuni DSM 4688T | Penner serovar 23, bovine isolate | DSMZ |
ETEC, enterotoxigenic E. coli; EAEC, enteroaggregative E. coli; EIEC, enteroinvasive E. coli; EPEC, enteropathogenic E. coli; EHEC, enterohemorrhagic E. coli.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig; Germany; RKI, Robert Koch Institute, Wernigerode, Germany; Matforsk, Norwegian Food Research Institute, Ås, Norway; H. Karch, Department of Hygiene, University of Münster, Münster, Germany.
Aggregation assays.
Autoaggregation was determined as described by Reniero et al. (37). Briefly, cells of overnight cultures were harvested by centrifugation at 5,000 ×g and 4°C for 20 min, washed three times with sterile distilled water, and suspended in the initial volume of phosphate-buffered saline (PBS) (39). Then 10% (vol/vol) of the culture's own filter-sterilized SCS was added, and the mixture was incubated at 20°C for 2 h. Coaggregation was determined as described by Reid et al. (35). Briefly, cells were prepared as described above and washed twice. Equal volumes of cells of the different strains were mixed and incubated at 20°C for 2 h without agitation. Aggregation of cells in the presence of the purified Cpf of L. coryniformis DSM 20001T or the SCS of another Lactobacillus strain was determined as described by Kmet and Lucchini (21). Briefly, cells were prepared as described above for the autoaggregation assay, mixed with 10% SCS or purified Cpf (added to a final concentration of 5 μg/ml), and incubated at 20°C for 2 h. For quantification of co/aggregation, the optical densities (OD600) of the mixtures described above were monitored during 4 h of incubation at the ambient temperature. The standard deviations derived from the co/aggregation values of three independent experiments did not exceed 10% of the mean value.
For characterization of coaggregation, cell suspensions of lactobacilli in PBS were treated with proteinase K (final concentration, 1 mg/ml) for 30 min or with heat (85°C) for 15 min. The enzyme was then removed by washing the preparation once with PBS. The cells were employed in the coaggregation assay by using both E. coli K88 strains as target strains (Table 1). To characterize aggregation, the SCS of L. coryniformis DSM 20001T was neutralized (pH 7.0), treated with proteinase K (final concentration, 1 mg/ml) at 37°C for 30 min or with heat (85°C) for 15 min and subjected to the aggregation assay with both E. coli K88 strains (Table 1). Proteinase K was inactivated by using Pefabloc SC (Roche Diagnostics). The survival of pathogens which had aggregated or which were included in coaggregates was investigated by determination of viable cell counts on LB agar after vortexing to break apart coaggregates.
Purification and characterization of Cpf.
A 1-liter culture of L. coryniformis DSM 20001T (late log phase) was centrifuged (10,000 ×g, 4°C, 30 min), and the proteins were precipitated from filter-sterilized SCS by addition of solid ammonium sulfate to 70% saturation. The precipitate was collected by centrifugation (10,000 ×g, 4°C, 30 min), dissolved in 10 mM Tris-HCl (pH 6.5), and dialyzed overnight at 4°C by using the same buffer. The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (25) by using a 12% acrylamide gel. Following electrophoresis, the gel was sliced into four pieces containing proteins having molecular masses of >45, 45 to 25, 25 to 15, and <15 kDa. The proteins in each slice were eluted by using a Bio-Rad 422 Electro-Eluter. After dialysis, the aggregation-promoting fraction was applied to a Mono Q HR column (0.5 by 5 cm; Amersham Biosciences) which had been previously equilibrated with 20 mM ethanolamine (pH 8.5). The protein was eluted at a flow rate of 0.5 ml/min with a linear gradient of 0 to 1 M sodium chloride in the same buffer.
The molecular weight of Cpf was determined by gel filtration by using a High Prep 10/30 Superose 200 column (Amersham Biosciences). The column was preequilibrated with 50 mM sodium phosphate containing 150 mM sodium chloride (pH 7.4). Purified Cpf was applied to the column and eluted with the same buffer at a flow rate of 1 ml/min. Isoelectric focusing was performed by the method of O'Farrell (33) by using a precast vertical Novex isoelectric focusing gel for pH 3 to 10 (Invitrogen), which was stained with Coomassie blue R250. The presence of carbohydrate side chains in Cpf was determined by staining the SDS-PAGE gel with periodic acid-Schiff (41) by using 2.5 to 7.5 μg of purified Cpf and a GelCode glycoprotein staining kit (Pierce Perbio Science). For visualization of total protein the gel was stained with GelCode blue stain reagent (Pierce Perbio Science).
Determination of amino acid sequences.
Cpf was digested with endoproteinase Lys-C in a Coomassie blue R250-stained gel and separated on a capillary high-performance liquid chromatography column (0.3 by 150 mm). The N-terminal amino acid sequences of two peptides were determined by automated Edman degradation with a Procise 492A sequencer (Applied Biosystems) equipped with an online model 140C PTH amino acid analyzer.
Genetic techniques.
DNA manipulations and electrotransformation of E. coli were performed by using standard protocols described by Sambrook et al. (39). Plasmid DNA from E. coli was extracted with a QIAprep Miniprep kit (QIAGEN). Chromosomal DNA of lactobacilli was isolated by using a High Pure PCR template preparation kit (Roche Diagnostics). Lysozyme (7,500 U/ml) and mutanolysin (250 U/ml) were used as the lysing enzymes, and the mixture was incubated at 37°C for 60 min. Southern hybridization was performed as described previously (7). DNA was digested with KpnI or HpaII. A 365-bp fragment of cpf was used as the DNA probe. This probe was amplified by PCR (95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 45 s and then 72°C for 2 min) by using primers Vec494G (5′-GCACCTACACCACTACCAGCTGCA-3′) and Vec494J (5′-GCTACGGCTGAAATAA CCTGG-3′) and chromosomal DNA of L. coryniformis DSM 20001T as the template. The probe was labeled by using the Alkaphos Direct labeling reagents (Amersham Biosciences). Hybridization was performed according to the manufacturer's instructions. Stringency washes corresponding to 70% stringency were carried out at 64°C. Detection was performed with the CDP-Star reagent (Amersham Biosciences).
Sequence analysis.
Degenerate primers PrimF494Inosin (5′-ACIGTIACIGTIGCIGCIWSICC-3′) and PrimR494Inosin (5′-TTRTTRTTIARIATIARYTGIGC-3′) were used to PCR amplify an internal region of cpf (95°C for 2 min, followed by 35 cycles of 95°C for 20 s, 45°C for 30 s, and 72°C for 30 s and then 72°C for 2 min). The amplified fragment was inserted into pGEM-T (Promega), and the resulting plasmid was introduced into E. coli JM109. Sequencing was performed with the IRD800-labeled T7 primer by using a SequiTherm EXEL II DNA sequencing kit LC (EPICENTRE) in combination with a LI-COR automated sequencing system (Li-Cor). To determine flanking DNA sequences, the Vectorette system (Sigma Genosys) was used. Amplified fragments were sequenced by using IRD800-labeled primer VecSeq (5′-CATGGGGTTCTCCTG-3′). Homology searches were performed against the GenBank database by using the BLASTX and BLASTP algorithms (http://www.ncbi.nlm.nih.gov/BLAST). The putative cleavage site of the leader sequence of Cpf was determined by using the TMHMM 2.0 program (31).
RNA techniques.
Total RNA of L. coryniformis DSM 20001T grown in MRS medium to an OD600 of 0.8 was isolated by the hot phenol-chloroform procedure described by Obst et al. (32). Northern hybridization analyses were performed as described previously (16), with the following modifications. Blotting was performed with the VacuGene XL vacuum blotting system (Amersham Biosciences). Digoxigenin-labeled probes were generated by PCR (95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 60 s and then 72°C for 3 min) by using primers Vec494G and Vec494J for the cpf-specific probe or primers Transp-F (5′-GCGGAATTTAAGCAGCGATAC-3′) and Transp-R (5′-CCTGGAAAGCATTAAATCAGG-3′) for the ISLco1 gene-specific probe, as well as digoxigenin-labeled dUTP (Roche Diagnostics).
The transcription start site was determined by primer extension analysis by using Moloney murine leukemia virus reverse transcriptase (Amersham Bioscience). The reaction was performed under the conditions for the first-strand reaction by using 6 pM specific IRD800-labeled primer Ex1-seq (5′-TGTTGTTGGTGTCGTTGGTTC-3′).
Expression of Cpf in E. coli.
The pET-30 Xa/LIC system (Novagen) was used according to the manufacturer's instructions. Briefly, primers XaLIC-F (5′-GGTATTGAGGGTCGCGCAACGACACCAGAACCAACG-3′) and XaLIC-R (5′-AGAGGAGAGTTAGAGCCTGCAACAATTAAGTTACAAG-3′) contained vector-compatible overhangs (underlined) and were designed according to the coding sequence of cpf without a leader peptide. Amplification (95°C for 2 min, followed by 37 cycles of 95°C for 45 s, 66°C for 45 s, and 72°C for 90 s and then 72°C for 3 min) was carried out by using Pwo polymerase (Roche Diagnostics). The amplified fragment was introduced into pET-30 Xa/LIC, and the resulting plasmid, pMS110, was transferred into the expression host E. coli BL21(DE3). The recombinant strain was grown in 250 ml of LB broth containing 30 μg of kanamycin per ml under noninducing conditions at 37°C to an OD600 of 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cells were incubated for 1 h. Cytoplasmic proteins were isolated and purified with a His · Bind resin column (Novagen).
Removal of Cpf from the cell surface and its reattachment.
Surface proteins of L. coryniformis DSM 20001T cells were extracted with 5 M LiCl by the method of Lortal et al. (26). One aliquot of the cells was suspended in PBS. To investigate the reattachment of Cpf, another aliquot was suspended in SCS or PBS containing recombinant Cpf (200 μg/ml) and incubated at 20°C for 1 h. After this, the cells were washed once with distilled water to remove residual Cpf. LiCl-treated cells, as well as reattached cells, were subjected to the quantitative coaggregation assay. In the control experiment cells were treated with water at each step. For investigation of heterologous attachment of Cpf, cells of lactobacilli were washed twice with distilled water, resuspended in SCS, and incubated at 20°C for 30 min. After this, the cells were washed once with distilled water and subjected to the quantitative coaggregation assay.
Nucleotide sequence accession number.
The DNA sequence of cpf has been deposited in the EMBL database under accession no. AJ605769.
RESULTS
Characterization of aggregation phenotypes.
Screening of 62 strains of 20 Lactobacillus species revealed that only the SCS of L. coryniformis subsp. coryniformis DSM 20001T and Lactobacillus bifermentans DSM 20003T aggregated cells of E. coli K88 (unpublished data). The potential of these strains to exhibit various aggregation phenotypes (autoaggregation, coaggregation, and aggregation) was investigated by using various aggregation assays. L. coryniformis subsp. torquens DSM 20004T and the food isolate L. coryniformis LTH 1750 were included in this study. In addition, the Apf producers L. gasseri 4B2 and L. johnsonii La1 were used as control strains. Except for L. gasseri 4B2, which has been described as an autoaggregating strain (37), neither the lactobacilli nor the pathogens exhibited autoaggregation. Investigation of coaggregation between lactobacilli and pathogens revealed that cells of L. coryniformis strains DSM 20001T and LTH 1750, as well as L. gasseri 4B2, coaggregate with cells of both strains of E. coli K88, Campylobacter coli, and Campylobacter jejuni. Aggregation of cells of the latter organisms occurred exclusively in the presence of the SCS of L. bifermentans DSM 20003T and L. coryniformis DSM 20001T. No co/aggregation took place with strains of Clostridium perfringens, Salmonella enterica serovar Typhimurium, and Listeria monocytogenes. Figure 1 shows the co/aggregation phenotype mediated by L. coryniformis DSM 20001T. Moreover, the aggregation of lactobacillus cells by the SCS of other lactobacilli was investigated. It was observed that the SCS of L. coryniformis DSM 20001T did not mediate aggregation of cells of L. gasseri 4B2 and L. johnsonii La1 and vice versa. On the other hand, cells of L. gasseri 4B2 aggregated in the presence of the SCS of L. johnsonii La1, but cells of L. johnsonii La1 did not aggregate in the presence of the SCS of L. gasseri 4B2.
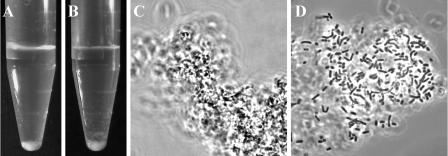
FIG. 1.
Macro- and microscopic manifestation of the co/aggregation property of L. coryniformis DSM 20001T. Coaggregation of cells of strain DSM 20001T with E. coli LTH 1577 resulted in the formation of coaggregates (C) which sedimented to the bottom of reaction tubes (A). Addition of SCS of DSM 20001T to cells of E. coli LTH 1577 led to clumping of the cells (D) into sedimenting aggregates (B).
The co/aggregation property of L. coryniformis DSM 20001T was further investigated by using E. coli K88 as the target. Incubation of the lactobacillus cells and SCS at 85°C for 15 min or with proteinase K abolished co/aggregation, revealing the proteinaceous nature of the Cpf. Remarkably, L. gasseri 4B2 (control) exhibited a heat- and proteinase K-resistant coaggregation phenotype. In addition, the co/aggregation phenotype of L. coryniformis DSM 20001T occurred within a pH range from 3.5 to 7.5, which coincides with the range for the coaggregation of L. gasseri 4B2 with E. coli K88. The E. coli K88 strains were used to investigate the survival of pathogens included in the co/aggregates. It was found that the cell counts of E. coli remained at the same level for up to 24 h (data not shown).
Identification and sequence analysis of cpf.
The Cpf was isolated from the SCS of L. coryniformis DSM 20001T and purified. In SDS-PAGE experiments, the co/aggregation activity was found in the fraction containing proteins with molecular masses of 15 to 25 kDa. After purification by anion-exchange chromatography, Cpf was investigated in more detail. Determination of the molecular mass by gel filtration revealed a single peak at approximately 18 kDa. The isoelectric point was determined to be 4.2, and carbohydrate residues were not detected. Pure Cpf was digested with endoproteinase Lys-C, and two internal amino acid sequences (TVTVAASPFTAK and NAQLILNNK) were obtained. Degenerate primers were deduced from these amino acid sequences and used to amplify an internal DNA fragment of the cpf gene. To determine the complete nucleotide sequence of cpf, flanking DNA regions were sequenced by the genomic walking technique.
Sequence analysis revealed a 2,713-bp DNA fragment containing an insertion sequence designated ISLco1, a complete open reading frame (ORF) (603 bp) located 402 bp downstream, followed by an incomplete 387-bp orfX (Fig. 2). The sequence of ISLco1 contained 37-bp imperfect terminal repeats. A homology search with ISLco1 revealed significant similarity at the nucleotide sequence level to several transposase genes belonging to the IS3 family of insertion elements. ISLco1 contained two out-of-phase and overlapping ORFs, orfA (249 bp) and orfB (952 bp), producing the putative proteins OrfA (83 amino acids) and OrfB (280 amino acids), as well as the transframe protein OrfAB (382 amino acids), which results from a −1 translational frameshift between orfA and orfB. The amino acid sequence of OrfA exhibited 100 and 96.4% identity to the OrfA sequences of ISLc3 of Lactobacillus casei (accession no. AF445084) and IS153 of Lactobacillus sanfransiscensis (accession no. CAB63122), respectively, whereas the corresponding OrfB sequences (accession no. AF445084 and CAB63123) exhibited 100 and 66.2% identity to the OrfB sequence of strain DSM20001T, respectively.
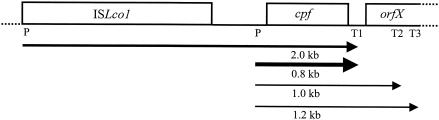
FIG. 2.
Schematic representation of the cpf gene of L. coryniformis DSM 20001T and its flanking regions. The boxes indicate the insertion element ISLco1, the open reading frames of cpf, and incomplete orfX. The localization of promoters (P) and transcriptional terminators (T) is shown. Transcripts of ISLco1 and cpf identified by Northern hybridization analysis are indicated by arrows.
The 603-bp ORF coded for the 19.9-kDa Cpf, which contains 201 amino acids. Analysis of the amino acid sequence revealed that the preprotein is preceded by a signal sequence with the predicted cleavage site AQA-AT between amino acids 27 and 28, resulting in a mature extracellular protein with an isoelectric point of 4.4. The amino acid sequence did not show any significant similarity to proteins with known functions. Low levels of similarity were found to the sequences of putative extracellular proteins of Lactobacillus plantarum WCSF 1 (accession no. NP785062 and NP786668) (27.4 and 35.1% identity) and to the sequences of putatively secreted proteins of Listeria innocua (accession no. NP469937) and L. monocytogenes (accession no. NP464113) (28.9 and 29.8% identity, respectively). Comparison of these sequences with Cpf revealed that the greatest similarity was at the C terminus having the amino acid sequence T(L/I)TW(T/S)L.
The amino acid sequence encoded by orfX located downstream of cpf exhibited 38.5% identity with the sequence of an extracellular protein of L. plantarum WCSF 1 (accession no. NP78666).
Analysis of cpf transcription.
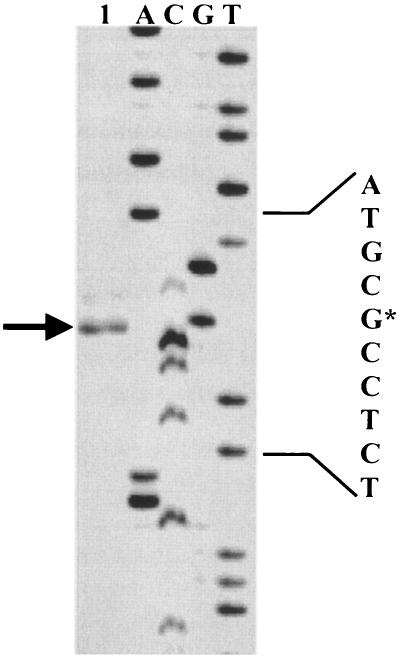
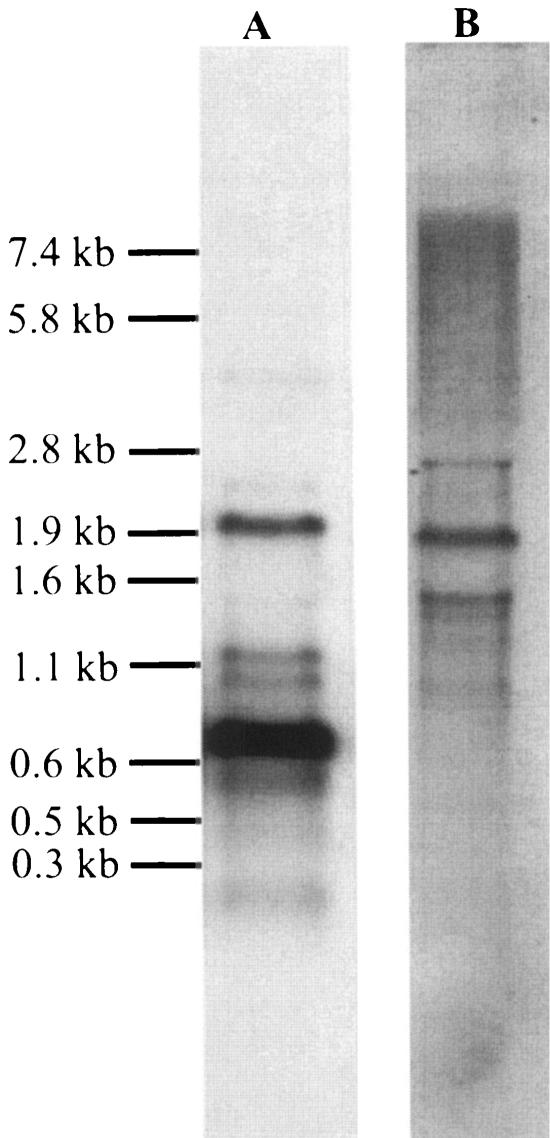
To characterize transcription of cpf, a primer extension reaction and Northern analysis were carried out by using a cpf-specific primer and DNA probe, respectively. As shown in Fig. 3, transcription of cpf started at a G residue located 121 bases upstream of the ATG start codon. Upstream of this transcription start site a typical −10 region (TATAAT) and a −35 region (TTGCAA) were identified in the cpf promoter region and were separated by 17 bp. Northern analysis revealed four transcripts that were approximately 2.0, 1.2, 1.0, and 0.8 kb long (Fig. 4A). The 0.8-kb RNA fragment represented the main transcript, began at the transcription start site determined, and terminated at terminator T1 (−3.5 kcal mol−1), which was located 51 bp downstream of cpf (Fig. 2). The 1.2- and 1.0-kb minor transcripts may have terminated at T2 (−9.2 kcal mol−1) and T3 (−10.9 kcal mol−1) located in the incomplete orfX sequence at 235 and 339 bp, respectively, downstream of cpf. To determine the composition of the 2.0-kb transcript, Northern analysis was repeated with a DNA probe specific for ISLco1 (Fig. 4B). The size of the main transcript was identical, indicating that the 2.0-kb transcript originated from readthrough of orfA/orfB of ISLco1 (Fig. 2). Minor 1.5- and 2.8-kb transcripts might have been caused by readthrough of genes located upstream of ISLco1, as no corresponding hybridization signals were obtained with the cpf-specific DNA probe.
FIG. 3.
Mapping of the 5′ end of mRNA of cpf by primer extension. Transcripts were generated from total RNA isolated from L. coryniformis DSM 20001T (lane 1). The arrow indicates the position of the product obtained; the asterisk indicates the transcription start site.
FIG. 4.
Northern hybridization analysis of RNA isolated from L. coryniformis DSM 20001T. The analysis was performed with a cpf-specific probe (A) and an ISLco1-specific probe (B). The sizes of the marker fragments are indicated on the left.
Distribution of cpf among other co/aggregating lactobacilli.
As the other strains of L. coryniformis, as well as the type strain of L. bifermentans, also showed the coaggregation and/or aggregation phenotype, the presence of cpf-like genes in the genomes of these lactobacilli was investigated by Southern blotting and hybridization. When a 365-bp fragment of cpf was used as the DNA probe, hybridization signals were obtained exclusively with the chromosomal DNA of L. coryniformis DSM 20001T at a stringency of 70% (data not shown).
Characterization of Cpf activity.
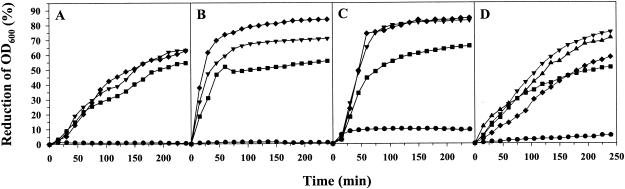
To obtain pure Cpf for further investigations, the corresponding gene was expressed in E. coli BL21(DE3) by using the pET-30 Xa/LIC expression system. Recombinant His-tagged fusion protein was isolated from the cytoplasm and purified with a His · Bind resin column. The His-tagged Cpf was digested with factor Xa to obtain recombinant Cpf without a leader sequence, starting with the amino acid residues AT. The molecular mass of the recombinant Cpf was determined by gel filtration to be 18 kDa and agreed with that of the protein purified from the SCS. The ability of the recombinant Cpf, as well as SCS of L. coryniformis DSM 20001T, to aggregate cells of pathogens was quantitatively determined. For purposes of comparison, quantification of the coaggregation ability of L. coryniformis DSM 20001T was included in the experiment. As shown in Fig. 5A, similar reductions in OD600 were observed for aggregation of E. coli LTH 1577 cells in the presence of recombinant Cpf or SCS and for coaggregation of E. coli LTH 1577 with strain DSM 20001T. When campylobacter cells were used, the co/aggregation resulted in greater and faster reduction of the OD600 compared to that observed with E. coli LTH 1577 cells (Fig. 5B and C). However, the recombinant Cpf showed reduced aggregation activity with the campylobacter cells.
FIG. 5.
Characterization of the Cpf activity of L. coryniformis DSM 20001T by quantification of co/aggregation. (A to C) Aggregation of E. coli LTH 1577 (A), C. coli DSM 4689T (B), and C. jejuni DSM 4688T (C) in the presence of recombinant Cpf (▪) or SCS (▴) of L. coryniformis DSM 20001T, as well as coaggregation of L. coryniformis DSM 20001T (▾) with the pathogens. The controls (•) consisted of cells of the corresponding pathogens only. (D) Coaggregation of E. coli LTH 1577 with cells of L. coryniformis DSM 20001T extracted with 5 M LiCl (•) and subsequently incubated in SCS (▴) or a solution containing recombinant Cpf (▪). Coaggregation of E. coli LTH 1577 with cells of L. curvatus LTH 684 incubated in SCS of strain DSM 20001T (▴) was also determined. The control (▾) consisted of lactobacillus cells that were treated with only water.
The binding of Cpf to the cell surface was investigated in more detail by extracting the protein from the cell surface with a 5 M LiCl solution and then reattaching it to the surface of LiCl-treated cells. As shown in Fig. 5D, treatment of cells with the LiCl solution abolished coaggregation of L. coryniformis DSM 20001T with E. coli LTH 1577, indicating removal of Cpf from the cell surface. The coaggregation phenotype was restored by addition of SCS or recombinant Cpf. Cells treated in parallel experiments with water (control) exhibited coaggregation activity similar to that of untreated cells in overnight cultures of L. coryniformis DSM 20001T (Fig. 5D).
Finally, the possibility of heterologous attachment of Cpf to untreated cell surfaces of four noncoaggregating lactobacilli was investigated. Incubation of cells of Lactobacillus curvatus LTH 684 resulted in acquisition of the coaggregation phenotype, and the activity was comparable to that of L. coryniformis DSM 20001T (Fig. 5D). No attachment of Cpf was observed for L. johnsonii La1, L. coryniformis subsp. torquens DSM 20004T, and L. plantarum LTH 1389.
DISCUSSION
We showed that Cpf of L. coryniformis DSM 20001T not only is a novel surface protein of lactobacilli but also mediates coaggregation with and aggregation among pathogenic bacteria, such as E. coli K88, C. coli, and C. jejuni. Analysis of the amino acid sequence of Cpf revealed no noticeable similarity to known bacterial surface proteins and homology (35.1% identity) only to extracellular proteins with unknown functions. Cpf exhibited features such as an absence of sulfur-containing amino acids and high contents of hydrophobic amino acids (50%) and hydroxy amino acids (28%), which have been described as characteristics of S-layer proteins (3, 44). This finding is consistent with the observation that the overall amino acid compositions of bacterial S-layer proteins are rather similar, although the amino acid sequences exhibit very low levels of similarity (44). The highest levels of similarity are restricted mostly to C-terminal sequences containing cell wall-targeting or sorting signals, surface layer homology domains, or cell membrane anchors (30). Cpf also exhibited the highest levels of similarity to the C termini of the extracellular proteins with unknown functions containing the putative sequence motif T(L/I)TW(T/S)L. However, this motif could not be allotted to any known sorting signal or motif involved in cell wall targeting, and, in addition, no membrane-spanning segments were identified. Moreover, Cpf is a weakly acidic protein with a pI of 4.4, which falls into the range of pIs for most bacterial S-layer proteins (44). However, this pI is unique among the pIs described for S-layer proteins of lactobacilli, which generally have pIs of >9.5. The small size of Cpf (17 kDa) is also uncommon among bacterial S-layer proteins, whose molecular masses usually range from 40 to 200 kDa, except for the 13-kDa S-layer protein of Flexibacter polymorphus (43).
Our studies of removal from and reattachment to the cell surface further supported the assumption that Cpf acts as a surface protein in L. coryniformis DSM 20001T. Cpf was removed from the surface by treatment with 5 M LiCl, a procedure that has been used successfully to isolate S-layer or surface proteins from various lactobacilli (26, 45, 48). Furthermore, we found that large amounts of Cpf were released into the culture medium. This finding is consistent with the observation that several bacteria produce an excess of S-layer protein to ensure complete coverage of the surface during cell growth (3). It has also been reported that isolated S-layer subunits from gram-positive bacteria can be reattached to surfaces of cell wall layers (43). In our study we showed that Cpf can be reattached to the cell surface of L. coryniformis DSM 20001T, resulting in restoration of the coaggregation phenotype, and thus the reattached Cpf regained its native function on the cell surface. Remarkably, coaggregation activity could also be achieved by reattaching recombinant Cpf produced by E. coli (Fig. 5D). This indicated that E. coli produces the recombinant Cpf in a correctly folded conformation. Furthermore, all attempts to inactivate cpf and heterologously express cpf in L. reuteri or L. sakei failed (data not shown), indicating that Cpf may play an important role in cell surface composition, as recently demonstrated for Apf of L. gasseri (19).
Characterization of the aggregation phenotypes revealed that L. coryniformis DSM 20001T exhibits a phenotype which differs from that of L. gasseri 4B2 with respect to aggregation and autoaggregation. Using recombinant Cpf, we obtained strong evidence that the co/aggregation phenotype of strain DSM 20001T is mediated by Cpf. Reniero et al. (37) showed that for L. gasseri 4B2 Apf is involved in autoaggregation, as well as aggregation of LAB cells, and that both events are sensitive to proteinase K. We observed that the coaggregation of strain 4B2 with pathogens is resistant to heat and proteinase K, showing that a nonproteinaceous surface component mediates the coaggregation of L. gasseri 4B2 with pathogens. Remarkably, both L. coryniformis DSM 20001T and L. gasseri 4B2 coaggregated exclusively with E. coli K88, C. coli, and C. jejuni. Therefore, the proteinaceous (Cpf) and nonproteinaceous surface components can specifically interact with a surface structure of these pathogens. For E. coli the K88 antigen was described to be involved not only in coaggregation with intestinal lactobacilli of pigs (46) but also in adhesion to porcine enterocytes and mucus glycoproteins of pigs (10, 28). For both E. coli K88 and C. jejuni, the adhesion to eukaryotic cells was found to be mannose resistant (17, 34).
There is great diversity among the surface components of lactobacilli involved in aggregation phenomena. For example, the DEAD box helicase AggH of L. reuteri 1063 (38), the S-layer protein CbsA of L. crispatus JCM 5810 (42), and the Apf of L. gasseri 4B2 involved in maintenance of the cell shape (19) were found to mediate autoaggregation. Thus, the aggregation property of lactobacilli may be a side activity of surface components involving random interaction with another surface component. This hypothesis is supported by our finding that the phenotypes differ with respect to the different types of aggregation not only among species but also among strains, which is also consistent with the observations of Jankovic et al. (19) and Kmet et al. (20). Furthermore, the results of hybridization with DNAs of other co/aggregating lactobacillus strains indicate that cpf appears to be exclusively located on the chromosome of strain DSM 20001T. Remarkably, the insertion element ISLco1 is located upstream of cpf and triggers additional transcription of cpf (Fig. 4). ISLco1 belongs to the IS3 family, whose members are widely distributed among lactobacilli (9). Involvement of this insertion element in horizontal transfer of cpf, especially the search for homologues in related lactobacilli, requires further investigation.
Cpf is the first example of a surface protein from lactobacilli which mediates co/aggregation with the human pathogens C. coli and C. jejuni, the major species involved in food-borne Campylobacter infections (29). Food-associated lactobacilli that coaggregate numerous pathogens are of special interest with regard to potential applications, and Boris et al. (5) discussed the finding that coaggregation is a mechanism which prevents adherence of pathogens to the host tissue. Although the co/aggregation spectrum of Cpf is rather limited, this molecule is still a promising candidate for application due to its ability to act in a heterologous system. We showed that Cpf attaches to the cell surface of the food-fermenting organism L. curvatus LTH 684, resulting in a coaggregation phenotype similar to that of L. coryniformis DSM 20001T (Fig. 5D). Furthermore, our studies revealed that the co/aggregation activity of Cpf is rather pH independent. Thus, coaggregates or aggregates may be formed in the food matrix and have a protective effect for their passage through the gastrointestinal tract by preventing the entrapped pathogens from adhering to host cells. This mechanism is assumed for the yeast Saccharomyces boulardii, which aggregates E. coli strains expressing type 1 fimbriae (12).
Acknowledgments
We thank R. Reissbrodt and L. Axelsson for providing strains of E. coli and L. monocytogenes, respectively. We are indebted to M. Jostameling for determining the isoelectric point of Cpf. We are grateful to C. Franz for critical reading of the manuscript.
This work was financed in part by Nestec Ltd., Vevey, Switzerland.
REFERENCES
- 1.Abo-Elnaga, I. G., and O. Kandler. 1965. Zur Taxonomie der Gattung Lactobacillus Beijerinck. I. Das Subgenus Streptobacterium Orla Jensen. Zentbl. Bakteriol. Parasitenkd. Infektionskr. Abt. II 119:1-36. [PubMed] [Google Scholar]
- 2.Bensing, B. A., and G. M. Dunny. 1993. Cloning and molecular analysis of genes affecting expression of binding substances, the recipient-encoded receptor(s) mediating mating aggregate formation in Enterococcus faecalis. J. Bacteriol. 175:7421-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boot, H. J., and P. H. Pouwels. 1996. Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol. Microbiol. 21:1117-1123. [DOI] [PubMed] [Google Scholar]
- 4.Boris, S., J. E. Suarez, and C. Barbes. 1997. Characterization of the aggregation promoting factor from Lactobacillus gasseri, a vaginal isolate. J. Appl. Microbiol. 83:413-420. [DOI] [PubMed] [Google Scholar]
- 5.Boris, S., J. E. Suarez, F. Vazquez, and C. Barbes. 1998. Adherence of human lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect. Immun. 66:1985-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bujnakova, D., and V. Kmet. 2002. Aggregation of animal lactobacilli with O157 enterohemorrhagic Escherichia coli. J. Vet. Med. Ser. B 49:152-154. [DOI] [PubMed] [Google Scholar]
- 7.Bunte, C., C. Hertel, and W. P. Hammes. 2000. Monitoring and survival of Lactobacillus paracasei LTH 2579 in food and human intestinal tract. Syst. Appl. Microbiol. 23:260-266. [DOI] [PubMed] [Google Scholar]
- 8.Cesena, C., L. Morelli, M. Alander, T. Siljander, E. Tuomola, S. Salminen, T. Mattila-Sandholm, T. Vilpponen-Salmela, and A. von Wright. 2001. Lactobacillus crispatus and its nonaggregating mutant in human colonization trails. J. Dairy Sci. 84:1001-1010. [DOI] [PubMed] [Google Scholar]
- 9.Ehrmann, M. A., and R. F. Vogel. 2001. Characterization of IS153, an IS3-family insertion sequence isolated from Lactobacillus sanfranciscensis and its use for strain differentiation. Syst. Appl. Microbiol. 24:443-450. [DOI] [PubMed] [Google Scholar]
- 10.Gaastra, W., and F. K. de Graaf. 1982. Host-specific fimbrial adhesins of noninvasive enterotoxigenic Escherichia coli strains. Microbiol. Rev. 46:129-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasson, M. J., S. Swindell, S. Maeda, and H. M. Dodd. 1992. Molecular rearrangement of lactose plasmid DNA associated with high-frequency transfer and cell aggregation in Lactococcus lactis 712. Mol. Microbiol. 6:3213-3223. [DOI] [PubMed] [Google Scholar]
- 12.Gedek, B. R. 1999. Adherence of Escherichia coli serogroup O 157 and the Salmonella typhimurium mutant DT 104 to the surface of Saccharomyces boulardii. Mycoses 42:261-264. [DOI] [PubMed] [Google Scholar]
- 13.Hammes, W. P., and C. Hertel. 2003. The genera Lactobacillus and Carnobacterium. .In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.15. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 14.Hancioglu, O., and M. Karapinar. 1997. Microflora of Boza, a traditional fermented Turkish beverage. Int. J. Food Microbiol. 35:271-274. [DOI] [PubMed] [Google Scholar]
- 15.Hegazi, F. Z., and I. G. Abo-Elnaga. 1980. Characters of Lactobacillus coryniformis, isolated from an Iraqi cheese. Zentrbl. Bakteriol. Naturwiss. 135:205-211. [DOI] [PubMed] [Google Scholar]
- 16.Hertel, C., G. Schmidt, M. Fischer, K. Oellers, and W. P. Hammes. 1998. Oxygen-dependent regulation of the expression of the catalase gene katA of Lactobacillus sakei LTH 677. Appl. Environ. Microbiol. 64:1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, L., and D. J. Kopecko. 2000. Interactions of Campylobacter with eukaryotic cells: gut and luminal colonization and mucosal invasion mechanisms, p. 191-216. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. American Society for Microbiology, Washington, D.C.
- 18.Huis in't Veld, J. H., R. Havenaar, and P. Marteau. 1994. Establishing a scientific basis for probiotic R&D. Trends Biotechnol. 12:6-8. [DOI] [PubMed] [Google Scholar]
- 19.Jankovic, I., M. Ventura, V. Meylan, M. Rouvet, M. Elli, and R. Zink. 2003. Contribution of aggregation-promoting factor to maintenance of cell shape in Lactobacillus gasseri 4B2. J. Bacteriol. 185:3288-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kmet, V., M. L. Callegari, V. Bottazzi, and L. Morelli. 1995. Aggregation-promoting factor in pig intestinal Lactobacillus strains. Lett. Appl. Microbiol. 21:351-353. [DOI] [PubMed] [Google Scholar]
- 21.Kmet, V., and F. Lucchini. 1997. Aggregation-promoting factor in human vaginal Lactobacillus strains. FEMS Immunol. Med. 19:111-114. [DOI] [PubMed] [Google Scholar]
- 22.Kolenbrander, P. E. 1988. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Annu. Rev. Microbiol. 42:627-656. [DOI] [PubMed] [Google Scholar]
- 23.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-439. [DOI] [PubMed] [Google Scholar]
- 24.Kos, B., J. Suskovic, S. Vukovic, M. Simpraga, J. Frece, and S. Matosic. 2003. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94:981-987. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lortal, S., J. van Heijenoort, K. Gruber, and U. B. Sleytr. 1992. S-layer of Lactobacillus helveticus ATCC 12046: isolation, chemical characterization and re-formation after extraction with lithium chloride. J. Gen. Microbiol. 138:611-618. [Google Scholar]
- 27.Mastromarino, P., P. Brigidi, S. Macchia, L. Maggi, F. Pirovano, V. Trinchieri, U. Conte, and D. Matteuzzi. 2002. Characterization and selection of vaginal Lactobacillus strains for the preparation of vaginal tablets. J. Appl. Microbiol. 93:884-893. [DOI] [PubMed] [Google Scholar]
- 28.Metcalfe, J. W., K. A. Krogfelt, H. C. Krivan, P. S. Cohen, and D. C. Laux. 1991. Characterization and identification of a porcine small intestine mucus receptor for the K88ab fimbrial adhesin. Infect. Immun. 59:91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nachamkin, I., J. Engberg, and F. Møller Aarestrup. 2000. Diagnoses and antimicrobial susceptibility of Campylobacter species, p. 45-66. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. American Society for Microbiology, Washington, D.C.
- 30.Navarra, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Obst, M., E. R. Meding, R. F. Vogel, and W. P. Hammes. 1995. Two genes encoding the β-galactosidase of Lactobacillus sake. Microbiology 141:3059-3066. [DOI] [PubMed] [Google Scholar]
- 33.O'Farrell, P. H. 1975. High-resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 34.Ofek, I., and E. H. Beachey. 1978. Mannose binding and epithelial cell adherence of Escherichia coli. Infect. Immun. 22:247-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid, G., J. A. McGroarty, R. Angotti, and R. L. Cook. 1988. Lactobacillus inhibitor production against Escherichia coli and coaggregation ability with uropathogens. Can. J. Microbiol. 34:344-351. [DOI] [PubMed] [Google Scholar]
- 36.Reid, G., J. A. McGroarty, P. A. G. Domingue, A. W. Chow, A. W. Bruce, A. Eisen, and J. W. Costerton. 1990. Coaggregation of urogenital bacteria in vitro and in vivo. Curr. Microbiol. 20:47-52. [Google Scholar]
- 37.Reniero, R., P. Cocconcelli, V. Bottazzi, and L. Morelli. 1992. High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J. Gen. Microbiol. 138:763-768. [Google Scholar]
- 38.Roos, S., S. Lindgren, and H. Jonsson. 1999. Autoaggregation of Lactobacillus reuteri is mediated by a putative DEAD-box helicase. Mol. Microbiol. 32:427-436. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Samelis, J., F. Maurogenakis, and J. Metaxopoulos. 1994. Characterisation of lactic acid bacteria isolated from naturally fermented Greek dry salami. Int. J. Food Microbiol. 23:179-196. [DOI] [PubMed] [Google Scholar]
- 41.Segrest, J. P., and R. L. Jackson. 1972. Molecular weight determination of glycoproteins by polyacrylamide gel electrophoresis in sodium dodecyl sulfate. Methods Enzymol. 28:54-63. [Google Scholar]
- 42.Sillanpää, J., B. Martinez, J. Antikainen, T. Toba, N. Kalkkinen, S. Tankka, K. Lounatmaa, L. Keränen, M. Höök, B. Westerlund-Wikström, P. H. Pouwels, and T. K. Korhonen. 2000. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 182:6440-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sleytr, U. B., and P. Messner. 1983. Crystalline surface layers on bacteria. Annu. Rev. Microbiol. 37:311-339. [DOI] [PubMed] [Google Scholar]
- 44.Sleytr, U. B. 1997. Basic and applied S-layer research: an overview. FEMS Microbiol. Rev. 20:5-12. [Google Scholar]
- 45.Smit, E., F. Oling, R. Demel, B. Martinez, and P. H. Pouwles. 2001. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J. Mol. Biol. 305:245-257. [DOI] [PubMed] [Google Scholar]
- 46.Spencer, R. J., and A. Chesson. 1994. The effect of Lactobacillus spp. on the attachment of enterotoxigenic Escherichia coli to isolated porcine enterocytes. J. Appl. Bacteriol. 77:215-220. [DOI] [PubMed] [Google Scholar]
- 47.Vandevoorde, L., H. Christiaens, and W. Verstraete. 1992. Prevalence of coaggregation reactions among chicken lactobacilli. J. Appl. Microbiol. 72:214-219. [DOI] [PubMed] [Google Scholar]
- 48.Ventura, M., I. Jankovic, D. C. Walker, R. D. Pridmore, and R. Zink. 2002. Identification and characterization of novel surface proteins in Lactobacillus johnsonii and Lactobacillus gasseri. Appl. Environ. Microbiol. 68:6172-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]