Abstract
The polyphosphate kinase gene from Pseudomonas aeruginosa was overexpressed in its native host, resulting in the accumulation of 100 times the polyphosphate seen with control strains. Degradation of this polyphosphate was induced by carbon starvation conditions, resulting in phosphate release into the medium. The mechanism of polyphosphate degradation is not clearly understood, but it appears to be associated with glycogen degradation. Upon suspension of the cells in 1 mM uranyl nitrate, nearly all polyphosphate that had accumulated was degraded within 48 h, resulting in the removal of nearly 80% of the uranyl ion and >95% of lesser-concentrated solutions. Electron microscopy, energy-dispersive X-ray spectroscopy, and time-resolved laser-induced fluorescence spectroscopy (TRLFS) suggest that this removal was due to the precipitation of uranyl phosphate at the cell membrane. TRLFS also indicated that uranyl was initially sorbed to the cell as uranyl hydroxide and was then precipitated as uranyl phosphate as phosphate was released from the cell. Lethal doses of radiation did not halt phosphate secretion from polyphosphate-filled cells under carbon starvation conditions.
Biological methods for the removal of heavy metals and actinides have recently been investigated due to their cost effectiveness at moderate metal concentrations. Many strains have been isolated that sorb (14, 23, 51, 59, 61), reduce (10, 18, 52, 56), and precipitate (4, 33, 41, 44, 45) metals, usually on the outer membrane of the cell. Sorption is generally achieved in a reversible process and is dependent on the composition of the water being treated, as other chelating agents compete with the complexing moieties on the cell. Thus, the applicability of this method in more complex waste streams or environments may be somewhat suspect due to the relatively low binding affinities of cellular components for metals compared to chelators, such as humic or organic acids. Reduction is generally limited to anaerobic processes and is ineffective against single-oxidation-state metals. Precipitation has the advantage of producing chemically stable forms of metal, and its use is not limited to reducible metals.
With respect to the biological precipitation of metals, both metal sulfides (12, 57, 58, 60) and metal phosphates (4, 33, 45) have been investigated due to their low solubilities. While some work has been done to produce sulfide aerobically (57), its production is generally an anaerobic process, limiting its applicability. The release of phosphate via the hydrolysis of an organic phosphate has been shown to be an effective method for the precipitation of metals on cell membranes. Macaskie and Dean (30) isolated a cadmium-resistant strain of Citrobacter that precipitated numerous metals, such as uranyl ion, as metal phosphates through the use of a membrane-bound acid phosphatase (26, 31, 32). Using glycerol-2-phosphate as a phosphate source, the organism cleaved phosphate from the source in the periplasmic space, leaving it to bind with metals that had been complexed in solution. Complexation was required, as the acid phosphatase was sensitive to the presence of free metal in solution (43). The insoluble metal phosphates precipitated on the surface of the cell.
Polyphosphate, a phosphate polymer with chain lengths of two to a few hundred, has been implicated in metal tolerance and removal in many microorganisms (1, 42, 49). Polyphosphate is reversibly and processively synthesized by polyphosphate kinase (PPK) with the addition of a phosphate from a high-energy phosphoryl donor, such as ATP, and irreversibly and processively hydrolyzed by exopolyphosphatase (PPX) (22). While heavy metal resistance has been reported to be correlated with intracellular polyphosphate levels, work with genetically engineered Escherichia coli indicates that the ability to turn over polyphosphate reserves was more important for metal resistance than simply the ability to accumulate polyphosphate: E. coli harboring multiple copies of the genes for PPK (ppk) and PPX (ppx) was able to tolerate increased concentrations of heavy metals, such as cadmium, whereas E. coli engineered with only ppk (so that it accumulated significant amounts of polyphosphate) had little more tolerance to heavy metals than polyphosphate-free strains (20). Furthermore, when ppk and ppx were overexpressed under the control of independently inducible promoters in an E. coli ppk-ppx deletion mutant, this organism accumulated large amounts of polyphosphate when ppk only was induced and then degraded polyphosphate and released phosphate into the medium when ppx was induced, simulating the release of phosphate seen in the metal-binding Citrobacter sp. We suspected that the phosphate generated by polyphosphate hydrolysis and secreted from the cell precipitated heavy metals on the outside of the cell, thereby reducing their toxicity, and that this mechanism could be used to remove metal and actinides from aqueous waste streams. Due to E. coli's intolerance of uranyl, this engineered organism could not be used to examine polyphosphate metabolism and biological uranyl removal.
Two examples of the use of polyphosphate cycling organisms, which were isolated from wastewater treatment processes, for heavy metal precipitation have been reported in the literature. The use of Acinetobacter johnsonii, isolated from an enhanced biological phosphorus removal (EBPR) reactor, was effective in removing lanthanum from solution (4, 5). In EBPR, biomass is cycled between an aerobic phase, in which phosphate is accumulated and stored as polyphosphate inside cells, and an anaerobic phase, in which phosphate is released from the microorganisms. While Acinetobacter is not believed to be the major phosphate cycling organism in the EBPR process, it does accumulate and degrade polyphosphate in aerobiosis and anaerobiosis, respectively. Approximately 0.5 g (dry cell weight) of Acinetobacter/liter was able to remove up to 0.3 mM lanthanum from solution through polyphosphate cycling. A full EBPR consortium, isolated from a wastewater treatment plant and subsequently enriched for its ability to cycle phosphate, was able to remove over 98% of a 1.5 mM solution of uranyl nitrate, with reported loading of over 0.5 g of uranium/g of dry-cell weight (45).
In this work, a two-stage system is presented for the removal of heavy metals from aqueous streams. First, large amounts of polyphosphates were accumulated in Pseudomonas aeruginosa, which was chosen for its native metal tolerance and apparent ability to accumulate larger amounts of polyphosphates than many other organisms. The metal-binding effects shown with the Citrobacter sp. and the polyphosphate-accumulating organisms from EBPR were then mimicked via the degradation of large amounts of polyphosphate and the concomitant release of phosphate from the cell. Phosphate was therefore available to bind and precipitate uranyl out of solution in a manner similar to that shown with the Citrobacter sp., while doing so in a manner that requires neither an organic phosphate source nor the presence of a chelation system nor, in fact, the existence of a live cell.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa strain HN854 (24) was chosen for its antibiotic susceptibility and ease in engineering. Experiments were conducted in minimal MOPS (morpholinepropanesulfonic acid)-buffered minimal medium (40). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 50 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 5 μg/ml. Excess phosphate, when added, was supplemented to a final concentration of 13.2 mM. Phosphate starvation conditions were achieved by providing 0.132 mM phosphate and growing until stationary phase (to an optical density at a wavelength of 600 nm [OD600] of approximately 0.4).
Molecular biological methods.
The genes encoding PPK and PPX in P. aeruginosa were identified (16, 62); the sequence for the ppk-ppx gene cluster was obtained from GenBank. PCR amplification of ppk from the chromosome was performed with primers PAO1 M.2 (GGAAGCTTTGACCCCCTCGGGAAGATGAATG) and PAO1E.2 (GGAAGCTTGGCTACAGCCTCAACGTGCGGT); homologous regions are underlined. The resulting 2.2-kb PCR product was digested with HindIII and ligated into the HindIII site of the broad-host-range plasmid pMMB206 (37) behind the Ptac-lac promoter to make pNSR20.
Plasmids were transformed into E. coli by using electroporation with a Bio-Rad Gene Pulser electroporator at 1.8 kV. Plasmids were transformed into P. aeruginosa HN854 by triparental mating (46) by using E. coli DH5α as the donor strain, E. coli DH5α pRK2013 (9) as a helper strain, and streptomycin resistance in selection for P. aeruginosa HN854.
The ppk and ppx genes were knocked out of HN854 through a homologous deletion-insertion, thereby creating strain P. aeruginosa NSR1. The region containing both ppk and ppx (3′ of ppk, but on the complementary strand) were PCR amplified with primers PAO1 M.2 and PPXUP3.S (GAGAGGTACCGCAGGATGGCGCAGTTTTC; the homologous region is underlined) and cloned into pUC19. The 2.1-kb region bound by PstI sites was excised and replaced with a tetracycline resistance cassette (8). This replacement removed 1.0 kb of the 2.2-kb ppk gene and 1.1 kb of the 1.3-kb ppx gene, both at the 3′ ends of the genes. The ppk′-ppx′ region was then subcloned into the suicide vector pEX100T (47) and transformed into P. aeruginosa HN854, where it was homologously recombined into the chromosome. The first and second crossover events were selected for with tetracycline resistance and sucrose tolerance, respectively. Presence of the construct in the chromosome was confirmed by PCR.
PCR was performed by using an Extend High Fidelity system (Boehringer Mannheim). Oligonucleotides were purchased from Genemed Synthesis (South San Francisco, Calif.) and QIAGEN Operon (Alameda, Calif.).
Polyphosphate degradation in crude cell lysates.
P. aeruginosa culture samples of 1 ml were taken in stationary phase, harvested by centrifugation at 12,000 × g for 5 min, and resuspended in 50 mM Tris (pH 7.5)-10% sucrose to give a final OD600 of 20. The samples were frozen in liquid nitrogen and stored at −87°C prior to lysis. The samples were thawed on ice, and 50 μl of cells was combined with 50 μl of lysis buffer containing 50 mM Tris (pH 7.5), 10% sucrose, 300 mM NaCl, 90 mM EDTA, and 3 mg of lysozyme/ml. The resulting mixture was incubated on ice for 1 h. Samples were subjected to repeated freeze-thaw cycles, alternating between liquid nitrogen and 37°C, followed by sonication twice at approximately 100 W for 4 s at 4°C with a Branson Sonifier (Branson Ultrasonics Corp., Danbury, Conn.) fitted with a microtip. The samples were then frozen in liquid nitrogen and stored at −87°C prior to the assays.
Radiolabeled polyphosphate was prepared in 50 mM HEPES (pH 7.2), 40 mM (NH4)2SO4, 4 mM MgCl2, 1 mM ATP, 8 mM phosphoenolpyruvate, 2 U of pyruvate kinase/ml, and 15 μCi of γ-labeled ATP/ml. Purified PPK was added to the mixture, and the reaction was run at 37°C for 45 min. Radiolabeled polyphosphate was extracted from the reaction mixture by binding and eluting from Glassmilk (silica glass; Bio 101) (2).
Radiolabeled polyphosphate degradation experiments were performed with 50 mM HEPES (pH 7.2)-40 mM (NH4)2SO4-4 mM MgCl2 supplemented with ATP, ADP, AMP, glucose, glucose-1-phosphate, or potassium phosphate (at 20 mM) or with glycogen (at 20 mg/ml), all at pH 7.2. Approximately 200 μmol of radiolabeled polyphosphate (in phosphate equivalents)/liter and 20 μl of crude cell lysate per ml of reaction mixture were added. The reaction was run at 37°C for 1 h. Samples of the reaction mixtures (2 μl) were spotted onto cellulose-polyethyleneimine thin-layer chromatography plates and developed in a solution of 1 M formic acid and 0.4 M LiCl. Plates were imaged with a PhosphorImager (Molecular Dynamics). The intensities of the polyphosphate spots (at the origins) were recorded by using IPLab Gel software, and pseudo-first order polyphosphate decay constants were estimated from these data.
Controlled polyphosphate accumulation and degradation and uranyl-binding experiments.
Cells were grown under phosphate starvation conditions in minimal medium for 24 h. At the end of the 24-h period, excess phosphate and IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) were added, and cells were incubated for an additional 24 h. At the end of the incubation period, cells were centrifuged at 10,000 × g for 10 min, washed with a 0.5 M NaCl solution, and then centrifuged again. In polyphosphate-degradation experiments, cells were suspended in 40 mM MOPS (pH 7.2). In metal-binding experiments, cells were suspended in a 1 mM uranyl nitrate solution at pH 4, below the solubility limit of U(VI) (50, 13).
Assays of uranyl, polyphosphate, and phosphate.
Uranyl concentration was assayed by using the Arsenazo III reagent (11). A sample of 20 μl was added to 50 μl of Arsenazo reagent (60 mg/ml) and 450 μl of 0.5 M HCl. The absorbance at a wavelength of 652 nm was recorded. Concentrations were calculated by comparing to a standard curve.
Polyphosphate was measured using one of two assays. In the absence of heavy metals, polyphosphate was measured by binding polyphosphate to Glassmilk (2). In the presence of uranyl, this assay does not work, due to the interference of uranyl with the binding of polyphosphate to Glassmilk. In this case, polyphosphates were precipitated selectively from cell lysates (48), hydrolyzed with 0.1 M HCl at 95°C for 1 h, and analyzed for phosphate.
Phosphate concentration was measured with a Calcium/Phosphorus Measurement Kit (Sigma). A fixed-volume sample was added to 500 μl of aluminum molybdate reagent. The absorbance at a wavelength of 340 nm was measured and compared to a set of similar-volume standards.
Electron microscopy and energy-dispersive X-ray spectroscopy (EDXS).
One-milliliter samples were fixed in a 2% glutaraldehyde solution at 4°C for 12 h and then with a 1% osmium tetroxide solution for 2 h. Samples were dehydrated with a series of 5-min acetone washes of increasing concentrations. Samples were infiltrated with an epoxy resin and left to harden overnight. Samples were sectioned by using an MT-6000 microtome, placed on 100 mesh carbon-coated copper grids, and analyzed by a JEOL transmission electron microscope.
Energy-dispersive X-ray spectroscopy was performed by using a JEOL 200CX microscope with two Kevex EDX detectors coupled to a computer running Emispec analytical software. Elemental analysis was performed by the comparison of uranium m-peaks at 3.16 keV and phosphorus k-peaks at 2.01 keV. Lack of microscope drift during the EDX scan was confirmed after the scan by reacquiring the image and observing the location of the path of material eliminated by the scan.
Time-resolved laser-induced fluorescence spectroscopy (TRLFS).
To excite U(VI), a pulsed Nd-YAG laser (GCR-3; Spectra Physics) operating at a wavelength of 355 nm was used. The wavelength of 355 nm is achieved by third harmonic generation of the 1,064-nm emission of the Nd-YAG laser with two KD*P crystals. The laser was operated at a pulse energy of about 1 mJ. Laser energy was monitored by a calibrated energy meter (1812C; Newport) placed in the reflex of the laser beam from a quartz plate placed in the beam at a 45° angle to the beam axis. The laser beam then passes through a rectangular quartz cuvette with a 3-ml volume containing the samples. The fluorescence emission, perpendicular to the laser beam axis, is focused by a lens system onto the entrance slit of a spectrograph (Spectra Pro 500i; Acton Research). The spectrograph provides three gratings. For our experiments, a grating with 300 lines per mm and a spectral resolution of 0.2 nm was used. The fluorescence emission was detected by an intensified gated charge-coupled device camera system (PI-Max; Princeton Instruments). A combined camera controller and a pulser and timing generator (ST-133; Princeton Instruments) can set a variable time delay and time gate, respectively, to the laser pulse for the measurements. A delay time of 2 μs after the lamp trigger signal of the laser was found to be optimal to discriminate light scattering. The gate width was set to 1 μs. One hundred spectra were accumulated for every measurement. All functions of the spectrograph, controller, pulser, timing generator, and charge-coupled device camera, as well as the data collection, were controlled with a personal computer by using the program Winspec 2.4 (Princeton Instruments).
Prior to analysis by TRLFS, biomass samples were centrifuged, washed in 50 mM potassium nitrate, centrifuged again, and suspended in 50 mM potassium nitrate to remove any remaining uranyl from solution. The final pH after the wash and suspension is roughly 4.5. Therefore, all comparisons were made to solutions which were also at pH 4.5.
Phosphate release in irradiated cells.
Cells were grown as described above to accumulate polyphosphate. Cells were then washed, suspended in 50 mM MOPS (pH 7.4), and exposed to 35 Gy/h from Co-60 (1.2 and 1.1 MeV gamma rays) source while being shaken. The dose rate was measured by using a Fricke dosimeter at the same distance from the Co source as the samples. Control samples were shaken in the same room behind a lead-shielded 2-ft concrete wall. The samples remained at the same temperature, approximately 25°C. Samples were taken intermittently and soluble phosphate was measured.
RESULTS
Polyphosphate accumulation in P. aeruginosa overexpressing ppk.
The ppk gene from P. aeruginosa was placed under control of the tac-lac promoter (Ptac-lac) on the broad-host-range plasmid pMMB206, and the resulting plasmid was transformed into P. aeruginosa HN854 and NSR1. Cells were grown to an OD600 of approximately 0.3 in MOPS minimal medium with various phosphate concentrations. Inducer was then added, and polyphosphate content within the cell was monitored.
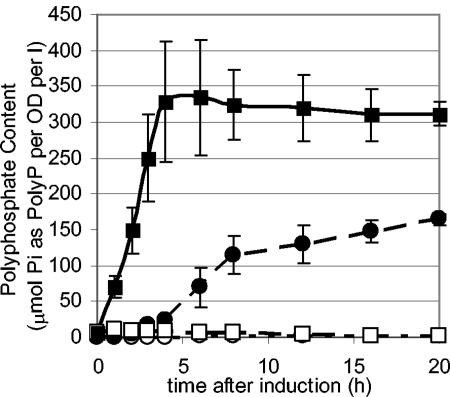
Polyphosphate accumulation in P. aeruginosa HN854 overexpressing ppk was significant, resulting in a 100-fold increase in polyphosphate levels compared to those for wild-type cells (no overexpression of ppk) and a nearly 10,000-fold increase compared to those for the ppk-ppx knockout strain (NSR1) (Fig. 1). Deletion of chromosomal ppk and ppx did not affect the final polyphosphate content of cells overexpressing ppk. P. aeruginosa was able to accumulate approximately three times the polyphosphate compared to a similarly engineered E. coli strain (55). Phosphate starvation of the cells prior to induction resulted in a twofold increase in polyphosphate content over cells that were similarly engineered but not phosphate starved, likely attributed to an increase in active phosphate import into the cell.
FIG. 1.
Polyphosphate accumulation in P. aeruginosa HN854. Cells were grown to an OD of 0.3 and then induced for ppk overexpression. Filled squares: initially phosphate starved, ppk overexpressed. Filled circles: phosphate rich, ppk overexpressed. Empty squares: initially phosphate starved, control (no ppx overexpressed). Empty circles: phosphate rich, control. Cells overexpressing ppk accumulated 100-fold more polyphosphate (PolyP) than control cells. Phosphate starvation conditions prior to induction resulted in a faster accumulation of polyphosphate and a higher end content of polyphosphate. l, liter.
Phosphate release from polyphosphate-packed P. aeruginosa.
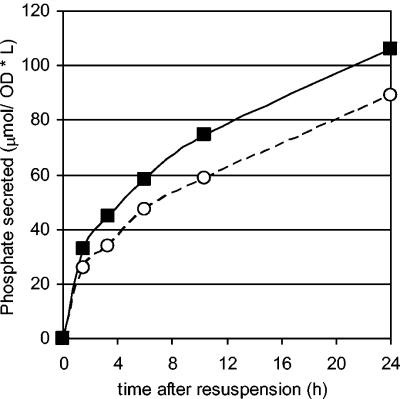
To study the degradation of polyphosphate and the subsequent release of phosphate, P. aeruginosa cells were filled with polyphosphate by growing cultures under phosphate-starvation conditions to an OD600 of 0.3 and subsequently inducing for ppk overexpression along with the addition of excess phosphate to the medium. After 20 h, cultures were washed and suspended in 40 mM MOPS (pH 7.2), and phosphate release from the cell was observed.
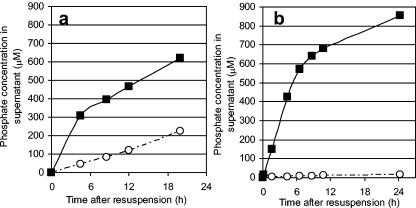
A P. aeruginosa ppk-ppx deletion mutant (NSR1) showed that polyphosphate degradation was independent of exopolyphosphatase (encoded by ppx) and was induced via carbon starvation (Fig. 2). In the mutant, the addition of any carbon source to the medium halted all polyphosphate degradation, while the parent strain (HN854) maintained a slow release in the presence of a carbon source, attributed to the native activity of PPX.
FIG. 2.
Effect of carbon on phosphate release from (a) P. aeruginosa HN854 and (b) the P. aeruginosa ppk-ppx deletion mutant (NSR1). Cells were filled with polyphosphate and resuspended in 40 mM MOPS (pH 7.2) with and without a carbon source (glycerol). Filled squares: glycerol fed. Open circles: no carbon addition. Phosphate release from cells was significantly faster for cells that were carbon starved.
Polyphosphate degradation activity.
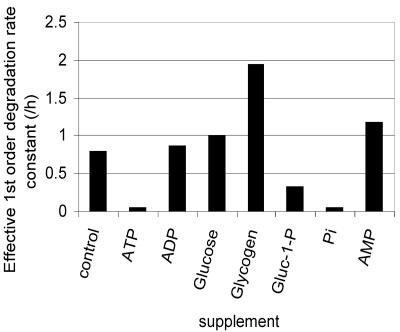
In vivo polyphosphate results indicated that PPX is not responsible for the majority of the polyphosphate degradation activity. To determine the nature of the polyphosphate degradation activity, the activity of crude cell lysates on radiolabeled polyphosphate was investigated (Fig. 3). Various metabolites (ATP, ADP, AMP, glucose, glucose-1-phosphate, potassium phosphate, and glycogen) were added to this reaction mixture to determine their effect on the polyphosphate degradation rate in vitro. When ATP was added to the reaction mixture, polyphosphate degradation was almost completely inhibited, potentially implicating the reverse activity of PPK in polyphosphate degradation. However, upon addition of ADP to the crude lysate reaction mixture, no enhancement of the polyphosphate degradation rate was observed, indicating that the polyphosphate degradation activity was due neither to the reverse activity of PPK nor to a similar activity that could use ADP as a phosphate acceptor.
FIG. 3.
Polyphosphate degradation in crude cell lysates of HN854. Radiolabeled polyphosphate was added to crude cell lysate, and polyphosphate degradation was monitored after the addition of various supplements. All supplements were added to a final concentration of 20 mM, except for glycogen, which was added to a final concentration of 20 mg/ml. Water was added to the control sample to mimic the dilution effect of the addition of a supplement.
Because polyphosphate degradation appeared to be stimulated by carbon starvation, several glycogen-related intermediates were tested for their effect on polyphosphate degradation in vitro. While the addition of glucose gave no increase in the rate of polyphosphate degradation, the addition of glycogen increased the rate nearly threefold, and the addition of the product of glycogen degradation, glucose-1-phosphate, reduced the rate of degradation by a factor of 2. These results imply that polyphosphate degradation is associated with the conversion of glycogen into glucose-1-phosphate, perhaps by the enzyme glycogen phosphorylase. Repeated attempts to knock out glycogen phosphorylase by the same method used to knock out ppk and ppx were unsuccessful, making it difficult to test this hypothesis in vivo.
Additionally, polyphosphate degradation in vitro was not dependent on the conditions from which the cell lysate was prepared. That is, lysates taken from cells prior to resuspension in carbon-free medium degraded polyphosphate at the same rate as those taken postresuspension, indicating that the production of enzymes required to degrade polyphosphate are not induced by carbon starvation, but rather that they may be allosterically regulated by energy-related molecules in the cell, as is the regulation of E. coli glycogen phosphorylase by AMP (7).
Uranyl precipitation by P. aeruginosa releasing phosphate from polyphosphate.
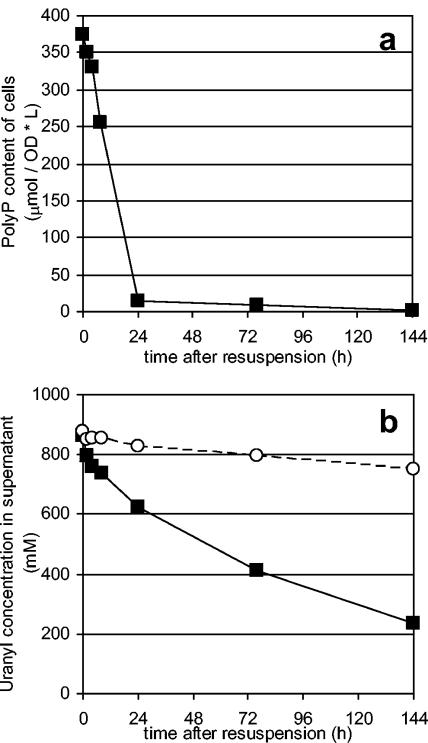
Noting that efficient phosphate release had been achieved simply by suspending the cells into carbon-free medium, uranyl removal from solution via precipitation was attempted. Cells that had been induced for polyphosphate accumulation as before were suspended to an OD600 of 1.5 and challenged with a 1 mM uranyl nitrate solution (Fig. 4). A significant amount of the removed uranyl (20%) was apparently sorbed to the cell. An additional 60% was removed from solution upon the degradation of polyphosphate and subsequent release of phosphate from the polyphosphate-filled cells. The resulting cells accumulated over 40% of their dry cell weight as uranyl. More densely resuspended cultures were able to completely remove uranyl from solution. In order to confirm the presence of the metal on the cell, samples were passed through a 0.22-μm-pore-size filter. While cells are too large to pass through the pores, uranyl phosphate crystals formed in solution are small enough to pass through and remain in solution. In the presence of cells secreting phosphate due to polyphosphate degradation, nearly all of the uranyl remained with the cells, indicating that the uranyl removed from solution was cell associated. In control experiments under identical conditions with no cells but with inorganic phosphate added to the medium, no uranyl was removed from solution by filtration, indicating the need for cells. In addition, in control experiments under identical conditions but with no cells and in solutions around pH 4.5, no uranyl was removed from solution, indicating that uranyl did not precipitate as a hydroxide complex (13, 50). These filtration experiments also highlight the distinct advantages that precipitation in or on the cell membrane has over purely chemical precipitation, as cell attachment allows faster settling over the fine precipitate formed chemically. Furthermore, the resulting metal-loaded biomass is less bulky than precipitates formed with the addition of chemical flocculants traditionally used to aid precipitation and settling.
FIG. 4.
Polyphosphate degradation (a) and concomitant uranyl removal from solution (b). Polyphosphate (PolyP) and control samples were resuspended in 1 mM uranyl nitrate. As a result of polyphosphate degradation and phosphate release, additional uranyl (550 μM) was removed from solution. L, liter.
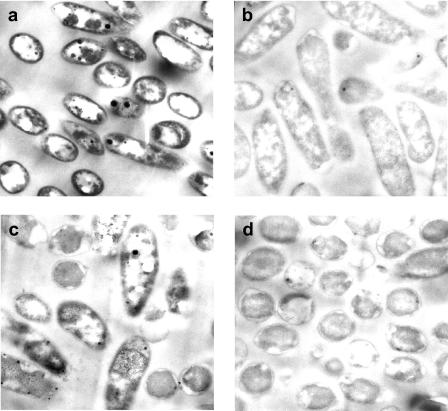
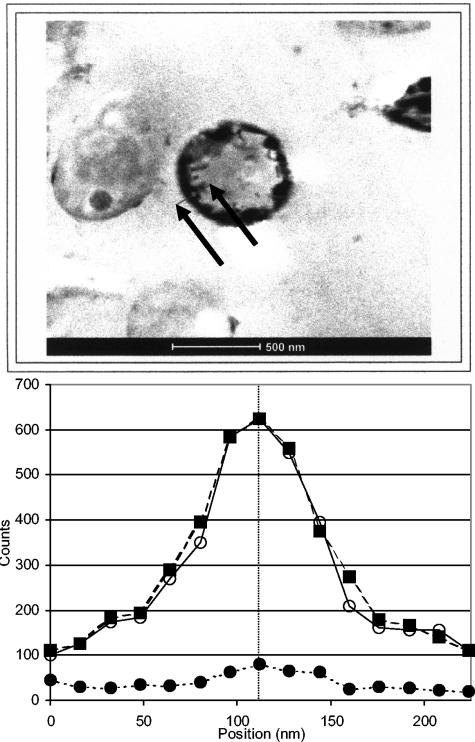
Transmission electron micrographs (Fig. 5) show electron-dense materials surrounding polyphosphate-packed, metal-challenged cells that are not seen in control micrographs. It is assumed that much of the uranyl initially sorbed to the cells had been washed off the cells during the preparation for TEM. More stable precipitates, however, would likely remain on the cells. Energy-dispersive X-ray spectroscopy was used to identify the nature of the precipitate surrounding the cell (Fig. 6). A line scan taken through the membrane region of the cell showed the colocalization of uranium and phosphorus at the cell membrane. Stoichiometric analysis showed that the electron-dense material proved to be composed of a 1:1 stoichiometric ratio of uranyl to phosphorous.
FIG. 5.
Transmission electron micrographs of uranyl-challenged HN854 8 h after resuspension. (a) Polyphosphate-packed, uranyl-challenged cells. (b) Control uranyl-challenged cells. (c) Polyphosphate-packed control cells. (d) Polyphosphate-free control cells. Note the dark, electron-dense membranes in panel A, which are not observed in any of the control samples. The electron-dense regions inside cells as well as the completely electron-free regions are polyphosphate granules.
FIG. 6.
EDXS of metal-associated membrane. The results of a line scan through the cell membrane, as highlighted by the arrows on the micrograph. Closed circles, background. Closed squares, phosphorus. Open circles, uranium. The line on the graph corresponds to the small dot on the line in the micrograph. The phosphorus and uranium are colocated in a 1:1 stoichiometric ratio, suggesting the presence of uranyl phosphate (UO2HPO4).
Uranium in the oxidation state U(VI) dissolved in aqueous solutions forms the uranyl cation (UO22+). The uranyl cation can be excited with UV light, and the relaxation of excited electronic levels of uranyl results in fluorescent light emission. A time- and wavelength-resolved fluorescence spectrum of uranyl is measured by TRLFS. The lifetime of the fluorescence, the wavelength of maximum fluorescence intensity, and the width of fluorescence peaks depend on the chemical environment of the uranyl cation. Therefore, the exact chemical speciation of uranyl complexes can be performed by TRLFS (3, 38). Furthermore, the presence of fluorescent species removes any doubt that the uranium may have been reduced to uranyl(IV) as has been seen with other biological cases (10, 25, 52), since it is the excitation of the axial oxygen bonds that results in the fluorescence.
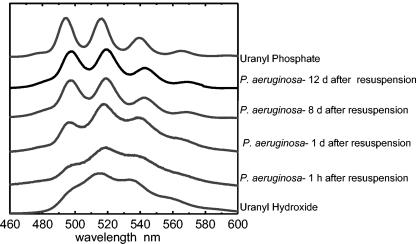
Changes in the inner sphere complexation of the uranyl cation can be easily monitored by changes in the fluorescence spectrum (Fig. 7). At pH 4.5, various hydrolysis products of uranyl [UO2OH+, (UO2)2(OH)22+, and (UO2)3(OH)5+] are dominant (19, 21, 39), having been formed by complexation with hydroxyl anions. Samples from polyphosphate-packed cells shortly after uranyl challenge and polyphosphate-lacking cells challenged with uranyl showed similar spectra. These spectra closely resemble those of uranyl hydroxide species at the relevant pH. This finding implies that upon resuspension in uranyl, hydroxides on the surface of the cell sorb the metal.
FIG. 7.
Time-resolved laser fluorescence spectra of uranyl bound to P. aeruginosa. Polyphosphate-filled cells were resuspended in 1 mM uranyl nitrate, and samples were subsequently taken for TRLFS analysis. Uranyl appears to sorb to cells as a uranyl hydroxide species and then, as phosphate is secreted from the cell, is precipitated to the surface of the cell as uranyl phosphate. d, days.
For uranyl phosphates, changes in spectra can be seen with changes in the atom directly bound to the phosphate. For example, uranyl AMP gives a different spectrum than does uranyl ATP, which gives the same spectrum as inorganic uranyl phosphate (45). As cells remain challenged in a uranyl nitrate solution for longer periods of time, the resulting TRLF spectra shift from being similar to those of the hydroxide species to being similar to that of uranyl phosphate. After 2 days, the spectrum from previously polyphosphate-packed, metal-challenged cells appears nearly identical to a standard of uranyl phosphate at the relevant pH, indicating that the uranyl precipitated on the surface of the cell is uranyl phosphate. The slight broadening of the peaks compared to the uranyl phosphate standard suggests another slight influence, likely that of localization of the precipitate on the cell. This broadening may be the result of the presence of uranyl hydroxide or uranyl bound to surface active sites (e.g., carboxylic acid species).
Since the polyphosphate degradation machinery apparently exists constantly within the cell, a live cell may not be necessary for the release of phosphate and the subsequent removal of metals from solution. Thus, it may be possible to remove radionuclide from solution in situations where the resulting radiation would quickly kill cells. To test this hypothesis, engineered P. aeruginosa was subjected to lethal doses of irradiation from a 60Co source in a buffered medium, and phosphate secretion was monitored (Fig. 8). Little difference in phosphate release between irradiated and nonirradiated cells was observed. It should be noted that since polyphosphate is stable under the levels of radiation used, phosphate release was due solely to enzymatic degradation of internal phosphate stores. This finding indicates that live cells—that is, cells that are actively synthesizing new protein—are not required for the degradation of polyphosphate and subsequent precipitation of metal phosphates, increasing the possible applicability of such a technology.
FIG. 8.
Phosphate secretion in irradiated P. aeruginosa. Cells were filled with polyphosphate and resuspended in MOPS-buffered medium and shaken at 25°C. Closed squares: exposed to 60 Gy/h from a 60Co source. Open circles: nonirradiated samples. Exposure to lethal doses of radiation did not affect phosphate release, implying that the enzymes responsible for the degradation of polyphosphate were present in the cell prior to suspension. L, liter.
DISCUSSION
We have demonstrated here that controlled metabolism of long-chain polyphosphates can be used to remove uranyl from solution and immobilize the uranyl in a relatively insoluble form on the cell wall. When ppk was induced, the cells accumulated a large store of polyphosphate, which was depleted when ppx was induced or when the cells were starved for carbon. Uranyl in solution initially sorbed to the cell, most likely as a hydroxide, and then was transformed to a phosphate complex upon secretion of phosphate from the cell.
Unlike results with E. coli in previous works on engineering the polyphosphate metabolism (53-55), P. aeruginosa is able to survive and metabolize polyphosphate when exposed to relatively high levels of uranyl. Even in the presence of ionizing radiation, the enzymes present in P. aeruginosa were able to metabolize polyphosphate and remove uranyl from solution. This finding may be particularly important for concentrating uranyl in radioactive waste sites; dilute solutions of radioactive uranyl could be concentrated on the outside of polyphosphate-engineered cells or even natural cells whose polyphosphate metabolism could be controlled by changes in environmental conditions.
This report is not the first documentation of uranyl phosphate immobilization on the outside of the cell. It has been shown that metals and actinides can be precipitated on the Citrobacter cell wall by phosphate starving the cells and adding organophosphates and chelating agents (15, 27-29, 31-33, 36). A membrane-bound acid phosphatase hydrolyzes the organophosphate, releasing orthophosphate, which precipitates metal phosphate complexes. Because organophosphates and chelating agents can be expensive to add to wastewater, and the ability to degrade organophosphates in the presence of metal does not appear to be universal, another biological source for phosphate might be more practical; one such source is polyphosphate. While the addition of chemical inducers to uranyl-contaminated wastewater to induce polyphosphate synthesis and degradation in the engineered strain would also be expensive, promoters that respond to oxygen or carbon deprivation may be realistic and economical alternatives to chemically inducible promoters.
Alternatives to genetically modified organisms are organisms that naturally accumulate polyphosphate, such as those found in wastewater treatment plants designed to biologically remove phosphate (34, 35). These organisms synthesize polyphosphate under aerobic conditions and degrade it under anaerobic conditions. Indeed, we have recently shown that wastewater sludge enriched for organisms that accumulate and degrade polyphosphate can remove uranyl from solution (45). Others have shown the ability for a single isolate from EBPR to accumulate metals through phosphate cycling (4, 5). Unfortunately, the specific organism responsible for polyphosphate metabolism in wastewater has neither been isolated nor characterized. While much work has been done in the analysis of Acinetobacter as the polyphosphate-accumulating organism, recent studies have shown that it represents a small proportion of the EBPR consortium. Here, we have shown that a single organism can be engineered to accumulate and degrade polyphosphate and subsequently immobilize uranyl phosphate on the outside of the cell. The use of a genetically engineered microorganism in conjunction with electron microscopy, EDXS, and TRLFS allowed us to examine the mechanism of uranyl association with the cell and subsequent precipitation on the outside of the cell.
Interestingly, polyphosphate degradation was largely independent of ppx expression and dependent on carbon starvation. In vitro enzyme assays using cell lysates indicated that polyphosphate degradation was tied to degradation of glycogen. Unfortunately, the link between polyphosphate and glycogen metabolism could not be confirmed in vivo because of the inability to delete the glycogen phosphorylase gene, indicating that the gene may be essential for cell viability.
Recently, a second polyphosphate kinase (PPK2) in P. aeruginosa and several other bacteria was discovered (17, 63). PPK2 can synthesize polyphosphate by using both ATP and GTP but prefers to use GDP to degrade polyphosphate, whereas PPK1 prefers to use ATP for synthesis and ADP for degradation. In addition, the reverse PPK2-catalyzed reaction rate is 75-fold higher than the forward reaction rate, whereas the reverse PPK1-catalyzed reaction rate is one-fourth of the forward reaction rate. The combination of the in vitro reaction results presented here and the kinetics of the PPK1- and PPK2-catalyzed reactions implicated PPK2 in the in vivo polyphosphate degradation observed here. A possible link between polyphosphate and glycogen metabolisms is alginate biosynthesis. Alginate synthesis is induced during starvation and requires both mannose-1-phosphate and GTP (6). Polyphosphate may supply the GTP (via PPK2) and glycogen may supply the mannose-1-phosphate (e.g., via glycogen phosphorylase to form glucose-1-phosphate, phosphoglucomutase, and phosphoglucose isomerase to form fructose-6-phosphate and via mannose-6-phosphate isomerase and phosphomannomutase to form mannose-1-phosphate). However, neither the regulation of PPK2 activity nor its connection to glycogen degradation and carbon starvation is known.
Regardless of the mechanism of polyphosphate degradation, we have demonstrated that controlled polyphosphate metabolism can be used to sequester large quantities of phosphate in the form of intracellular polyphosphate, degrade the polyphosphate, secrete the resulting phosphate from the cell, and precipitate uranyl phosphate on the cell wall. Furthermore, we demonstrated that uranyl ion initially sorbs to the cell, most likely to hydroxyls on the outside of the cell, and then forms a uranyl phosphate complex when polyphosphate is degraded and phosphate is secreted from the cell. The initial sorption to the cell may be critical for uranyl removal from solution. If there were no sorption to the cell, phosphate secreted from the cell could form a complex with the uranyl in solution rather than on the cell wall. These fine uranyl phosphate complexes do not settle well and are too small to be filtered, making uranyl removal difficult or impossible. Precipitation of uranyl phosphate on the cell wall is an efficient method for uranyl removal from solution, as uranyl phosphate-plated cells are easily separated from the medium by filtration or settling.
Acknowledgments
We thank Hiroshi Nikaido for donation of strains, Chuck Echer for assistance with EDXS, and Cynthia Gong for assistance with the 60Co source.
We thank the NABIR program of the Department of Energy for funding.
REFERENCES
- 1.Aiking, H., A. Stijnman, C. van Garderen, H. van Heerikhuizena, and J. van't Riet. 1984. Inorganic phosphate accumulation and cadmium detoxification in Klebsiella aerogenes NCTC 418 growing in continuous culture. Appl. Environ. Microbiol. 47:374-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ault-Riche, D., C. D. Fraley, C. M. Tzeng, and A. Kornberg. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard, G., G. Geipel, V. Brendler, and H. Nitsche. 1996. Speciation of uranium in seepage waters of a mine tailing pile studied by time-resolved laser-induced fluorescence spectroscopy (TRLFS). Radiochim. Acta 74:87-91. [Google Scholar]
- 4.Boswell, C. D., R. E. Dick, H. Eccles, and L. E. Macaskie. 2001. Phosphate uptake and release by Acinetobacter johnsonii in continuous culture and coupling of phosphate release to heavy metal accumulation. J. Ind. Microbiol. Biotechnol. 26:333-340. [DOI] [PubMed] [Google Scholar]
- 5.Boswell, C. D., R. E. Dick, and L. E. Macaskie. 1999. The effect of heavy metals and other environmental conditions on the anaerobic phosphate metabolism of Acinetobacter johnsonii. Microbiology 145:1711-1720. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarty, A. M. 1998. Nucleoside diphosphate kinase: role in bacterial growth, virulence, cell signalling and polysaccharide synthesis. Mol. Microbiol. 28:875-882. [DOI] [PubMed] [Google Scholar]
- 7.Chen, G. S., and I. H. Segel. 1968. Purification and properties of glycogen phosphorylase from Escherichia coli. Arch. Biochem. Biophys. 127:175-186. [DOI] [PubMed] [Google Scholar]
- 8.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredrickson, J. K., H. M. Kostandarithes, S. W. Li, A. E. Plymale, and M. J. Daly. 2000. Reduction of Fe(III), Cr(VI), U(VI), and Tc(VII) by Deinococcus radiodurans R1. Appl. Environ. Microbiol. 66:2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritz, J. S., and E. C. Bradford. 1958. Detection of thorium and uranium. Anal. Chem. 30:1021-1022. [Google Scholar]
- 12.Glombitza, F. 2001. Treatment of acid lignite mine flooding water by means of microbial sulfate reduction. Waste Manag. 21:197-203. [DOI] [PubMed] [Google Scholar]
- 13.Grenthe, I., J. Fuger, R. J. M. Konings, R. J. Lemire, A. B. Muller, C. Nguyen-Trung, and H. Wanner 1992. Chemical thermodynamics of uranium. OECD Publications, Paris, France.
- 14.Gupta, V. K., A. K. Shrivastava, and N. Jain. 2001. Biosorption of chromium(VI) from aqueous solutions by green algae Spirogyra species. Water Res. 35:4079-4085. [DOI] [PubMed] [Google Scholar]
- 15.Hambling, S. G., L. E. Macaskie, and A. C. R. Dean. 1987. Phosphatase synthesis in a Citrobacter sp. growing in continuous culture. J. Gen. Microbiol. 133:2743-2749. [Google Scholar]
- 16.Ishige, K., A. Kameda, T. Noguchi, and T. Shiba. 1998. The polyphosphate kinase gene of P. aeruginosa. DNA Res. 5:157-162. [DOI] [PubMed] [Google Scholar]
- 17.Ishige, K., H. Zhang, and A. Kornberg. 2002. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. USA 99:16684-16688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kashefi, K., and D. R. Lovley. 2000. Reduction of Fe(III), Mn(IV), and toxic metals at 100°C by Pyrobaculum islandicum. Appl. Environ. Microbiol. 66:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, Y., G. Meinrath, T. Kimura, and Z. Yoshida. 1994. A study of U(Vi) hydrolysis and carbonate complexation by time-resolved laser-induced fluorescence spectroscopy (TRLFS). Radiochim. Acta 64:107-111. [Google Scholar]
- 20.Keasling, J. D., and G. A. Hupf. 1996. Genetic manipulation of polyphosphate metabolism affects cadmium tolerance in Escherichia coli. Appl. Environ. Microbiol. 62:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitamura, A., T. Yamamura, H. Hase, T. Yamamoto, and H. Moriyama. 1998. Measurement of hydrolysis species of U(VI) by time-resolved laser induced fluorescence spectroscopy. Radiochim. Acta 82:147-152. [Google Scholar]
- 22.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 23.Leung, W. C., H. Chua, and W. Lo. 2001. Biosorption of heavy metals by bacteria isolated from activated sludge. Appl. Biochem. Biotechnol. 91-93:171-184. [DOI] [PubMed] [Google Scholar]
- 24.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovley, D. R. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Ind. Microbiol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 26.Macaskie, L. E. 1990. An immobilized cell bioprocess for the removal of heavy metals from aqueous flows. J. Chem. Technol. Biotechnol. 49:357-379. [DOI] [PubMed] [Google Scholar]
- 27.Macaskie, L. E., K. M. Bonthrone, and D. A. Rouch. 1994. Phosphatase-mediated heavy metal accumulation by a Citrobacter sp. and related enterobacteria. FEMS Microbiol. Lett. 121:141-146. [DOI] [PubMed] [Google Scholar]
- 28.Macaskie, L. E., and A. C. R. Dean. 1984. Cadmium accumulation by a Citrobacter sp. J. Gen. Microbiol. 130:53-62. [DOI] [PubMed] [Google Scholar]
- 29.Macaskie, L. E., and A. C. R. Dean. 1984. Cadmium accumulation by immobilized cells of a Citrobacter sp. Environ. Technol. Lett. 5:177-186. [Google Scholar]
- 30.Macaskie, L. E., and A. C. R. Dean. 1982. Cadmium removal by micro-organisms. Environ. Technol. Lett. 3:49-56. [Google Scholar]
- 31.Macaskie, L. E., A. C. R. Dean, A. K. Cheetham, R. J. B. Jakeman, and A. J. Skarnulis. 1987. Cadmium accumulation by a Citrobacter sp.: the chemical nature of the accumulated metal precipitate and its location on the bacterial cells. J. Gen. Microbiol. 133:539-544. [Google Scholar]
- 32.Macaskie, L. E., R. M. Empson, A. K. Cheetham, C. P. Grey, and A. J. Skarnulis. 1992. Uranium bioaccumulation by a Citrobacter sp. as result of enzymatically mediated growth of polycrystalline HUO2PO4. Science 257:782-784. [DOI] [PubMed] [Google Scholar]
- 33.Macaskie, L. E., B. C. Jeong, and M. R. Tolley. 1994. Enzymically accelerated biomineralization of heavy metals: application to the removal of americium and plutonium from aqueous flows. FEMS Microbiol. Rev. 14:351-367. [DOI] [PubMed] [Google Scholar]
- 34.McMahon, K. D., M. A. Dojka, N. R. Pace, D. Jenkins, and J. D. Keasling. 2002. Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal. Appl. Environ. Microbiol. 68:4971-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon, K. D., D. Jenkins, and J. D. Keasling. 2002. Polyphosphate kinase genes from activated sludge carrying out enhanced biological phosphorus removal. Water Sci. Technol. 46:155-162. [PubMed] [Google Scholar]
- 36.Montgomery, D. M., A. C. R. Dean, P. Wiffen, and L. E. Macaskie. 1995. Phosphatase production and activity in Citrobacter freundii and a naturally occurring heavy-metal-accumulating Citrobacter sp. Microbiology 141:2433-2441. [DOI] [PubMed] [Google Scholar]
- 37.Morales, V. M., A. Baekman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 38.Moulin, C., P. Decambox, L. Couston, and D. Pouyat. 1994. Time-resolved laser-induced fluorescence of UO22+ in nitric acid solutions—comparison between nitrogen and tripled Nd-Yag laser. J. Nucl. Sci. Technol. 31:691-699. [Google Scholar]
- 39.Moulin, C., P. Decambox, V. Moulin, and J. G. Decaillon. 1995. Uranium speciation in solution by time-resolved laser-induced fluorescence. Anal. Chem. 67:348-353. [Google Scholar]
- 40.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nies, D. H. 2000. Heavy metal-resistant bacteria as extremophiles: molecular physiology and biotechnological use of Ralstonia sp. CH34. Extremophiles 4:77-82. [DOI] [PubMed] [Google Scholar]
- 42.Pettersson, A., L. Kunst, B. Bergman, and G. M. Roomans. 1985. Accumulation of aluminum by Anabaena cylindrica into polyphosphate granules and cell walls: an X-ray energy-dispersive microanalysis study. J. Gen. Microbiol. 131:2545-2548. [Google Scholar]
- 43.Plummer, E. J., and L. E. Macaskie. 1990. Actinide and lanthanum toxicity towards a Citrobacter sp.: uptake of lanthanum and a strategy for the biological treatment of liquid wastes containing plutonium. Bull. Environ. Contam. Toxicol. 44:173-180. [DOI] [PubMed] [Google Scholar]
- 44.Podda, F., P. Zuddas, A. Minacci, M. Pepi, and F. Baldi. 2000. Heavy metal coprecipitation with hydrozincite [Zn5(CO3)2(OH)6] from mine waters caused by photosynthetic microorganisms. Appl. Environ. Microbiol. 66:5092-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renninger, N., K. D. McMahon, R. Knopp, H. Nitsche, D. S. Clark, and J. D. Keasling. 2001. Uranyl precipitation by biomass from an enhanced biological phosphorus removal reactor. Biodegradation 12:401-410. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 48.Sharfstein, S. T., S. J. Van Dien, and J. D. Keasling. 1996. Modulation of the phosphate-starvation response in E. coli by genetic manipulation of the polyphosphate pathways. Biotechnol. Bioeng. 51:434-438. [DOI] [PubMed] [Google Scholar]
- 49.Sicko-Goad, L., and D. Lazinsky. 1986. Quantitative ultrastructural changes associated with lead-coupled luxury phosphate uptake and polyphosphate utilization. Arch. Environ. Contam. Toxicol. 15:617-627. [Google Scholar]
- 50.Silva, R. J. 1992. Solubility of UO22+ and uranium carbonate complexes, Symp. Proc. 257. Materials Research Society, Warrendale, Pa.
- 51.Travieso, L., R. O. Cañizares, R. Borja, F. Benítez, A. R. Domínguez, R. Dupeyrón, and V. Valiente. 1999. Heavy metal removal by microalgae. Bull. Environ. Contam. Toxicol. 62:144-151. [DOI] [PubMed] [Google Scholar]
- 52.Tucker, M. D., L. L. Barton, and B. M. Thomson. 1998. Reduction of Cr, Mo, Se and U by Desulfovibrio desulfuricans immobilized in polyacrylamide gels. J. Ind. Microbiol. Biotechnol. 20:13-19. [DOI] [PubMed] [Google Scholar]
- 53.Van Dien, S. J., and J. D. Keasling. 1998. Control of polyphosphate metabolism in genetically engineered Escherichia coli. Enz. Microb. Technol. 24:21-25. [Google Scholar]
- 54.Van Dien, S. J., and J. D. Keasling. 1998. Optimization of polyphosphate degradation and phosphate secretion using hybrid metabolic pathways and engineered host strains. Biotechnol. Bioeng. 59:754-761. [DOI] [PubMed] [Google Scholar]
- 55.Van Dien, S. J., S. Keyhani, C. Yang, and J. D. Keasling. 1997. Manipulation of independent synthesis and degradation of polyphosphate in Escherichia coli for investigation of phosphate secretion from the cell. Appl. Environ. Microbiol. 63:1689-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wakatsuki, T. 1995. Metal oxidoreduction by microbial cells. J. Ind. Microbiol. 14:169-177. [DOI] [PubMed] [Google Scholar]
- 57.Wang, C. L., P. D. Maratukulam, A. M. Lum, D. S. Clark, and J. D. Keasling. 2000. Metabolic engineering of an aerobic sulfate reduction pathway and its application to precipitation of cadmium on the cell surface. Appl. Environ. Microbiol. 66:4497-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, C. L., P. C. Michels, S. C. Dawson, S. Kitisakkul, J. A. Baross, J. D. Keasling, and D. S. Clark. 1997. Cadmium removal by a new strain of Pseudomonas aeruginosa in aerobic culture. Appl. Environ. Microbiol. 63:4075-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, T. C., J. C. Weissman, G. Ramesh, R. Varadarajan, and J. R. Benemann. 1998. Heavy metal binding and removal by phormidium. Bull. Environ. Contam. Toxicol. 60:739-744. [DOI] [PubMed] [Google Scholar]
- 60.White, C., A. K. Sharman, and G. M. Gadd. 1998. An integrated microbial process for the bioremediation of soil contaminated with toxic metals. Nat. Biotechnol. 16:572-575. [DOI] [PubMed] [Google Scholar]
- 61.Wong, M. F., H. Chua, W. Lo, C. K. Leung, and P. H. Yu. 2001. Removal and recovery of copper (II) ions by bacterial biosorption. Appl. Biochem. Biotechnol. 91-93:447-457. [DOI] [PubMed] [Google Scholar]
- 62.Zago, A., S. Chugani, and A. M. Chakrabarty. 1999. Cloning and characterization of polyphosphate kinase and exopolyphosphatase genes from Pseudomonas aeruginosa 8830. Appl. Environ. Microbiol. 65:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, H., K. Ishige, and A. Kornberg. 2002. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. USA 99:16678-16683. [DOI] [PMC free article] [PubMed] [Google Scholar]