Abstract
The genus Propionibacterium has a wide range of probiotic activities that are exploited in dairy and fermentation systems such as cheeses, propionic acid, and tetrapyrrole compounds. In order to improve production of tetrapyrrole compounds, we expressed the hemA gene, which encodes δ-aminolevulinic acid (ALA) synthase from Rhodobacter sphaeroides, and the hemB gene, which encodes porphobilinogen (PBG) synthase from Propionibacterium freudenreichii subsp. shermanii IFO12424, either monocistronically or polycistronically in strain IFO12426. The recombinant strains accumulated larger amounts of ALA and PBG, with resultant 28- to 33-fold-higher production of porphyrinogens, such as uroporphyrinogen and coproporphyrinogen, than those observed in strain IFO12426, which harbored the shuttle vector pPK705.
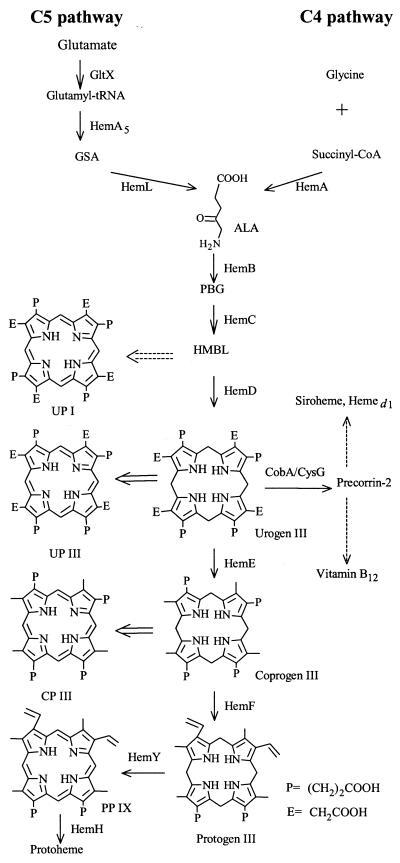
Members of the genus Propionibacterium have been used industrially for the production of cheeses, vitamin B12 (cyanocobalamin), and propionic acid (29). In efforts to clarify details of the biosynthesis of cobalamin and tetrapyrrole derivatives in Propionibacterium, several genes for enzymes involved in these cobalamin biosynthetic pathways have been identified (5, 20, 28). In Propionibacterium, δ-aminolevulinic acid (ALA) is a precursor of tetrapyrrole derivatives and cobalamin and is the first committed compound in the biosynthesis of cobalamin, tetrapyrrole derivatives, and chlorophylls (Fig. 1). ALA is produced via the Shemin pathway (C4 pathway) (19) and the C5 pathway (1). The Shemin pathway has been found in animals, fungi (including yeasts), and α-proteobacteria, such as the photosynthetic genera Rhodobacter and Bradyrhizobium. In the Shemin pathway, ALA is synthesized by the condensation of glycine and succinyl coenzyme A (succinyl-CoA), which is catalyzed by ALA synthase encoded by hemA. The C5 pathway has been found in plants (including algae) and in all other bacteria examined to date. In this pathway, glutamate is coupled with a cognate tRNA in a reaction catalyzed by glutamyl-tRNA synthase and then it is reduced to glutamate-1-semialdehyde (GSA) in a reaction catalyzed by glutamyl-tRNA reductase. Finally, the transamination of GSA, catalyzed by GSA aminomutase, yields ALA. Porphobilinogen (PBG) is formed by the condensation of two molecules of ALA, in a reaction catalyzed by δ-aminolevulinic acid dehydratase (HemB), and the immediate precursor of the tetrapyrrole uroporphyrinogen (urogen) III. Urogen III is the common precursor to all tetrapyrroles, and thus it represents the major branching point in pathways to protoheme, vitamin B12, heme d1, and siroheme (Fig. 1). However, little is known about the regulation of the synthesis of ALA and protoheme in gram-positive bacteria (8). Most information is limited to details of the regulation of the biosynthesis of porphyrins via the C5 pathway in bacteria, such as Bacillus subtilis, Escherichia coli, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium. One major regulatory target in these organisms is the formation of ALA. The rate-limiting nature of the initial step in the pathway was documented convincingly by ALA-feeding experiments, in which the production of porphyrins and the accumulation and excretion of coproporphyrin (CP) III were enhanced (23). The finding of the hemL gene encoding GSA aminomutase in Propionibacterium freudenreichii (20) and the recent genome sequence of P. freudenreichii (Pel et al., Int. Symp. Propionibacteria Bifidobacteria, St. Malo, France, 2 to 4 June, 2004, p. 1) indicate that ALA is synthesized via the C5 pathway in Propionibacterium.
FIG. 1.
Overview of the biosynthesis of tetrapyrroles with gene products indicated. Dashed lines denote multistep pathways, and broad arrows indicate nonenzymic steps.
We developed a host-vector system for propionibacteria using a shuttle vector, pPK705 (12), constructed an expression vector based on pPK705 and endogenous promoters (14), and then successfully expressed the hemA gene encoding ALA synthase from Rhodobacter sphaeroides in Propionibacterium (13). Our success in the overproduction of ALA in P. freudenreichii harboring the hemA gene led us to investigate the production of end products of the metabolism of ALA in recombinant cells of P. freudenreichii subsp. shermanii IFO12426 that harbored a series of expression vectors. We found that exogenous ALA affected the production of porphyrins in P. freudenreichii and ALA that was synthesized de novo in the recombinant P. freudenreichii had a more considerable stimulatory effect on the production of porphyrins than exogenous ALA.
Construction of expression vectors.
The expression vectors pKHEM01 and pKHEM04 (13), including the hemA gene from R. sphaeroides, was constructed previously to overproduce ALA via the Shemin pathway, which is a one-step process for synthesis from glycine. Since HemA from R. sphaeroides shows high affinity of substrates, i.e., glycine and succinyl coenzyme A (2), the hemA gene from R. sphaeroides was used. The DNA fragment that included hemB, which encodes PBG synthase, had been amplified by PCR using 5′-CCATGGCTGATATCTCCATCGATCC-3′ and 5′-CATATGAATCAGGAACGCACCTCGTTG-3′ as the forward and reverse primers, respectively, and pFRB003 (6) as a template DNA. The amplified DNA fragment containing the hemB gene was digested with NcoI and NdeI, and the resultant DNA fragment was inserted into pKHEM01 and pKHEM04 by replacing the hemA gene with the hemB gene. The newly generated plasmids, in which hemB was driven by promoters P138 and P4, were designated pKHEM03 and pKHEM06, respectively. We also subcloned a DNA fragment that included hemB, which had been amplified by PCR, downstream of the hemA gene. The resulting plasmids, pKHEM02 and pKHEM05, contained both hemA and hemB. E. coli DH5α was used as a host to construct various plasmid DNAs and grown in Luria-Bertani broth (26) at 37°C. When necessary, the medium was supplemented with 100-μg/ml ampicillin for E. coli. All expression vectors were introduced into P. freudenreichii subsp. shermanii IFO12426 by electroporation (12). After the pulse, an appropriate portion of the cell suspension recovered was plated on NLB medium, which contained 1% sodium lactate, 1% yeast extract, and 1% Trypticase soy broth, plus 1.5% agar with hygromycin B (250 μg/ml), and incubated anaerobically at 30°C for 4 days. The isolation of plasmid DNA, digestion by restriction endonucleases, and other treatments of DNA fragments and plasmids were performed by standard methods (26).
Expression of the hemA gene from R. sphaeroides and the hemB gene from P. freudenreichii in Propionibacterium.
Crude cell extracts were prepared as described previously (13) with some modifications. The recombinant strains of Propionibacterium were grown anaerobically at 30°C for 48 h in GYT medium, which contained 1% glucose, 1% yeast extract, and 1% Trypticase soy broth. Harvested cells were washed with 50 mM potassium phosphate buffer (pH 7.0), which contains 10 mM β-mercaptoethanol, for assays of ALA synthase activity, or they were washed with 100 mM Bis-Tris-propane-HCl (pH 6.8), which contained 10 mM β-mercaptoethanol, for assays of PBG synthase activity. The cells were disrupted with a Mini-BeadBeater (BioSpec Products, Inc.). After centrifugation of each lysate for 15 min at 8,000 × g, the supernatants were used immediately for measurements of enzymatic activities. The assay of ALA synthase was based on the method described by Lien and Beattie (16). The activity of PBG synthase was determined by a colorimetric assay with PBG and modified Ehrlich's reagent (27). Concentrations of protein were measured by Bradford's method (3) with a commercial kit (Bio-Rad Laboratories), with bovine serum albumin as the standard.
The ALA synthase activity in strain IFO12426 that carried the hemA expression vector pKHEM01, pKHEM02, pHEM04, or pKHEM05 was 2.7- to 4.6-fold higher than that in strain IFO12426 that harbored a control plasmid DNA, pPK705 (Table 1). ALA synthase in the transformed cells without a cloned hemA gene, namely cells transformed with pKHEM03 or pKHEM06, was low; it was almost same level in strain IFO12426 that harbored pPK705.
TABLE 1.
Activities of ALA synthase and PBG synthase and production of ALA and PBG by P. freudenreichii harboring various expression vectorsa
| Plasmid | Genotype | ALA synthase activity (μmol/min/mg of protein)b | Production of ALA (μM)c | PBG synthase activity (μmol/min/mg of protein)b | Production of PBG (μM)c |
|---|---|---|---|---|---|
| pPK705 | 0.26 | 35 | 0.15 | 2 | |
| pKHEM01 | hemA+ | 0.71 | 62 | 0.20 | 12 |
| pKHEM02 | hemA+hemB+ | 0.88 | 99 | 0.45 | 34 |
| pKHEM03 | hemB+ | 0.28 | 27 | 0.63 | 5 |
| pKHEM04 | hemA+ | 1.04 | 160 | 0.18 | 55 |
| pKHEM05 | hemA+hemB+ | 1.19 | 159 | 0.56 | 72 |
| pKHEM06 | hemB+ | 0.26 | 35 | 0.74 | 4 |
Enzymatic activities and products were quantitated in triplicate under the conditions described in the text, and deviators were within 15%.
Cells were grown in GYT medium for 48 h.
Cells were grown in GYT medium for 72 h.
In the strains that carried pPK705, pKHEM01, and pHEM04 without a cloned hemB gene, the specific activity of PBG synthase was low (Table 1). As might be expected, the specific activity of PBG synthase in recombinant propionibacteria that carried plasmid pKHEM02, pKHEM03, pKHEM05, or pHEM06 with a cloned hemB gene was three- to fivefold higher than that in the cells that harbored pPK705. These results suggest that the constructs pKHEM02 and pKHEM05 allowed the overexpression of both genes as a novel operon in P. freudenreichii.
Production of ALA and PBG in recombinant Propionibacterium.
Since levels of ALA and PBG are considered to depend on the activities of ALA synthase and PBG synthase, we determined the levels of ALA and PBG in recombinant propionibacteria that harbored an expression vector for either hemA, hemB, or both. The supernatant from culture of recombinant strains of Propionibacterium that were grown in GYT medium at 30°C for 72 h was used for separation of ALA and PBG on a chromatography system containing two different types of ion-exchange column (ALA/PBG by column test kit; Bio-Rad Laboratories) (18). Amounts of ALA and PBG were quantified on the basis of their reactivity with Ehrlich's reagent according to the instructions from Bio-Rad Laboratories.
In strains that harbored a plasmid with a cloned hemA gene, ALA was produced at a two- to fivefold-higher level than in strains that harbored a plasmid without a cloned hemA gene as a consequence of the overexpression of hemA (Table 1). Although the level of PBG in strains that harbored pKHEM01, pKHEM02, pKHEM04, or pKHEM05 was 6- to 36-fold higher than that in the strain that harbored pPK705, cells harboring pKHEM03 or pKHEM06 produced almost the same amount of PBG as cells that harbored pPK705. The amounts of ALA and PBG were significantly larger in recombinant strains that expressed the hemA gene from R. sphaeroides than those in the cells without a cloned hemA gene. Moreover, while cells carrying a cloned homologous hemB gene had significantly increased HemB activity, levels of PBG were not elevated. These results suggest that the synthesis of ALA might be the rate-limiting step in the biosynthesis of PBG or, at least, an important step in the ALA metabolic pathway. Thus, a low level of PBG in strains harbored a plasmid with a cloned hemB gene is probably due to a low level of ALA because ALA is a substrate of HemB enzyme.
Identification of porphyrins in strains of P. freudenreichii.
When a culture of recombinant IFO12426 cells that harbored a cloned hemA gene was incubated at 30°C for 24 h, the culture and cells became pinkish in color, as did cultures of E. coli cells with a cloned hemA gene (21). Therefore, we examined the production of porphyrins in P. freudenreichii IFO12426.
Porphyrinogens are colorless, but the oxidization of porphyrinogens yields porphyrins, which are photosensitizing moieties. Porphyrin compounds are strong absorbers of light from 400 to 405 nm and from 600 to 650 nm (22).
Analyses of porphyrinogens commonly involve the esterification of porphyrins, multisolvent partitioning, and solid-phase extraction (17). Since such procedures for preparing samples that are oxidation derivatives of corresponding porphyrinogens are time-consuming, we analyzed porphyrinogens indirectly as oxidized porphyrins, such as uroporphyrin (UP), CP, and protoporphyrin (PP), which are fluorescent compounds with distinct spectral properties. Porphyrins in the culture and cells of P. freudenreichii IFO12426 that harbored pPK705 or pKHEM04 with a cloned hemA gene were extracted as described by Cox and Charles (4). In order to examine the fluorescence characteristics of porphyrins, the fluorescence spectra in the 550- to 750-nm range were recorded with an excitation wavelength of 405-nm spectra in a fluorescence spectrophotometer (model F-2000; Hitachi, Tokyo, Japan). We found that more than 98% of the porphyrins produced by P. freudenreichii IFO12426 were present in the culture supernatant. Therefore, we extracted porphyrins from culture supernatants for further examination. Both samples yielded a shoulder and specific peaks at 638 and 653 nm, respectively, which corresponded to UP and CP (data not shown). There were no specific peaks at 648 nm and 660 nm corresponding to PP.
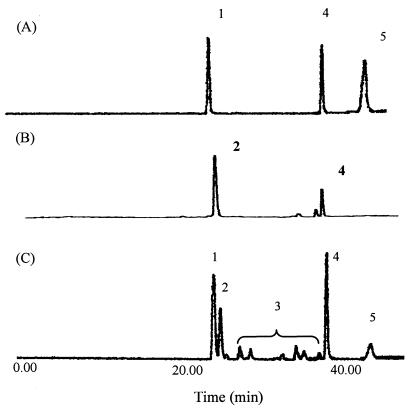
The detailed identification and quantitation of the production of porphyrins were performed with a high-performance liquid chromatography system (LC Module I; Nihon Waters K.K., Tokyo, Japan), equipped with an Intelligent spectrofluorometer (821-FP; JASCO Co., Tokyo, Japan), with a porphyrin chromatographic marker kit, which was purchased from Frontier Scientific, Inc. (Logan, Utah), as a source of standards. Samples were fractionated on a reverse-phase C18 column (4.6-mm inside diameter by 250 mm; 5-μm particle size; TSK-gel ODS-80TM; Tosoh Co., Tokyo, Japan) with a linear gradient from 100% solvent A (10% acetonitrile in 1 M ammonium acetate, pH 5.16) to 100% solvent B (10% acetonitrile in methanol) over the course of 40 min, at a flow rate of 1.0 ml per min. The detection was performed with an excitation wavelength of 405 nm and an emission wavelength of 638 nm. Results were analyzed with a Chromatocorder 21 (System Instruments, Inc., Tokyo, Japan). We found two prominent peaks (Fig. 2B) and three prominent peaks plus some small peaks (Fig. 2C) in the analysis of samples from supernatants of cultures of IFO12426 cells that harbored pPK705 and pKHEM04, respectively. These peaks were identified as UP III (peak 2) and CP III (peak 4) in the sample from cells transformed with pPK705 and as UP I (peak 1), UP III (peak 2) and CP III (peak 4) in the sample from cells transformed with pKHEM04 by HPLC analysis, spectrophotometric analysis, and mass spectrometry (data not shown). Minor peaks that corresponded to derivatives (peak 3) of porphyrin, namely 7-carboxy-, 6-carboxy-, and 5-carboxyporphyrin (25) and PP (Fig. 2, peak 5), were also detected.
FIG. 2.
Profiles after HPLC of porphyrins from (A) a standard mixture of porphyrins, (B) a sample extracted from a culture of P. freudenreichii IFO12426 that harbored pPK705, and (C) a sample extracted from a culture of P. freudenreichii IFO12426 that harbored pKHEM04. Peaks: 1, UP I; 2, UP III; 3, various porphyrin derivatives; 4, CP III; 5, PP. Conditions for HPLC are described in the text.
Biosynthesis of porphyrins with and without exogenous ALA in P. freudenreichii.
We examined the profile of porphyrins in an extract of P. freudenreichii IFO12426 cells that harbored pPK705 at 12-h intervals after inoculation into fresh GYT medium until 120 h. The cells were incubated with a final concentration of glucose of 1%, and cultures were adjusted to pH 7.0 with 30% NaOH daily during time course studies. During the early phase of growth (up to 24 h after inoculation), we found UP III and CP III but not PP. Levels of UP III and CP III increased gradually, reaching maxima of 0.14 and 0.11 mg/liter, respectively, after 96 h and then decreasing slightly (data not shown). UP III was the dominant porphyrin in oxidized samples during the entire incubation. Traces of UP I, PP, and other porphyrin derivatives were also detected in oxidized samples (data not shown).
Since ALA is a key substrate in the production of porphyrins, we examined the effects of exogenous ALA on the production of porphyrins. We added ALA at 0.2 mM to GYT medium 12 h after the start of incubation and examined the porphyrin profile of a culture of P. freudenreichii IFO12426 at 24-h intervals (data not shown). Levels of UP III and CP III increased with time. After 120 h, UP III and CP III had accumulated to approximately 0.24 and 0.32 mg/liter, respectively. Addition of ALA stimulated the production of total porphyrin in P. freudenreichii IFO12426, causing a 2.2-fold increase during the course of incubation. In addition, minor peaks of UP I, PP, and other porphyrin derivatives were detected in oxidized samples by HPLC (data not shown).
Kinetics of the production of porphyrin by recombinant propionibacteria.
We monitored the growth of P. freudenreichii IFO12426 that harbored an expression vector that included the cloned hemA and/or hemB gene and examined the production of UPs and CP III in the culture supernatant of GYT medium at 30°C for 120 h. All cultures reached an optical density (600 nm; path length, 1 cm) of 9.5 to 10.5. The growth curves were very similar irrespective of the nature of each expression vector (data not shown).
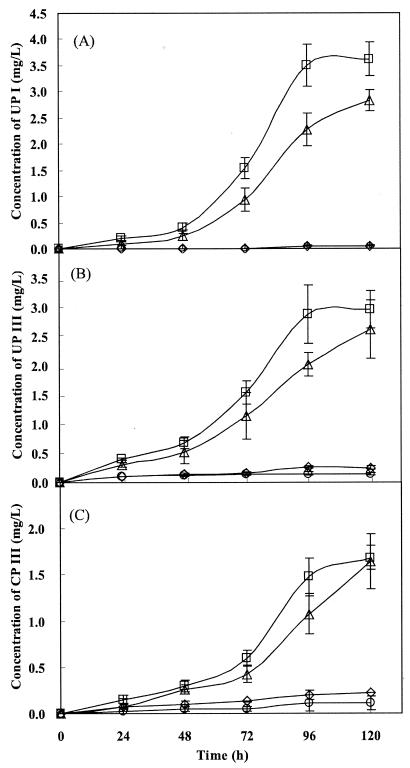
The levels of UPs and CP III in P. freudenreichii IFO12426 that harbored various expression vectors with the biosynthetic gene under the control of the P4 promoter are shown in Fig. 3. In the hemA recombinant strains, increases in the production of UPs and CP III occurred with time. In particular, strain IFO12426, which harbored pKHEM05 that included cloned hemA and hemB genes, produced a maximum level of total porphyrins of 8.3 mg/liter (UP I, 3.6 mg/liter; UP III, 3.0 mg/liter; and CP III, 1.7 mg/liter), as determined at 120 h. Expression of the hemA gene stimulated the synthesis of porphyrin in recombinant strains harboring pKHEM04 and pKHEM05 when biosynthetic genes were under the control of the P4 promoter, with 28- to 33-fold increases, respectively, in levels during the 120-h fermentation. However, levels of porphyrins were low when P. freudenreichii IFO12426 harbored only a plasmid that encoded a cloned hemB gene, despite the overexpression of HemB (Table 1). These results suggest that since ALA is a limiting factor in the production of porphyrins, porphyrins are produced depending on the amount of ALA even if there is overproduction of HemB.
FIG. 3.
Time course of the production of porphyrins by P. freudenreichii IFO 12426 that harbored various expression vectors. (A) Production of UP I; (B) production of UP III; (C) production of CP. Open circles, pPK705; open triangles, pKHEM04; open squares, pKHEM05; open diamonds, pKHEM06. Each point is the mean of three experiments ± standard deviation.
During culture of the recombinant propionibacteria, we confirmed by HPLC that UP and CP were the most abundant porphyrins in the culture broth. In addition, we detected minor or trace amounts of PP. The analysis of oxidized samples revealed high levels of porphyrinogens, which were mainly coprogen and urogens, with minor levels of 7-carboxy-, 6-carboxy-, and 5-carboxyporphyrin. These results indicate that the synthesized ALA was converted mainly into uroporphyrinogens I and III and coproporphyrinogen III.
Urogen III is the precursor to all metabolic tetrapyrroles. The biosynthesis of urogen III from PBG requires the activities of two enzymes, PBG deaminase and urogen III synthase, which are known also as cosynthetase. Four molecules of PBG are condensed by PBG deaminase to yield 1-hydroxymethylbilane (HMBL; also called pre-uroporphyrinogen) (11). HMBL is unstable and is converted to urogen I in the absence of the urogen III synthase at neutral pH. In the presence of cosynthetase, HMBL is rapidly converted into urogen III (9). This reaction involves inversion of the D ring of HMBL and cyclization, with the release of one water molecule. In E. coli, cosynthetase is encoded by the hemD gene (10). The level of UP I was greatly elevated in the recombinant strains of P. freudenreichii that carried a hemA gene. This result suggests that ALA is rapidly converted to HMBL even in the absence of overproduction of PBG deaminase. High levels of HMBL in the recombinant strain, in the presence of insufficient HemD, may be converted at a slightly higher rate to UP I because of the neutral pH of the fermentation culture in this study (Fig. 1 and 3). The propionibacteria that harbored a plasmid with a hemA gene also accumulated elevated levels of UP and CP. This result indicates that porphyrinogens were synthesized at high levels. However, high levels of urogen and coprogen did not lead to high levels of protogen, suggesting that the synthesis of ALA might be the rate-limiting step in the biosynthesis of UP and CP, but not for the entire heme-biosynthetic pathway. Kjeldstd et al. reported that CP was synthesized as the predominant porphyrin in Propionibacterium acnes, but the amounts of CP and PP differed with the pH of the growth medium and fermentation time (15). The conversion of coprogen to protogen is the rate-limiting step in the bacterial biosynthesis of heme (7, 24). Taken together, there appears to be some other rate-limiting step in this pathway.
Exogenous ALA affected the production of porphyrins in P. freudenreichii. ALA that was synthesized de novo in the recombinant P. freudenreichii that harbored pKHEM04 had a more considerable stimulatory effect on the production of porphyrins (in total, 7.1 mg/liter) than exogenous ALA (in total, 0.56 mg/liter), even when exogenous ALA was added at a very high concentration (data not shown). One plausible explanation is that exogenous ALA in the medium might not diffuse efficiently into the cells.
In conclusion, we have described an attempt to stimulate the production of porphyrins by genetic engineering. The construct with homogenic hemB downstream of heterogenic hemA allowed the overexpression of both genes as a novel operon. As compared to levels in wild-type cells, larger amounts of ALA were synthesized via the C4 pathway and larger amounts of porphyrins were generated to a maximum level of 8.3 mg/liter of culture. Our investigations provide new opportunities for controlled multigene expression in Propionibacterium, which should be useful in attempts to produce elevated levels of vitamin B12 in Propionibacterium.
Acknowledgments
Y. Piao gratefully acknowledges the support of a Yoneyama Scholarship.
We thank Hisayo Ono for many comments and helpful suggestions about experimental procedures.
REFERENCES
- 1.Beale, S. I., and P. A. Castelfranco. 1974. The biosynthesis of 5-aminolevulinic acid in higher plants. II. Formation of 14C 5-aminolevulinic acid from labeled precursors in greening plant tissues. Plant Physiol. 53:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolt, E. L., L. Kryszak, J. Zeilstra-Ryalls, P. M. Shoolingin-Jordan, and M. J. Warren. 1999. Characterization of the Rhodobacter sphaeroides 5-aminolaevulinic acid synthase isoenzymes, HemA and HemT, isolated from recombinant Escherichia coli. Eur. J. Biochem. 265:290-299. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Cox, R., and H. P. Charles. 1973. Porphyrin-accumulating mutants of Escherichia coli. J. Bacteriol. 113:122-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimoto, Y., M. Yamashita, and Y. Murooka. 1997. The Propionibacterium freudenreichii hemYHBXRL gene cluster, which encodes enzymes and a regulator involved in the biosynthetic pathway from glutamate to protoheme. Appl. Microbiol. Biotechnol. 47:385-392. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto, Y., M. Yamashita, H. Ono, and Y. Murooka. 1996. Characterization of the hemB gene encoding δ-aminolevulinic acid dehydratase from Propionibacterium freudenreichii. J. Ferment. Bioeng. 82:93-100. [Google Scholar]
- 7.Javor, G. T., and E. F. Febre. 1992. Enzymatic basis of thiol-stimulated secretion of porphyrins by Escherichia coli. J. Bacteriol. 174:1072-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson, P., and L. Hederstedt. 1999. Organization of genes for tetrapyrrole biosynthesis in Gram-positive bacteria. Microbiology 145:529-538. [DOI] [PubMed] [Google Scholar]
- 9.Jordan, P. M. 1982. Uroporphyrinogen III cosynthetase: a direct assay method. Enzyme 28:158-169. [DOI] [PubMed] [Google Scholar]
- 10.Jordan, P. M., B. I. Mgbeje, S. D. Thomas, and A. F. Alwan. 1988. Nucleotide sequence for the hemD gene of Escherichia coli encoding uroporphyrinogen III synthase and initial evidence for a hem operon. Biochem. J. 249:613-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan, P. M., and J. S. Seehra. 1979. The biosynthesis of uroporphyrinogen III: order of assembly of the four porphobilinogen molecules in the formation of the tetrapyrrole ring. FEBS Lett. 104:364-366. [DOI] [PubMed] [Google Scholar]
- 12.Kiatpapan, P., Y. Hashimoto, H. Nakamura, Y.-Z. Piao, H. Ono, M. Yamashita, and Y. Murooka. 2000. Characterization of pRGO1, a plasmid from Propionibacterium acidipropionici, and its use for development of a host-vector system in propionibacteria. Appl. Environ. Microbiol. 66:4688-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiatpapan, P., and Y. Murooka. 2001. Construction of an expression vector for propionibacteria and its use in production of 5-aminolevulinic acid by Propionibacterium freudenreichii. Appl. Microbiol. Biotechnol. 56:144-149. [DOI] [PubMed] [Google Scholar]
- 14.Kiatpapan, P., M. Yamashita, N. Kawaraichi, T. Yasuda, and Y. Murooka. 2001. Heterologous expression of a gene encoding cholesterol oxidase in probiotic strains of Lactobacillus plantarum and Propionibacterium freudenreichii under the control of native promoters. J. Biosci. Bioeng. 95:459-465. [DOI] [PubMed] [Google Scholar]
- 15.Kjeldstad, B., A. Johnsson, and S. Sandberg. 1984. Influence of pH on porphyrin production in Propionibacterium acnes. Arch. Dermatol. Res. 276:396-400. [DOI] [PubMed] [Google Scholar]
- 16.Lien, L. F., and D. S. Beattie. 1982. Comparisons and modifications of the colorimetric assay for delta-aminolevulinic acid synthase. Enzyme 28:120-132. [DOI] [PubMed] [Google Scholar]
- 17.Lim, C. K., F. M. Li, and T. J. Peters. 1988. High-performance liquid chromatography of porphyrins. J. Chromatogr. 429:123-153. [DOI] [PubMed] [Google Scholar]
- 18.Mauzerall, D., and S. Granick. 1956. The occurrence and determination of 5-aminolevulinic acid and porphobilinogen in urine. J. Biol. Chem. 219:435-446. [PubMed] [Google Scholar]
- 19.Menon, I. A., and D. Shemin. 1967. Concurrent decrease of enzymic activities concerned with the synthesis of coenzyme B12 and of propionic acid in propionibacteria. Arch. Biochem. Biophys. 121:304-310. [DOI] [PubMed] [Google Scholar]
- 20.Murakami, K., Y. Hashimoto, and Y. Murooka. 1993. Cloning and characterization of the gene encoding glutamate 1-semialdehyde 2,1-aminomutase, which is involved in δ-aminolevulinic acid synthesis in Propionibacterium freudenreichii. Appl. Environ. Microbiol. 59:347-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neidle, E. L., and S. Kaplan. 1993. 5-Aminolevulinic acid availability and control of spectral complex formation in HemA and HemT mutants of Rhodobacter sphaeroides. J. Bacteriol. 175:2304-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver, I. T., and W. A. Rawlinson. 1955. The absorption spectra of porphyrin alpha and derivatives. Biochem. J. 61:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philipp-Dormston, W. K., and M. Doss. 1973. Comparison of porphyrin and heme biosynthesis in various heterotrophic bacteria. Enzyme 16:57-64. [DOI] [PubMed] [Google Scholar]
- 24.Rompf, A., C. Hungerer, T. Hoffmann, M. Lindenmeyer, U. Romling, U. Gross, M. O. Doss, H. Arai, Y. Igarashi, and D. Jahn. 1998. Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol. Microbiol. 29:985-997. [DOI] [PubMed] [Google Scholar]
- 25.Sailer, R., W. S. L. Strauss, K. Konig, A. Ruck, and R. Steiner. 1997. Correlation between porphyrin biosynthesis and photodynamic inactivation of Pseudomonas aeruginosa after incubation with 5-aminolaevulinic acid. J. Photochem. Photobiol. B Biol. 39:236-242. [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Sassa, S. 1982. Delta-aminolevulinic acid dehydratase assay. Enzyme 28:133-145. [DOI] [PubMed] [Google Scholar]
- 28.Sattler, I., C. A. Roessner, N. J. Stolowich, S. H. Hardin, L. W. Harris-Haller, N. T. Yokubaitis, Y. Murooka, Y. Hashimoto, and A. I. Scott. 1995. Cloning, sequencing, and expression of the uroporphyrinogen III methyltransferase cobA gene of Propionibacterium freudenreichii (shermanii). J. Bacteriol. 177:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vorobjeva, L. I. 1999. Propionibacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.