Abstract
The development of freshwater multispecies biofilms at solid-liquid interfaces occurs both in quiescent waters and under conditions of high shear rates. However, the influence of hydrodynamic shear rates on bacterial biofilm diversity is poorly understood. We hypothesized that different shear rates would significantly influence biofilm diversity and alter the relative proportions of coaggregating and autoaggregating community isolates. In order to study this hypothesis, freshwater biofilms were developed at five shear rates (<0.1 to 305 S−1) in a rotating concentric cylinder reactor fed with untreated potable water. Eubacterial diversity was assessed by denaturing gradient gel electrophoresis (DGGE) and culturing on R2A agar. Fifty morphologically distinct biofilm strains and 16 planktonic strains were isolated by culturing and identified by partial 16S rRNA gene sequencing, and their relatedness was determined by the construction of a neighbor-joining phylogenetic tree. Phylogenetic and DGGE analyses showed an inverse relationship between shear rate and bacterial diversity. An in vitro aggregation assay was used to assess the relative proportions of coaggregating and autoaggregating species from each biofilm. The highest proportion of autoaggregating bacteria was present at high shear rates (198 to 305 S−1). The intermediate shear rate (122 S−1) selected for the highest proportion of coaggregating bacteria (47%, or 17 of a possible 36 coaggregation interactions). Under static conditions (<0.1 S−1), 41 (33%) of a possible 125 coaggregation interactions were positive. Few coaggregation (3.3%) or autoaggregation (25%) interactions occurred between the 16 planktonic strains. In conclusion, these data show that shear rates affect biofilm diversity as well as the relative proportions of aggregating bacteria.
Microorganisms colonize a wide range of environments as spatially organized, taxonomically diverse, multispecies biofilm communities (1, 20, 44). In potable water distribution systems, many bacterial species, including members of the Proteobacteria, Actinobacteria, low-G+C-content gram-positive bacteria, and Cytophaga-Flavobacterium-Bacterioides group, readily adhere to surfaces to form multispecies biofilms (29, 41). The abilities of these taxonomically diverse organisms to attach to surfaces and codevelop within multispecies biofilms are imperative for their survival and persistence within flowing environments (4, 33, 44).
Few published articles have described the effect of shear on the bacterial composition and diversity of freshwater biofilms. Evidence is emerging, however, that at high fluid velocities with associated high shear rates, multispecies communities are less diverse than those developed at lower shear rates (9, 23, 35, 42). In order to adhere to surfaces or surface-attached cells and subsequently to form biofilms, bacteria in high-velocity flowing systems must overcome shear stress at the fluid-surface interface (7, 11). Attachment is facilitated by the expression of cell surface polymers that alter cell surface properties (3, 15). Properties that enhance attachment include increases in whole-cell hydrophobicity (14) and the abilities to coaggregate (25, 36) and autoaggregate (17). Recently, it was shown, using a simple recirculating tank model (35), that freshwater biofilms formed at high shear rates contained a high proportion of bacterial species that were not detectable in the surrounding fluid phase. In addition, a larger proportion of biofilm strains than of their planktonic counterparts were able to coaggregate and/or autoaggregate.
It is clear that hydrodynamic shear forces have the potential to profoundly affect microbial diversity, a property that is important with regard to the potential of opportunistic pathogens to integrate into freshwater biofilms. Opportunistic pathogens that have been isolated from biofilms in potable water systems include Mycobacterium avium, Pseudomonas aeruginosa, Klebsiella spp., Legionella spp., and Flavobacterium spp. (29, 39, 50). Payment et al. (31) suggested that approximately 35% of gastrointestinal infections could be associated with the consumption of apparently potable water. Other problems associated with multispecies biofilms formed within potable water systems include microbially induced corrosion and unpleasant taste and odor (18). Shear forces vary throughout a potable water system (12). Thick biofilms are often observed at water-solid interfaces in stagnant regions, while much thinner biofilms are found on surfaces exposed to high shear forces (18). It is important to understand the impact of these different hydrodynamic conditions on biofilm diversity and colonization resistance, especially with regard to the integration of pathogens into biofilms.
The aim of the work presented here was to describe the effects of different hydrodynamic shear forces on the formation and diversity of freshwater biofilm communities. A concentric cylinder reactor (CCR) (52) was used to control flow velocity and thereby shear, and community diversity was monitored by culturing and other methods. The surface properties of the bacteria from each community were also assessed by measuring whole-cell hydrophobicity and coaggregation and autoaggregation abilities.
MATERIALS AND METHODS
CCR.
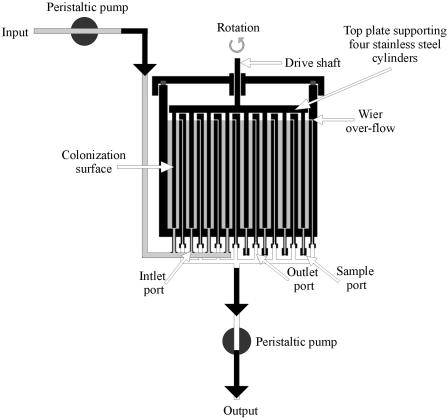
The CCR (52) (Fig. 1) consists of four stainless steel cylindrical pipe sections that are contained and rotated within concentric reaction chambers. The chambers have a defined constant volume and can be fed medium or water at controlled flow rates. In this instance, potable water was delivered directly to the reactor from an intermediate supply reservoir. Wiers within the chamber walls maintained a constant volume in each chamber. Rates of water flow through the chambers gave D values of >3 h−1, which were higher than the maximum D values in all instances. Planktonic cells therefore would be washed from the system before division. Fluid flow over the test surfaces was a function of the rotational velocity (43 rpm) of the pipe sections and the diameter of each rotating unit (101, 76, 50, and 26 mm). Fluid velocity profiles between the stator and the rotator walls were calculated on the basis of computational fluid dynamics (2). From these fluid velocity profiles, shear rates were calculated 50 μm from each of the surfaces. Fluid velocities of 0.26, 0.19, 0.16, and 0.12 m/s corresponded to 101-, 77-, 50-, and 26-mm cylinders, and these corresponded to shear rates of 305, 198, 122, and 65 S−1, respectively. These shear rates are similar to those found in channels within biofilms (2). The temperature of the water that was fed into the unit was monitored over 3 months (mean and standard deviation, 7.0 ± 2.0°C), as was the pH (6.9 ± 0.2).
FIG. 1.
Diagrammatic representation of the CCR (52).
The volume of water in the intermediate supply reservoir was maintained at 2 liters by a constant-head device. Stainless steel disks were placed into this reservoir so that biofilms could be formed under conditions of minimal shear. The system was operated continuously for 12 weeks. At the end of the 12 weeks, biofilm samples were harvested by using sterile swabs; the samples included triplicate vertical swab samples obtained through the water line from the outer surfaces of the four rotators and triplicate samples obtained from stainless steel coupon surfaces on cylinders held within the intermediate supply reservoir.
Strains and growth conditions.
Biofilm samples were collected as swab samples from approximately 2 cm2 of the biofilm on each of the concentric cylinder surfaces in a vertical line from the bottom edge through the water line, and each sample was resuspended in 1 ml of autoclaved tap water. Planktonic samples were collected as 50 ml of water from the tap, reservoir, and CCR outlet. These samples were filtered through a Swinnex filter (Millipore, Stonehouse, Gloucestershire, United Kingdom) with a membrane pore size of 0.2 μm, and the cells on the membrane were resuspended in 1 ml of autoclaved tap water. Biofilm and planktonic samples were cultured on R2A agar (34) at 30°C for 5 days. All distinct colony morphotypes were separated and subcultured on R2A agar. Strains were stored at −70°C in 50% (vol/vol) glycerol for subsequent analyses.
Characterization of bacterial strains.
Strains isolated after growth on solid R2A or in liquid R2A medium at 30°C were subjected to Gram staining and catalase and oxidase reactions. Cell morphologies were observed by using a light microscope (Axioskop 2-MOT; Carl Zeiss, Esslingen, Germany).
Identification of isolated bacterial strains by partial 16S rRNA gene sequencing.
Strains were identified by partial 16S rRNA gene sequencing by a modification of the method of Rickard et al. (38). Prior to sequencing, partial 16S rRNA gene amplification was performed by taking one bacterial colony of each strain from R2A agar, boiling it in 100 μl of sterile double-processed tissue culture water (Sigma, Poole, Dorset, United Kingdom) for 10 min, and using 10 μl of the suspension as template DNA for PCR. Degenerate primers 806R (53) and 8FPL (51) were used to amplify a fragment of the 16S rRNA gene from each strain. The fragment corresponds to nucleotides 8 to 806 in the Escherichia coli gene sequence.
PCR was carried out with PCR buffer (Boehringer Mannheim, Indianapolis, Ind.) containing 3.2 μM each primer, 0.5 mM deoxynucleoside triphosphates, and 2 U of Taq DNA polymerase (Sigma) per 100 μl. PCR cycles consisted of 35 cycles at 94°C (1 min), 53°C (1 min), and 72°C (1 min) and a final 15-min chain elongation step at 72°C. Amplified products were purified by using a QIA-quick PCR purification system (Qiagen, Crawley, West Sussex, United Kingdom) according to the manufacturer's instructions. PCR products were subsequently sequenced with primers 806R and 8FPL. Sequencing reaction mixtures consisted of 50 to 100 ng of PCR product, 10 ng of primer, and 4 μl of Big Dye (Perkin-Elmer, Boston, Mass.) in a total volume of 20 μl. The samples were incubated at 94°C (4 min), followed by 25 cycles of 96°C (30 s), 50°C (15 s), and 60°C (4 min). Sequencing was performed with an ABI 377 sequencer (Perkin-Elmer, Cambridge, United Kingdom), and the sequences of each strain were compiled by using Inherit (PE Applied Biosystems) on the attached computer.
Unambiguous compiled sequences of approximately 600 to 700 bases were obtained for each strain and compared to known sequences in the EMBL database by using FASTA (http://www.ebi.ac.uk/FASTA33/). Based on criteria described by Stackebrandt and Goebel (43), gene sequences that were >97.0% identical to sequences of species in the EMBL database were assigned genus and species names. Sequences that possessed a sequence identity of between 94.0 and 97.0% were assigned a genus name, and sequences that were <94.0% identical to sequences in the EMBL database were described as unknown and possibly novel genera.
Tree construction.
CLUSTALX (version 1.81) (48) was used to align 500 bp of unambiguous partial 16S ribosomal DNA from each strain against sequences from related strains in the EMBL database. Neighbor-joining analysis was conducted with the correction of Jukes and Cantor (16) by using TREECON (version 1.3b) (49). Thermus thermophilus (EMBL accession number X07998) was used as the outgroup. The neighbor-joining tree was edited with Corel Draw (version 8.0) (Corel, Dallas, Tex.) to show the phylogenetic relationships among bacteria in biofilms developed under conditions of different flow velocities.
DGGE.
In order to obtain suitable cell numbers for denaturing gradient gel electrophoresis (DGGE) analysis, biofilms were removed from approximately 2 cm2 of each of the cylinders and resuspended in 1 ml of sterile water. To obtain adequate cell numbers for DGGE analysis of the planktonic populations, 50 ml of water was filtered through a Swinnex filter, with a membrane pore size of 0.2 μm, and the cells were collected in 500 μl. Tris-equilibrated phenol (pH 8.0) (150 μl) was added, and the suspensions were shaken three times in a Mini-Bead Beater (Biospec Products, Bartlesville, Okla.) for 80 s at maximum speed. Following 10 min of centrifugation at 13,000 × g, each supernatant was extracted three times with an equal volume of phenol-chloroform and once with chloroform-isoamyl alcohol (24:1 [vol/vol]). The DNA from each sample was precipitated from the aqueous phase with 3 volumes of ethanol, air dried, and resuspended in 100 μl of deionized water. The amount and quality of DNA extracted were estimated by electrophoresis of 5-μl aliquots on 0.8% agarose gels and by comparison to a molecular weight standard (stained with ethidium bromide). DNA extracts were stored at −60°C prior to analysis.
PCR was performed by using methods described by McBain et al. (27). The eubacterium-specific primers HDA1-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) were used to amplify the V2-V3 region of 16S ribosomal DNA (corresponding to positions 339 to 539 of E. coli). The reactions were carried out in 0.2-ml tubes by using a model 480 DNA thermal cycler (Perkin-Elmer). In all instances, the reactions were carried out with Red Taq DNA polymerase Ready Mix (25 μl) (Sigma), HDA primers (2 μl each, 5 μM), nanopure water (16 μl), and extracted community DNA (5 μl). Optimization studies, carried out as described by Muyzer (28), showed that a minimum of a 1:10 dilution of extracted community DNA was required to ensure a reliable PCR. Quantification and standardization of extracted DNA were achieved by using a fluorescence assay (DNA quantitation kit; Sigma) according to the manufacturer's instructions. The thermal program was as follows: 94°C (4 min); 30 thermal cycles of 94°C (30 s), 56°C (30 s), and 68°C (60 s); and a 7-min chain elongation step (68°C).
Biofilm samples were analyzed by DGGE with a D-Code universal mutation detection system (Bio-Rad, Hemel Hempstead, United Kingdom). Polyacrylamide (10%) gels (16 by 16 cm and 1 mm deep) were run with 1× TAE buffer diluted from 50× TAE buffer (40 mM Tris base, 20 mM glacial acetic acid, 1 mM EDTA). Initially, separation parameters were optimized by running PCR products from selected pure cultures of biofilm bacteria and PCR amplicons from extracted biofilm DNA on gels with a 0 to 100% denaturation gradient perpendicular to the direction of electrophoresis (100% denaturing solution contained 40% [vol/vol] formamide and 7.0 M urea). Denaturing gradients were formed with two 10% acrylamide (acrylamide-bisacrylamide, 37.5:1) stock solutions (Sigma). On this basis, a denaturaturing gradient ranging from 20 to 60% for a parallel DGGE analysis was selected for community analyses. DNA for loading onto gels was quantified and, when necessary, standardized between samples by using a fluorescence assay (see above). Electrophoresis was carried out at 150 V and 60°C for approximately 5 h. All gels were stained with SYBR Gold stain (diluted to 10−4 in 1× TAE buffer) [Molecular Probes (Europe), Leiden, The Netherlands] for 30 min. Gels were viewed and images were documented by using a BioDocit system (UVP, Upland, Calif.).
For analysis of the major resolved DGGE amplicons, selected resolved bands were cut out of the polyacrylamide gels by using a sterile scalpel under UV illumination and were incubated at 4°C for 20 h together with 20 μl of nanopure water in nuclease-free universal bottles. Portions (5 μl) were removed and used as PCR templates. PCR products were purified by using a QIA-quick PCR purification system and sequenced by using the reverse (non-GC clamp) primer (HDA2). The sequencing cycles were 94°C (4 min), followed by 25 cycles of 96°C (30 s), 50°C (15 s), and 60°C (4 min). Once chain termination was complete, sequencing was done with the Perkin-Elmer ABI 377 sequencer. DNA sequences were compiled by using Autoassembler (Applied Biosystems) to obtain consensus sequences or to check and edit unidirectional sequences. For PCR of excised DGGE bands, the presence of a GC clamp in sequence analyses confirmed that the correct target rather than a contaminant had been reamplified. FASTA searches were performed for each compiled sequence and the sequences in the EMBL prokaryote database. Closest relative species were assigned based on FASTA comparisons of compiled partial 16S rRNA gene sequences and sequences in the EMBL database.
Visual aggregation assay.
Prior to the visual coaggregation assay, strains were grown for 72 h in R2A broth at 30°C in a rotary shaker set at 200 rpm. A visual aggregation assay modified from that of Rickard et al. (37) was used to assess the abilities of strains to both coaggregate and autoaggregate. Cells were harvested, concentrated by centrifugation for 15 min at 5,000 × g, and washed three times in filter-sterilized distilled water. Cells were resuspended in distilled water to an optical density at 650 nm (OD650) of 1.0 and concentrated to give a calculated OD650 of 1.5. For coaggregation, pairs of strains at an OD650 of 1.5 were mixed in equal volumes (200 μl) in silica Durham tubes (6 by 50 mm; Scientific Lab Supplies, Wilford, Nottingham, United Kingdom) at room temperature. Mixtures were vortexed for 30 s and rolled gently for 1 min. The degree of coaggregation for each pair was scored by using a subjective assay (8). The scoring criteria were as follows: 0, no coaggregates in suspension; 1, small uniform aggregates in a turbid suspension; 2, easily visible coaggregates in a turbid suspension; 3, clearly visible coaggregates which settle, leaving a clear supernatant; 4, large flocs of coaggregates that settle almost instantaneously, leaving a clear supernatant. Control tubes containing each isolate on its own were also included to assess autoaggregation (self-aggregation). Autoaggregation was scored by using the same criteria as those used for coaggregation. When autoaggregation occurred, the autoaggregation score was deducted from the coaggregation score to give a corrected coaggregation score.
Whole-cell hydrophobicity.
The surface hydrophobicity of individual cells was determined by measuring bacterial adhesion to hexadecane (40). Cells were grown to stationary phase in R2A medium (72 h), washed three times with phosphate-buffered saline (PBS) by centrifugation (3,000 × g for 10 min), and resuspended to an OD650 of 1.0. The cell suspensions (0.8 ml) alone or with 0.4 ml of hexadecane (Sigma) added were transferred to 5-ml glass test tubes and preincubated for 15 min at 30°C with shaking at 200 rpm. The tubes were vortexed for 2 min, and the two phases were allowed to separate for 15 min. The aqueous PBS-cell phase was removed, and its OD650 was determined. Whole-cell hydrophobicity was expressed as the percent difference between the original OD650 of cells in PBS (1.0) and the OD650 of the PBS-cell suspension following the hexadecane treatment.
RESULTS
Culture-independent diversity of biofilm and planktonic communities.
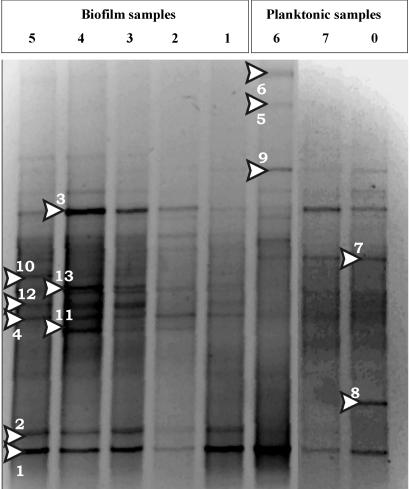
DGGE was used to determine the total diversity of biofilm communities developed at different shear rates. Furthermore, to compare the planktonic community composition to the biofilm community composition, samples from the potable water supply, the intermediate supply reservoir, and the CCR outlet were also analyzed. Figure 2 shows a negative image of the DGGE gel which indicates that significantly different communities have developed at different shear rates. The biofilm which developed at the lowest shear rate (<0.1 S−1) showed the highest diversity (10 clearly visible bands). Conversely, at the highest shear rate (305 S−1), the least diverse biofilm developed (five bands). Some bands were present in all planktonic and biofilm samples (Table 1). For example, Mycobacterium mucogenicum (band 1) and Streptomyces avermitilis (band 3) were present in all biofilm and planktonic populations. Some bands were, however, unique to each biofilm. For example, Mesorhizobium loti (band 10) and Bradyrhizobium japonicum (band 12) were found only in biofilms that developed at shear rates of <0.1 to 65 S−1. Some bands were unique to planktonic communities (e.g., Corynebacterium glutamicum) (Table 1.). Slight differences in community composition were also detected among planktonic communities (tap, storage tank, and CCR outlet). In particular, S. avermitilis and C. glutamicum (Fig. 2, bands 5 and 6) were detected only in water directly from the tap and not in the storage tank, outlet, or any of the biofilms. These two species were not detected by culturing (see below), and it is possible that they originated from a biofilm in the water supply line upstream of the tap and required specific environmental conditions in order to survive.
FIG. 2.
Negative image from the DGGE gel. From left to right, samples were as follows: 5, biofilm community exposed to a shear rate of <0.1 S−1; 4, biofilm community exposed to a shear rate of 65 S−1; 3, biofilm community exposed to a shear rate of 122 S−1; 2, biofilm community exposed to a shear rate of 198 S−1; 1, biofilm community exposed to a shear rate of 305 S−1; 6, planktonic population from the tap feed; 7, planktonic population from the storage tank; 0, planktonic population from the CCR outlet. Arrowheads indicate selected dense bands from which DNA was sequenced.
TABLE 1.
Sequenced bands from the DGGE gel
| Band | Sequence length (bp) | Closest relative | Community sample(s) in which band was presenta |
|---|---|---|---|
| 1 | 142 | Mycobacterium mucogenicum | 0, 1, 2, 3, 4, 5, 6, 7 (All) |
| 2 | 161 | Mycobacterium mucogenicum | 0, 1, 2, 3, 4, 5, 6, 7 (All) |
| 3 | 160 | Streptomyces avermitilis | 0, 1, 2, 3, 4, 5, 6, 7 (All) |
| 4 | 151 | Sphingomonas sp. | 0, 1, 2, 3, 4, 5 (All) |
| 5 | 153 | Streptomyces avermitilis | 6 (Planktonic only) |
| 6 | 156 | Corynebacterium glutamicum | 6 (Planktonic only) |
| 7 | 148 | Uncultured α-proteobacterium | 7, 6, 0 (Planktonic only) |
| 8 | 152 | Mycobacterium sp. | 7 (Planktonic only) |
| 9 | 162 | Rhodococcus sp. | 7, 6, 0 (Planktonic only) |
| 10 | 153 | Mesorhizobium loti | 5, 4, 3 (Biofilm only) |
| 11 | 140 | Brevundimonas vesicularis | 4, 3, 2 (Biofilm only) |
| 12 | 146 | Bradyrhizobium japonicum | 5, 4, 3 (Biofilm only) |
| 13 | 138 | Methylobacterium fujisawae | 1, 2, 3, 4, 5 (Biofilm only) |
As indicated by the presence or absence of bands in the DGGE gel shown in Fig. 2, which also shows band positions.
Overall, for biofilm communities, DGGE showed an inverse relationship between the shear rate and the number of genera and species present. In addition, the compositions of the biofilms at various shear rates differed in spite of the use of a common source of potable water as an inoculum and growth medium.
Diversity of culturable planktonic and biofilm communities.
The diversity within culturable biofilm communities was assessed following isolation on R2A agar at 30°C. The total number of culturable bacteria in each biofilm community decreased by approximately 10-fold as the shear rate increased from <0.1 S−1 to the maximum of 305 S−1 (data not shown). There was also an inverse relationship between the shear rates and the quantities of distinct colony morphotypes isolated (Table 2). For example, 17 distinct colony morphotypes were isolated from the biofilm developed at lentic water velocities (<0.1 S−1), while only 6 distinct colony morphotypes were isolated from the biofilm developed at a high shear rate (305 S−1). Gram-negative organisms dominated in all of the biofilms, and only 15 gram-positive strains were isolated. For comparison, the diversity within culturable planktonic communities was also analyzed. The three planktonic communities (tap feed, storage vessel, and CCR outlet) were less diverse with respect to distinct dominant colony morphotypes than were the five biofilm communities. Freshwater samples from the tap, reservoir, and CCR outlet contained five, five, and six morphologically distinct colony types, respectively. Like the strains in the biofilm communities, the majority of the planktonic strains were gram negative.
TABLE 2.
Strains isolated and identified from biofilm communities that developed in the CCR
| Strain | Shear rate (S−1) | Most closely related species or genus | Sequence length (bp) | % Sequence identity | Assigned EMBL accession no. |
|---|---|---|---|---|---|
| 1.1 | 305 | Variovorax paradoxus | 638 | 99.8 | AJ626891 |
| 1.3 | 305 | Mycobacterium mucogenicum | 606 | 100 | AJ626892 |
| 1.4 | 305 | Unknown | 524 | 91.8 | AJ626893 |
| 1.5 | 305 | Unknown | 553 | 92.7 | AJ626894 |
| 1.6 | 305 | Acidovorax sp. | 544 | 94.7 | AJ626895 |
| 1.7 | 305 | Acidovorax sp. | 551 | 96.4 | AJ626982 |
| 2.1 | 198 | Mycobacterium mucogenicum | 594 | 99.7 | AJ626983 |
| 2.2 | 198 | Methylobacterium fujisawae | 593 | 97.4 | AJ6269849 |
| 2.3 | 198 | Variovorax paradoxus | 631 | 99.8 | AJ626985 |
| 2.4 | 198 | Flavobacterium sp. | 630 | 98.5 | AJ626986 |
| 2.5 | 198 | Brevundimonas vesicularis | 624 | 99.8 | AJ626987 |
| 2.6 | 198 | Mycobacterium mucogenicum | 590 | 96.4 | AJ626999 |
| 2.7 | 198 | Mycobacterium mucogenicum | 613 | 99.1 | AJ627000 |
| 2.8 | 198 | Unknown | 635 | 92.9 | AJ627001 |
| 2.9 | 198 | Unknown | 609 | 92.5 | AJ627002 |
| 3.1 | 122 | Mycobacterium mucogenicum | 558 | 99.5 | AJ627003 |
| 3.3 | 122 | Sphingomonas melonis | 559 | 98.7 | AJ627004 |
| 3.4 | 122 | Flavobacterium sp. | 645 | 98.6 | AJ627005 |
| 3.5 | 122 | Variovorax paradoxus | 640 | 99.8 | AJ627006 |
| 3.6 | 122 | Variovorax paradoxus | 624 | 99.7 | AJ627007 |
| 3.8 | 122 | Dermacoccus nishinomiyaensis | 595 | 99.5 | AJ627008 |
| 3.9 | 122 | Sphingomonas yanoikuyae | 610 | 99.8 | AJ627008 |
| 3.11 | 122 | Brevundimonas vesicularis | 649 | 100 | AJ627010 |
| 3.12 | 122 | Mycobacterium mucogenicum | 532 | 100 | AJ627011 |
| 4.1 | 65 | Brevundimonas vesicularis | 491 | 100 | AJ627012 |
| 4.2 | 65 | Variovorax paradoxus | 640 | 99.8 | AJ627013 |
| 4.3 | 65 | Micrococcus luteus | 659 | 100 | AJ627014 |
| 4.4 | 65 | Micrococcus luteus | 655 | 99.8 | AJ627015 |
| 4.5 | 65 | Mycobacterium mucogenicum | 660 | 100 | AJ627393 |
| 4.6 | 65 | Brayrhizobium japonicum | 592 | 100 | AJ627394 |
| 4.7 | 65 | Bacillus sphaericus | 668 | 98.3 | AJ627395 |
| 4.8 | 65 | Methylobacterium fujisawae | 593 | 98.1 | AJ627396 |
| 4.9 | 65 | Sphingobium yanoikuyae | 640 | 99.4 | AJ627397 |
| 5.1 | <0.1 | Variovorax paradoxus | 608 | 99.7 | AJ627398 |
| 5.2 | <0.1 | Methylobacterium nodulans | 581 | 98 | AJ627399 |
| 5.3 | <0.1 | Sphingomonas sp. | 564 | 100 | AJ627400 |
| 5.5 | <0.1 | Sphingomonas sp. | 594 | 100 | AJ627401 |
| 5.6 | <0.1 | Brevundimonas vesicularis | 629 | 99.8 | AJ627402 |
| 5.7 | <0.1 | Methylobacterium sp. | 555 | 99.6 | AJ627403 |
| 5.8 | <0.1 | Sphingomonas sp. | 640 | 100 | AJ627404 |
| 5.9 | <0.1 | Bradyrhizobium japonicum | 592 | 100 | AJ627405 |
| 5.10 | <0.1 | Pseudoxanthomonas mexicana | 651 | 99.8 | AJ627406 |
| 5.11 | <0.1 | Mycobacterium mucogenicum | 563 | 99.3 | AJ627407 |
| 5.12 | <0.1 | Roseomonas gilardii | 590 | 99.6 | AJ627408 |
| 5.13 | <0.1 | Sphingomonas sp. | 640 | 100 | AJ627409 |
| 5.14 | <0.1 | Micrococcus luteus | 560 | 99.8 | AJ627410 |
| 5.15 | <0.1 | Mycobacterium mucogenicum | 521 | 99.4 | AJ627411 |
| 5.16 | <0.1 | Methylobacterium nodulans | 560 | 97.1 | AJ627412 |
| 5.17 | <0.1 | Methylobacterium fujisawae | 593 | 98.1 | AJ627413 |
| 5.20 | <0.1 | Staphylococcus hominis | 642 | 99.5 | AJ627414 |
In order to determine the identities and phylogenetic relationships of the planktonic and biofilms strains, partial 16S rRNA gene sequencing was performed. Bacteria from six major taxonomic groups (Actinobacteria, bacilli, Cytophaga-Flexibacter-Bacteroides, α-Proteobacteria, β-Proteobacteria, and γ-Proteobacteria) were identified (Tables 2 and 3). All but four strains were identified to at least the genus level by 16S rRNA sequencing, and these are candidate members of novel genera. Using the criteria of Stackebrandt and Goebel (43), the majority of the strains were identified to the species level, as their sequences showed >97% identity to 16S rRNA gene sequences in the EMBL database (Tables 2 and 3).
TABLE 3.
Strains isolated and identified from planktonic communities that developed in the CCR
| Strain | Planktonic community | Most closely related species or genus | Sequence length (bp) | % Sequence identity | Assigned EMBL accession no. |
|---|---|---|---|---|---|
| 0.1 | Outlet | Dyadobacter fermentens | 573 | 98.3 | AJ626875 |
| 0.2 | Outlet | Dyadobacter fermentens | 618 | 98.9 | AJ626876 |
| 0.3 | Outlet | Variovorax paradoxus | 560 | 99.8 | AJ626877 |
| 0.5 | Outlet | Methylobacterium extorquens | 560 | 99.5 | AJ626878 |
| 0.6 | Outlet | Hymenobacter sp. | 602 | 96.7 | AJ626879 |
| 0.7 | Outlet | Variovorax paradoxus | 606 | 99.7 | AJ626880 |
| 6.1 | Tap feed | Mycobacterium mucogenicum | 560 | 99.8 | AJ626881 |
| 6.2 | Tap feed | Bradyrhizobium japonicum | 589 | 100 | AJ626882 |
| 6.3 | Tap feed | Brevundimonas vesicularis | 572 | 100 | AJ626883 |
| 6.4 | Tap feed | Variovorax paradoxus | 531 | 97.2 | AJ626884 |
| 6.5 | Tap feed | Flavobacterium sp. | 654 | 100 | AJ626885 |
| 7.2 | Storage tank | Paenibacillus amylolyticus | 637 | 99.7 | AJ626886 |
| 7.3 | Storage tank | Sphingomonas aromaticivoran | 540 | 100 | AJ626886 |
| 7.4 | Storage tank | Sphingomonas abikonensis | 555 | 100 | AJ626888 |
| 7.5 | Storage tank | Methylobacterium sp. | 521 | 100 | AJ626889 |
| 7.6 | Storage tank | Variovorax paradoxus | 640 | 99.7 | AJ626890 |
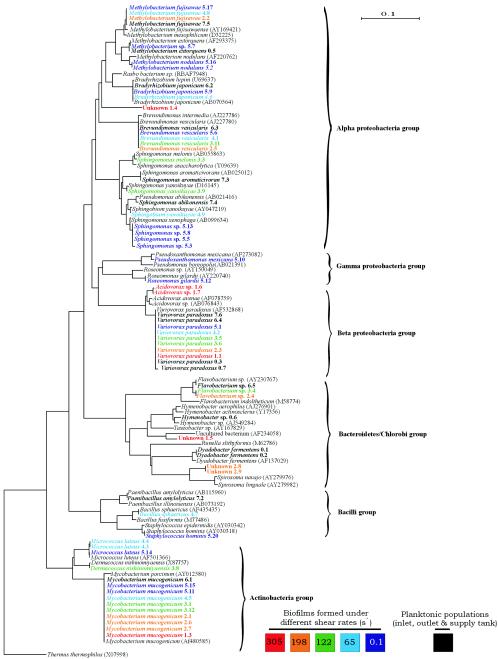
A neighbor-joining tree containing all of the isolated biofilm and planktonic strains was constructed (Fig. 3). Through color coding of each strain name according to the biofilm sample from which the strain originated, it is clear that each shear rate did not select for a single cluster of species, genera, or taxonomic group. Rather, it selected for groups of genetically unrelated strains to generate taxonomically diverse multispecies biofilms. It is evident, however, that high shear rates (122 to 305 S−1) selected for species from the Bacteroidetes-Chlorobi group and that low shear rates (<0.1 to 65 S−1) selected for species from the bacilli. Curiously, a comparison of the diversity of the biofilm populations to that of the planktonic populations showed that some species were unique to the planktonic phase and were not able to form or survive on biofilms. For example, Paenibacillus amylolyticus, Sphingomonas abikonensis, and Hymenobacter sp. were cultured from water running through the CCR unit but were not detected on biofilms. This ambiguity most likely reflects preferential colonization of upstream water piping or the inlet or outlet tubing linked to the device by these species. Conversely, many species of bacteria, such as Sphingomonas yanoikuyae and Acidovorax sp., were isolated from biofilm communities but were not detected in the planktonic phase. As also shown by DGGE (see above), each planktonic and biofilm community contained phylotypes that were exclusive to that community. Hence, each community was unique, in spite of the common inoculum and water feed.
FIG. 3.
Neighbor-joining phylogenetic tree of the cultured strains isolated from the CCR. Colors relate to the biofilm from which the strain was isolated (i.e., highest to lowest shear rates). The scale bar represents one substitution for every 10 nucleotides. The species used as the outgroup was T. thermophilus (EMBL accession number X07998).
Culturing versus culture-independent analysis.
An overall comparison of species identified by culturing (on R2A) and by culture-independent analysis (DGGE) indicated that DGGE identified species that were not identified by culturing. Only 7 of 13 sequenced bands (54%) from the DGGE gel were from species that were also isolated by culturing on R2A agar. For example, S. avermitilis (Table 1, band 3), which was detected by DGGE of biofilm and planktonic community samples, was not cultured on R2A agar. Additionally, Mesorhizobium loti (Table 1, band 10) was detected in biofilm communities by DGGE and not by culturing on R2A agar. The remaining four species that were detected by DGGE and not by culturing were from planktonic communities (Table 1, bands 5, 6, 7, and 9). Such differences in composition suggest that a proportion of each bacterial community was probably nonculturable. Conversely, many species of bacteria were detected by culturing and not by DGGE, probably because many of the bands that were detected by DGGE could not be sequenced because the quantity of DNA was too small or the bands were poorly resolved.
Aggregation abilities of bacteria within biofilm communities.
The phylogenetic relatedness of members of each biofilm community and the coaggregation and autoaggregation abilities of strains within each biofilm community were studied. The coaggregation ability, autoaggregation ability, and whole-cell hydrophobicity of each biofilm strain were determined (Table 4).
TABLE 4.
Visual coaggregation and autoaggregation scores for biofilm strains after growth on R2A agar for 73 h
| Strain | Coaggregation partner (scores)a | Total no. of coaggregating partners | Auto- aggregation score | % Whole-cell hydropho- bicity |
|---|---|---|---|---|
| Variovorax paradoxus 1.1 | 1.3 (1) | 1 | 0 | 12.1 |
| Mycobacterium mucogenicum 1.3 | 1.1 (1) | 1 | 3 | 99.1 |
| Unknown 1.4 | None | 0 | 2 | 88.7 |
| Unknown 1.5 | None | 0 | 2 | 99.3 |
| Acidovorax sp. strain 1.6 | None | 0 | 4 | 94.4 |
| Acidovorax sp. strain 1.7 | None | 0 | 4 | 99.9 |
| Mycobacterium mucogenicum 2.1 | None | 0 | 1 | 99.7 |
| Methylobacterium fujisawae 2.2 | 2.4 (1) | 1 | 3 | 78.7 |
| Variovorax paradoxus 2.3 | None | 0 | 0 | 4.2 |
| Flavobacterium sp. strain 2.4 | 2.2 (1), 2.5 (1), 2.6 (2), 2.5 (1) | 4 | 0 | 12.6 |
| Brevundimonas vesicularis 2.5 | 2.4 (1) | 1 | 0 | 9.4 |
| Mycobacterium mucogenicum 2.6 | 2.4 (2) | 1 | 1 | 74.8 |
| Mycobacterium mucogenicum 2.7 | 2.4 (1) | 1 | 3 | 99.7 |
| Unknown 2.8 | None | 0 | 1 | 5.4 |
| Unknown 2.9 | None | 0 | 1 | 5.3 |
| Mycobacterium mucogenicum 3.1 | 3.4 (2), 3.9 (2) | 2 | 2 | 99.1 |
| Sphingomonas melonis 3.3 | 3.4 (1), 3.6 (1), 3.9 (2), 3.11 (1), 3.12 (1) | 5 | 0 | 2.1 |
| Flavobacterium sp. strain 3.4 | 3.1 (2), 3.3 (1), 3.8 (1), 3.9 (3), 3.12 (1) | 5 | 0 | 1.6 |
| Variovorax paradoxus 3.5 | 3.9 (1), 3.12 (1) | 2 | 0 | 1.7 |
| Variovorax paradoxus 3.6 | 3.3 (1), 3.8 (1), 3.9 (2), 3.12 (1) | 4 | 1 | 5.0 |
| Dermacoccus nishinomiyaensis 3.8 | 3.4 (1), 3.6 (1), 3.12 (2) | 3 | 0 | 49.2 |
| Sphingomonas yanoikuyae 3.9 | 3.1 (2), 3.3 (2), 3.4 (3), 3.5 (1), 3.6 (2), 3.12 (1) | 6 | 0 | 0.3 |
| Brevundimonas vesicularis 3.11 | 3.3 (1) | 1 | 0 | 13.9 |
| Mycobacterium mucogenicum 3.12 | 3.3 (1), 3.4 (1), 3.5 (1), 3.6 (1), 3.8 (2), 3.9 (1) | 6 | 1 | 68.2 |
| Brevundimonas vesicularis 4.1 | 4.2 (3), 4.3 (1), 4.4 (2), 4.8 (1), 4.9 (1) | 5 | 0 | 3.6 |
| Variovorax paradoxus 4.2 | 4.1 (3), 4.4 (2), 4.8 (2), 4.9 (3) | 4 | 0 | 1.2 |
| Micrococcus luteus 4.3 | 4.1 (1), 4.6 (2), 4.8 (1), 4.9 (2) | 4 | 0 | 63.8 |
| Micrococcus luteus 4.4 | 4.1 (2), 4.2 (2), 4.6 (1), 4.9 (2) | 4 | 0 | 73.8 |
| Mycobacterium mucogenicum 4.5 | 4.6 (1) | 1 | 3 | 60.4 |
| Brayrhizobium japonicum 4.6 | 4.3 (2), 4.4 (1), 4.5 (1), 4.9 (4) | 4 | 0 | 6.9 |
| Bacillus sphaericus 4.7 | None | 0 | 0 | 7.7 |
| Methylobacterium fujisawae 4.8 | 4.1 (1), 4.2 (2), 4.3 (1) | 3 | 2 | 52.9 |
| Sphingobium yanoikuyae 4.9 | 4.1 (1), 4.2 (3), 4.3 (2), 4.4 (2), 4.6 (4) | 5 | 0 | 3.6 |
| Variovorax paradoxus 5.1 | 5.2 (1), 5.6 (2), 5.7 (3), 5.13 (1), 5.17 (3), 5.20 (1) | 6 | 0 | 1.2 |
| Methylobacterium nodulans 5.2 | 5.1 (1), 5.5 (2), 5.7 (3), 5.9 (2), 5.13 (1) | 5 | 0 | 3.4 |
| Sphingomonas sp. strain 5.3 | 5.6 (2), 5.7 (2), 5.9 (1), 5.10 (2), 5.12 (2), 5.13 (2) | 6 | 0 | 12.2 |
| Sphingomonas sp. strain 5.5 | 5.2 (2), 5.6 (2), 5.7 (4), 5.20 (2) | 4 | 0 | 5.2 |
| Brevundimonas vesicularis 5.6 | 5.1 (2), 5.3 (2), 5.5 (2), 5.7 (2), 5.8 (1), 5.10 (1), 5.13 (4) | 7 | 0 | 4.4 |
| Methylobacterium sp. strain 5.7 | 5.1 (3), 5.2 (3), 5.3 (2), 5.5 (3), 5.6 (2), 5.8 (3), 5.9 (2), 5.10 (2), 5.11 (2), 5.13 (1) | 10 | 1 | 88.5 |
| Sphingomonas sp. strain 5.8 | 5.6 (1), 5.7 (3), 5.9 (3), 5.10 (4), 5.17 (1) | 5 | 0 | 0.6 |
| Bradyrhizobium japonicum 5.9 | 5.2 (2), 5.3 (1), 5.7 (2), 5.8 (3), 5.10 (3), 5.11 (1), 5.13 (1), 5.17 (3) | 8 | 0 | 0.3 |
| Pseudoxanthomonas mexicana 5.10 | 5.3 (2), 5.6 (1), 5.7 (2), 5.8 (4), 5.9 (3), 5.11 (1), 5.20 (1) | 7 | 0 | 3.9 |
| Mycobacterium mucogenicum 5.11 | 5.7 (2), 5.9 (1), 5.10 (1), 5.17 (2) | 4 | 2 | 61.6 |
| Roseomonas gilardii 5.12 | 5.3 (2), 5.13 (2), 5.20 (2) | 3 | 0 | 2.1 |
| Sphingomonas sp. strain 5.13 | 5.1 (1), 5.2 (1), 5.3 (2), 5.7 (1), 5.9 (1), 5.12 (2) | 6 | 0 | 3.1 |
| Micrococcus luteus 5.14 | 5.6 (4), 5.20 (1) | 2 | 0 | 73.0 |
| Mycobacterium mucogenicum 5.15 | None | 0 | 2 | 99.0 |
| Methylobacterium nodulans 5.16 | None | 0 | 0 | 10.2 |
| Methylobacterium fujisawae 5.17 | 5.1 (3), 5.8 (1), 5.9 (3), 5.11 (2) | 4 | 1 | 26.4 |
| Staphylococcus hominis 5.20 | 5.1 (1), 5.5 (2), 5.10 (1), 5.12 (2), 5.14 (1) | 5 | 0 | 35.5 |
Coaggregation interactions were determined for strains from within each sampled community. If autoaggregation was present, the autoaggregation score was deducted to give the actual coaggregation score.
Of the 50 biofilm strains studied, a total of 20 (40%) autoaggregated. Species that autoaggregated included M. mucogenicum, Methylobacterium fujisawae, and Acidovorax sp. The proportion of biofilm bacteria that were able to autoaggregate was inversely related to the shear rate imposed on the biofilm community. In the biofilm that developed at the lowest shear rate (<0.1 S−1), 4 of 17 strains (24%) autoaggregated. In the biofilm that developed at the highest shear rate (305 S−1), five of six strains (83%) autoaggregated.
The surface hydrophobicity of each strain could also be related to the biofilm from which it was isolated. As the shear rate increased, so too did the proportion of bacteria that possessed a high level of surface hydrophobicity (>75%). In general, strains that autoaggregated also possessed a high level of surface hydrophobicity and included M. mucogenicum, Methylobacterium sp., and Acidovorax sp.
Unlike the findings for autoaggregation and surface hydrophobicity, the proportion of coaggregating biofilm strains did not increase consistently with increasing shear rate. Overall, 39 of the 50 biofilm strains studied coaggregated with at least one other partner strain. The biofilm community with the highest proportion of coaggregating bacteria developed at an intermediate shear rate (122 S−1). At 122 S−1, all of the culturable strains from this community were able to coaggregate with at least one other strain from this community (Table 4). The proportion of bacteria that coaggregated was smaller in biofilms developed at either high shear rates (2 of 6 strains at 305 S−1) or low shear rates (15 of 17 strains at <0.1 S−1). In the CCR unit, therefore, the optimum fluid velocity to select for coaggregating bacteria in freshwater biofilm communities was 122 S−1.
Few coaggregation and autoaggregation interactions were detected between bacteria from planktonic communities. Only 6 of 16 strains coaggregated to give a total of four coaggregation partnerships (Table 5). The most promiscuous coaggregating strains were B. japonicum 6.2 and Sphingomonas arimaticavoran 7.3. Four strains autoaggregated, and M. mucogenicum 6.1 gave the highest visual autoaggregation score (a score of 2). Like the observations for the strains from the biofilm communities, the planktonic strains that autoaggregated also possessed a high level of surface hydrophobicity.
TABLE 5.
Visual coaggregation and autoaggregation scores for planktonic strains after growth on R2A agar for 72 h
| Strain | Coaggregation partner (score)a | Total no. of coaggregating partners | Autoaggregation score | % Whole-cell hydrophobicity |
|---|---|---|---|---|
| Dyadobacter fermentens 0.1 | None | 0 | 0 | 11.1 |
| Dyadobacter fermentens 0.2 | None | 0 | 0 | 18.7 |
| Variovorax paradoxus 0.3 | 7.3 (2) | 1 | 1 | 14.5 |
| Methylobacterium extorquens 0.5 | None | 0 | 0 | 38.2 |
| Hymenobacter sp. strain 0.6 | None | 0 | 0 | 5.1 |
| Variovorax paradoxus 0.7 | 6.2 (1) | 1 | 0 | 6.4 |
| Mycobacterium mucogenicum 6.1 | 6.5 (2) | 1 | 2 | 87.4 |
| Bradyrhizobium japonicum 6.2 | 0.7 (1), 7.3 (1) | 2 | 0 | 4.0 |
| Brevundimonas vesicularis 6.3 | None | 0 | 0 | 8.8 |
| Variovorax paradoxus 6.4 | None | 0 | 1 | 23.5 |
| Flavobacterium sp. strain 6.5 | 6.1 (2) | 1 | 0 | 1.5 |
| Paenibacillus amylolyticus 7.2 | None | 0 | 0 | 18.1 |
| Sphingomonas aromaticivoran 7.3 | 0.3 (2), 6.2 (1) | 2 | 0 | 16.9 |
| Sphingomonas abikonensis 7.4 | None | 0 | 0 | 5.8 |
| Methylobacterium sp. strain 7.5 | None | 0 | 1 | 92.7 |
| Variovorax paradoxus 7.6 | None | 0 | 0 | 6.2 |
If autoaggregation was present, the autoaggregation score was deducted to give the actual coaggregation score.
Coaggregation interactions between strains from biofilms or the planktonic phase were predominantly intergeneric (occurring between members of two genera). Only three intrageneric coaggregations were detected, between Sphingomonas melonis 3.3 and S. yanoikuyae 3.9 (a visual score of 2), between Methylobacterium nodulans 5.2 and Methylobacterium sp. strain 5.7 (a visual score of 3), and between Sphingomonas sp. strain 5.3 and Sphingomonas sp. strain 5.13 (a visual score of 2). Intraspecies coaggregation (between members of the same species) was not detected.
DISCUSSION
Shear rates over surfaces directly influence the bacterial composition and diversity of multispecies biofilms that form on them. As biofilm diversity decreased with increasing fluid shear, the proportion of bacteria that autoaggregated and coaggregated was altered dramatically. The proportion of autoaggregating bacteria within a biofilm was shown to be inversely related to the shear rate. The proportion of coaggregating bacteria, however, was maximal only at an intermediate fluid flow velocity. Collectively, these findings support the suggestions by Cloete et al. (9) and Rickard et al. (35) that fluid flow velocity (and associated shear rate) moderates biofilm diversity and that the ability to aggregate is an important determinant of biofilm formation at high shear rates.
The experiments performed in the current study involved the use of a CCR to simulate shear rates 50 μm from the surface of a rotating stainless steel surface that were equivalent to those (50 to 300 s−1) suggested to occur within the infrastructure and channels of mature aquatic biofilms (2). The results obtained for coaggregation and community population dynamics therefore can be considered to relate to occurrences within polymicrobial freshwater communities. Shear rates have been suggested to be important in the development of biofilm community structure (3) and govern the abilities of individual species to immigrate to biofilms and to colonize new surfaces (9, 45). Other factors include substratum composition, concentrations of solutes, and nutrient availability (6, 19). Our data are in agreement with those of Soini et al. (42) and indicated that an increase in the fluid shear rate from approximately 0 to 305 S−1 reduced the total cell number by only 10-fold (from approximately 2.5 × 106 CFU/cm2 to 1 × 105 CFU/cm2). However, as shown by DGGE and culturing on R2A agar, the total diversity was also dramatically reduced. Biofilms were much less diverse at high shear rates (198 and 305 S−1) than at low shear rates (<0.1 S−1). Furthermore, the only strains that did not possess 16S rRNA gene sequences with significant identities (<94%) to known sequences in the EMBL database were isolated at the higher shear rates (strains 1.4, 1.5, 2.8, and 2.9; Fig. 3). From these data, it is clear that increasing fluid shear rates select for different species of biofilm-forming bacteria that would not necessarily be present elsewhere.
There are numerous mechanisms by which bacteria can attach to surfaces and integrate into biofilm communities in flowing environments (5, 22, 36, 46). Mechanisms that have recently received much interest include coaggregation and autoaggregation abilities and bacterial whole-cell hydrophobicity. The data presented here suggest that all three factors are important for colonization and biofilm development in flowing environments. However, autoaggregation ability and whole-cell hydrophobicity were the only two cellular properties that increased uniformly as the shear rate increased. At high shear rates (198 and 305 S−1), a larger proportion of bacteria possessed a high level of whole-cell hydrophobicity and were able to autoaggregate (Table 4). The largest proportion of bacteria that were able to coaggregate was seen at the intermediate fluid shear rate of 122 S−1. Therefore, it is likely that autoaggregation interactions, which are enhanced by whole-cell hydrophobicity (10, 21, 24), are stronger than coaggregation interactions and directly aid in cell-surface and cell-cell attachment and colonization at high shear rates. As such, the failure to integrate into a biofilm relates to shear conditions in which adhesive forces are weaker than dispersive ones. Consequently, more taxonomically diverse biofilms developed at lower shear rates, and these contained a high proportion of coaggregating bacteria (Table 4). It is possible that while coaggregation only weakly enhances cell-cell attachment (compared to autoaggregation), it mediates the juxtapositioning of species next to favorable partner species within taxonomically diverse biofilms. The recognition of specific species of bacteria through coaggregation is hypothesized to be crucial for improved growth and cellular division and can lead to luxuriant interdigitated growth (30). Such a mutualistic strategy is selected against at high shear rates; instead, an autoaggregative biofilm community develops.
An overall comparison of the bacterial diversity of the biofilm and planktonic communities by DGGE and culturing on R2A agar indicated that some strains were not cultured. As a consequence, the aggregation ability of the unculturable strains could not be studied. It is possible that these strains were able to coaggregate and/or autoaggregate. In support of this suggestion, it is clear that many of these strains that were clearly detected by DGGE, as shown by strongly stained bands (Fig. 2), were also detected on R2A agar as numerically dominant species within each biofilm community and possessed co- and/or autoaggregation properties that may have enhanced their ability to be members of each biofilm. For example, DGGE detected three Mycobacterium spp. in biofilm communities, and these were shown to be present by culturing and to possess autoaggregation and coaggregation abilities. The large numbers of genetically distinct aggregative Mycobacterium spp. that were detected by both culturing and culture-independent methods also highlight the importance of this genus with respect to its pathogenic ability and its ability to reside in biofilms (13).
Biofilms in water systems cause problems ranging from surface corrosion to support of the survival and growth of pathogens (18). Mycobacterium spp. and Flavobacterium spp. are known opportunistic pathogens in humans (26, 32), and members of both of these genera were isolated from biofilms exposed to different flow velocities (Fig. 2). M. mucogenicum has frequently been isolated from potable freshwater systems, and previous work by Falkinham et al. (13) indicated that Mycobacterium spp. (including M. mucogenicum) are common to freshwater biofilms exposed to a variety of flow velocities (and associated shear rates). In this study, it was shown that the majority of Mycobacterium spp. coaggregated with at least one partner and that all autoaggregated. As such, the abilities to autoaggregate and coaggregate may enhance the ability of the species to promiscuously integrate into biofilms developing at different fluid shear rates. Less is known about Flavobacterium spp. in freshwater systems, although Schmeisser et al. (41) and Tall et al. (47) identified strains in potable and dental water lines. The results shown here indicate that Flavobacterium spp. were present only in biofilms developing at intermediate and high shear rates of 122 and 198 S−1 (Fig. 2) and were able to coaggregate but not to autoaggregate (Table 3). It is possible that the inability of Flavobacterium spp. to autoaggregate contributed to their absence at high shear rates because there were fewer species with which to coaggregate and that the physiochemical interactions that mediate autoaggregation are stronger than those that mediate coaggregation. The strengths of coaggregation and autoaggregation interactions and their respective roles in biofilm development are currently being studied further.
In conclusion, we have demonstrated a relationship between shear rate and freshwater multispecies biofilm diversity. The proportions of bacteria that coaggregate and autoaggregate in biofilms also change in relation to shear rates. Circumstantial evidence suggests that it is likely that such cell-cell interactions aid in the integration of bacteria in flowing environments.
Acknowledgments
We thank the Society for Applied Microbiology (Bedford, United Kingdom) for financial assistance and support for Amy T. Stead.
We thank Sharon Lindsay and Ruth Ledder (University of Manchester, Manchester, United Kingdom) for technical assistance.
REFERENCES
- 1.Amann, R. I., J. Stromley, R. Devereux, R. Key, and D. A. Stahl. 1992. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl. Environ. Microbiol. 58:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakker, D. P., A. van der Plaats, G. J. Verkerke, H. J. Busscher, and H. C. van der Mei. 2003. Comparison of velocity profiles for different flow chamber designs used in studies of microbial adhesion to surfaces. Appl. Environ. Microbiol. 69:6280-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyenal, H., and Z. Lewandowski. 2002. Internal and external mass transfer in biofilms grown at various flow velocities. Biotechnol. Prog. 18:55-61. [DOI] [PubMed] [Google Scholar]
- 4.Bott, T. R., and D. M. Grant. 2001. Biofilms in flowing systems. Methods Enzymol. 337:88-103. [DOI] [PubMed] [Google Scholar]
- 5.Busscher, H. J., and H. C. van der Mei. 1997. Physico-chemical interactions in initial microbial adhesion and relevance for biofilm formation. Adv. Dent. Res. 11:24-32. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. C., M. Le Puil, J. Biggerstaff, A. A. Randall, A. Schulte, and J. S. Taylor. 2003. Direct estimation of biofilm density on different pipe material coupons using a specific DNA-probe. Mol. Cell. Probes 17:237-243. [DOI] [PubMed] [Google Scholar]
- 7.Christersson, C. E., R. G. Dunford, P. O. Glantz, and R. E. Baier. 1989. Effect of critical surface tension on retention of oral microorganisms. Scand. J. Dent. Res. 97:247-256. [DOI] [PubMed] [Google Scholar]
- 8.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24:742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloete, T. E., D. Westaard, and S. J. van Vuuren. 2003. Dynamic response of biofilm to pipe surface and fluid velocity. Water Sci. Technol. 47:57-59. [PubMed] [Google Scholar]
- 10.Del Re, B., B. Sgorbati, M. Miglioli, and D. Palenzona. 2000. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 31:438-442. [DOI] [PubMed] [Google Scholar]
- 11.Duddridge, J. E., C. A. Kent, and J. F. Laws. 1982. Effect of surface shear stress on the attachment of Pseudomonas fluorescens to stainless steel under defined flow conditions. Biotechnol. Bioeng. 24:153-164. [DOI] [PubMed] [Google Scholar]
- 12.Dumbleton, B. 1995. A question of scale and time. Water Waste Treat. 38:39-47. [Google Scholar]
- 13.Falkinham, J. O., C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiorina, J. C., M. Weber, and J. C. Block. 2000. Occurrence of lectins and hydrophobicity of bacteria obtained from biofilm of hospital catheters and water pipes. J. Appl. Microbiol. 89:494-500. [DOI] [PubMed] [Google Scholar]
- 15.Handley, P. S., A. H. Rickard, N. J. High, and S. A. Leach. 2001. Coaggregation: is it a universal biofilm phenomenon? p. 1-10. In P. Gilbert, D. Allison, M. Brading, J. Verran, and J. Walker (ed.), Biofilm interactions: chance or necessity? Bioline Press, Cardiff, United Kingdom.
- 16.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y.
- 17.Kang, Y., H. Liu, S. Genin, M. A. Schell, and T. P. Denny. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46:427-437. [DOI] [PubMed] [Google Scholar]
- 18.Kerr, C. J., K. S. Osborn, A. H. Rickard, G. D. Robson, and P. S. Handley. 2003. Biofilms in water distribution systems, p. 757-776. In D. Mara and N. J. Horan (ed.), Water and wastewater engineering. Academic Press Ltd., London, United Kingdom.
- 19.Kerr, C. J., K. S. Osborne, G. D. Robson, and P. S. Handley. 1998. The relationship between pipe material and biofilm formation in a laboratory model system. J. Appl. Microbiol. 85:29S-38S. [DOI] [PubMed] [Google Scholar]
- 20.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kos, B., J. Suskovic, S. Vukovic, M. Simpraga, J. Frece, and S. Matosic. 2003. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94:981-987. [DOI] [PubMed] [Google Scholar]
- 22.Lejeune, P. 2003. Contamination of abiotic surfaces: what a colonizing bacterium sees and how to blur it. Trends Microbiol. 11:179-184. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y., and J. H. Tay. 2002. The essential role of hydrodynamic shear force in the formation of biofilm and granular sludge. Water Res. 36:1653-1665. [DOI] [PubMed] [Google Scholar]
- 24.Ljungh, A., S. Hjerten, and T. Wadstrom. 1985. High surface hydrophobicity of autoaggregating Staphylococcus aureus strains isolated from human infections studied with the salt aggregation test. Infect. Immun. 47:522-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik, A., and K. Kakii. 2003. Intergeneric coaggregations among Oligotropha carboxidovorans and Acinetobacter species present in activated sludge. FEMS Microbiol. Lett. 224:23-28. [DOI] [PubMed] [Google Scholar]
- 26.Manfredi, R., A. Nanetti, M. Ferri, A. Mastroianni, O. V. Coronado, and F. Chiodo. 1999. Flavobacterium spp. organisms as opportunistic bacterial pathogens during advanced HIV disease. J. Infect. 39:146-152. [DOI] [PubMed] [Google Scholar]
- 27.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, A. H. Rickard, S. A. Symmons, and P. Gilbert. 2003. Microbial characterization of biofilms in domestic drains and the establishment of stable biofilm microcosms. Appl. Environ. Microbiol. 69:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muyzer, G. 1999. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 29.Norton, C. D., and M. W. LeChevallier. 2000. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl. Environ. Microbiol. 66:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payment, P., L. Richardson, J. Siemiatycki, R. Dewar, M. Edwardes, and E. Franco. 1991. A randomized trial to evaluate the risk of gastrointestinal disease due to consumption of drinking water meeting current microbiological standards. Am. J. Public Health 81:703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips, M. S., and C. F. von Reyn. 2001. Nosocomial infections due to nontuberculous mycobacteria. Clin. Infect. Dis. 33:1363-1374. [DOI] [PubMed] [Google Scholar]
- 33.Purevdorj, B., J. W. Costerton, and P. Stoodley. 2000. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 68:4457-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rickard, A. H., A. J. McBain, R. G. Ledder, P. S. Handley, and P. Gilbert. 2003. Coaggregation between freshwater bacteria within biofilm and planktonic communities. FEMS Microbiol. Lett. 220:133-140. [DOI] [PubMed] [Google Scholar]
- 36.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 37.Rickard, A. H., S. A. Leach, L. S. Hall, C. M. Buswell, N. J. High, and P. S. Handley. 2002. Phylogenetic relationships and coaggregation ability of freshwater biofilm bacteria. Appl. Environ. Microbiol. 68:3644-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rickard, A. H., S. A. Leach, C. M. Buswell, N. J. High, and P. S. Handley. 2000. Coaggregation between aquatic bacteria is mediated by specific-growth-phase-dependent lectin-saccharide interactions. Appl. Environ. Microbiol. 66:431-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridgway, H. F., and B. H. Olson. 1981. Scanning electron microscope evidence for bacterial colonization of a drinking-water distribution system. Appl. Environ. Microbiol. 41:274-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg, M., and E. Rosenberg. 1981. Role of adherence in growth of Acinetobacter calcoaceticus RAG-1 on hexadecane. J. Bacteriol. 148:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmeisser, C., C. Stockigt, C. Raasch, J. Wingender, K. N. Timmis, D. F. Wenderoth, H. C. Flemming, H. Liesegang, R. A. Schmitz, K. E. Jaeger, and W. R. Streit. 2003. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 69:7298-7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soini, S. M., K. T. Koskinen, M. J. Vilenius, and J. A. Puhakka. 2002. Effects of fluid-flow velocity and water quality on planktonic and sessile microbial growth in water hydraulic system. Water Res. 36:3812-3820. [DOI] [PubMed] [Google Scholar]
- 43.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 44.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 45.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1999. The formation of migratory ripples in a mixed species bacterial biofilm growing in turbulent flow. Environ. Microbiol. 1:447-455. [DOI] [PubMed] [Google Scholar]
- 46.Strevett, K. A., and G. Chen. 2003. Microbial surface thermodynamics and applications. Res. Microbiol. 154:329-335. [DOI] [PubMed] [Google Scholar]
- 47.Tall, B. D., H. N. Williams, K. S. George, R. T. Gray, and M. Walch. 1995. Bacterial succession within a biofilm in water supply lines of dental air-water syringes. Can. J. Microbiol. 41:647-654. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTALX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van de Peer, Y., and R. De Watcher. 1997. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput. Appl. Biosci. 13:227-230. [DOI] [PubMed] [Google Scholar]
- 50.Walker, J. T., A. Sonesson, C. W. Keevil, and D. C. White. 1993. Detection of Legionella pneumophila in biofilms containing a complex microbial consortium by gas chromatography-mass spectrometry analysis of genus-specific hydroxy fatty acids. FEMS Microbiol. Lett. 113:139-144. [DOI] [PubMed] [Google Scholar]
- 51.Weisberg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willcock, L., P. Gilbert, J. Holah, G. Wirtanen, and D. G. Allison. 2000. A new technique for the performance evaluation of clean-in-place disinfection of biofilms. J. Ind. Microbiol. Biotechnol. 25:235-241. [Google Scholar]
- 53.Wilson, K. H., R. B. Blitchington, and R. C. Greene. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]