Abstract
Low-sulfate, acidic (approximately pH 4) fens in the Lehstenbach catchment in the Fichtelgebirge mountains in Germany are unusual habitats for sulfate-reducing prokaryotes (SRPs) that have been postulated to facilitate the retention of sulfur and protons in these ecosystems. Despite the low in situ availability of sulfate (concentration in the soil solution, 20 to 200 μM) and the acidic conditions (soil and soil solution pHs, approximately 4 and 5, respectively), the upper peat layers of the soils from two fens (Schlöppnerbrunnen I and II) of this catchment displayed significant sulfate-reducing capacities. 16S rRNA gene-based oligonucleotide microarray analyses revealed stable diversity patterns for recognized SRPs in the upper 30 cm of both fens. Members of the family “Syntrophobacteraceae” were detected in both fens, while signals specific for the genus Desulfomonile were observed only in soils from Schlöppnerbrunnen I. These results were confirmed and extended by comparative analyses of environmentally retrieved 16S rRNA and dissimilatory (bi)sulfite reductase (dsrAB) gene sequences; dsrAB sequences from Desulfobacca-like SRPs, which were not identified by microarray analysis, were obtained from both fens. Hypotheses concerning the ecophysiological role of these three SRP groups in the fens were formulated based on the known physiological properties of their cultured relatives. In addition to these recognized SRP lineages, six novel dsrAB types that were phylogenetically unrelated to all known SRPs were detected in the fens. These dsrAB sequences had no features indicative of pseudogenes and likely represent novel, deeply branching, sulfate- or sulfite-reducing prokaryotes that are specialized colonists of low-sulfate habitats.
The dissimilatory reduction of sulfate is carried out exclusively by prokaryotic organisms and is one of the most important mineralization processes in anoxic aquatic environments, especially marine sediments (29, 30). In contrast to well-studied sulfate-reducing communities in marine (18, 19, 38, 41, 53, 56, 57, 72) and freshwater habitats (39, 40, 59, 60), relatively little is known about the distribution, diversity, and in situ activities of sulfate-reducing prokaryotes (SRPs) in terrestrial ecosystems. The contribution of terrestrial SRPs to the overall turnover of organic matter is likely of minor importance on a global scale. However, SRPs contribute to the biodegradation of pollutants in soils and subsurface environments (1, 15, 49, 71) and are important to the geomicrobiology of specialized terrestrial habitats that are subject to flooding, such as rice fields (68, 76, 77) and fens (3, 5).
δ34S values and 35S-labeling patterns indicate that the dissimilatory reduction of sulfate is an ongoing process in the acidic fens of a forested catchment in northern Bavaria, Germany (Lehstenbach, Fichtelgebirge) (3, 5). The deposition of sulfur that originated from the combustion of soft coal in Eastern Europe (10) led to accumulation of sulfur in the soils of this catchment (4). Although pollution controls have lessened the deposition in recent years, desorption of sulfate in aerated upland soils causes sulfate to enter fens at lower elevations. It was hypothesized that the dissimilatory reduction of sulfate in these mainly anoxic, waterlogged acidic fen soils (the pH of the fen soils is approximately 4) contributes to the retention of sulfur in this ecosystem (3, 4, 50). The reduction of sulfate in these fens is also a sink for protons and thus decreases the acidity of the soil solution and groundwater of this habitat.
The acidity and low sulfate content of some of the fens in the Lehstenbach catchment provide an unusual habitat for SRPs, and the occurrence and activity of these organisms in such habitats have received little attention. The main objectives of this study were (i) to assess the capacity of the fen soils to reduce sulfate along vertical soil profiles in the upper peat layers, (ii) to determine the vertical community profiles for all known SRP lineages that inhabit the fens by the use of a 16S rRNA-based oligonucleotide microarray (SRP-PhyloChip) (44), (iii) to resolve the possible existence of novel SRP lineages in the fens by retrieval of dsrAB, which are genes that encode the alpha and beta subunits of the siroheme dissimilatory (bi)sulfite reductase (EC 1.8.99.3) (34, 66, 74), and (iv) to deduce the possible in situ functional relationships that can be inferred from this collective information.
MATERIALS AND METHODS
Site description.
The two low-moor fens, designated Schlöppnerbrunnen I (50°08′14"N, 11°53′07"E) and Schlöppnerbrunnen II (50°08′38"N, 11°51′41"E), that were investigated are in the Lehstenbach catchment in the Fichtelgebirge mountains in northeastern Bavaria (Germany). The catchment has an area of 4.2 km2, and the highest elevation is 877 m above sea level. Ninety percent of the area is covered with Norway spruce (Picea abies [L.] Karst.) of different ages. Upland soils in the catchment are not water saturated, have developed from weathered granitic bedrock, and are predominantly cambisols and cambic podsols (according to the Food and Agriculture Organization system). Considerable parts of the catchment (approximately 30%) are covered by minerotrophic fens or intermittent seeps. The annual precipitation in the catchment is 900 to 1,160 mm, and the average annual temperature is 5°C.
Schlöppnerbrunnen I is covered with patches of Sphagnum moss and spruce, and the soil is a fibric histosol and is usually water saturated; in years with extremely hot summer months, the upper soil can become dry. Schlöppnerbrunnen II is permanently water saturated and completely overgrown by the grass Molinia caerula. The soil of Schlöppnerbrunnen II has a larger amount of bioavailable Fe3+ than the soil of Schlöppnerbrunnen I has. The soil pHs of Schlöppnerbrunnen I and II were approximately 3.9 and 4.2, respectively; the soil solution pH varied between 4 and 6.
Dialysis chambers.
A soil solution from the upper 40 cm of each site was sampled with dialysis chambers (27) every 2 months from July 2001 to November 2002. Each dialysis chamber consisted of 40 1-cm cells covered with a cellulose acetate membrane with a pore diameter of 0.2 μm. Prior to installation, the chamber was filled with anoxic, deionized water. The dialysis chambers were placed in the water-saturated fens for 2 weeks prior to sampling. On the sampling date, each chamber was closed (i.e., made airtight), transported to the laboratory, and sampled with argon-flushed syringes.
Collection of soil.
For microcosms, soil samples from three different depths (approximately 0 to 10, 10 to 20, and 20 to 30 cm) were obtained in December 2001 in sterile airtight vessels, transported to the laboratory, and processed within 4 h. For isolation of DNA, soil cores (diameter, 3 cm) from four different depths (approximately 0 to 7.5, 7.5 to 15, 15 to 22.5, and 22.5 to 30 cm) were collected on 24 July 2001 and immediately cooled on ice. Soil samples were brought to the laboratory, where they were diluted 1:1 (vol/vol) in phosphate-buffered saline (130 mM NaCl, 10 mM NaH2PO4, 10 mM Na2HPO4; pH 7.3), homogenized by vortexing, and stored at −20°C.
Anoxic microcosms.
Thirty-gram (fresh weight) portions of soil were placed into 125-ml infusion flasks (Merck ABS, Dietikon, Switzerland) inside an O2-free chamber (100% N2 gas phase), and 60-ml portions of anoxic, deionized water were added to facilitate sampling with sterile, argon-flushed syringes. The bottles were closed with rubber stoppers and screw caps and were incubated in the dark at 15°C. Sulfate was added from a sterile anoxic stock solution (0.5 M K2SO4) to a final concentration of 500 μM. Microcosms were prepared in triplicate and had initial pHs that ranged from 5 to 5.5. The rates of sulfate consumption and CH4 formation were determined for 17 days by linear regression analysis of the concentrations of sulfate and CH4, respectively. For CH4 formation rates, all concentration data were included for linear regression analysis. For sulfate consumption rates, only the parts of the curves for when part of the supplemented sulfate was still available for consumption were used.
Analytical methods.
pH was measured with a U457-S7/110 combination pH electrode (Ingold, Steinbach, Germany). The sulfate content was determined by ion chromatography (37). The concentration of CH4 in the headspace was measured with a 5980 series II gas chromatograph (Hewlett-Packard Co., Palo Alto, Calif.) (37). Total reduced inorganic sulfur (TRIS) and acid volatile sulfur (AVS) contents were determined by using previously described protocols (73). TRIS is assumed to be composed of pyrite (FeS2), amorphous FeS, and S0; AVS is amorphous FeS.
Extraction of DNA.
DNA was extracted from soil homogenates by a modification (44) of a previously described protocol (25). The amount of extracted DNA was determined spectrophotometrically by measuring the absorbance at 260 nm.
PCR amplification of 16S rRNA genes and dsrAB.
PCR amplification of genes was performed with 5 ng of environmental DNA. For subsequent microarray hybridization, bacterial 16S rRNA gene fragments from soil genomic DNA were amplified by using the primer pairs 616V-630R and 616V-1492R (Table 1), and PCR products were mixed prior to labeling. For confirmation of microarray results, 16S rRNA gene fragments of defined SRP groups were directly amplified from soil DNA by using previously described and newly designed primers (Table 1). In addition, an approximately 1.9-kb dsrAB fragment was amplified from fen soil DNA by using the degenerate primers DSR1Fmix (equimolar mixture of DSR1F, DSR1Fa, and DSR1Fb) and DSR4Rmix (equimolar mixture of DSR4R, DSR4Ra, DSR4Rb, and DSR4Rc) (Table 2).
TABLE 1.
16S rRNA gene-targeted primers
| Short namea | Full nameb | Annealing temp (°C) | Sequence (5′-3′) | Specificity | Reference |
|---|---|---|---|---|---|
| 616V | S-D-Bact-0008-a-S-18 | 52 | AGA GTT TGA TYM TGG CTC | Most Bacteria | 32 |
| 630R | S-D-Bact-1529-a-A-17 | 52 | CAK AAA GGA GGT GAT CC | Most Bacteria | 32 |
| 1492R | S-*-Proka-1492-a-A-19 | 52, 60c | GGY TAC CTT GTT ACG ACT T | Most Bacteria and Archaea | 33 |
| ARGLO36F | S-G-Arglo-0036-a-S-17 | 52 | CTA TCC GGC TGG GAC TA | Archaeoglobus spp. | 44 |
| DSBAC355F | S-*-Dsb-0355-a-S-18 | 60 | CAG TGA GGA ATT TTG CGC | Most “Desulfobacterales” and “Syntrophobacterales” | 61 |
| DSMON85F | S-G-Dsmon-0085-a-S-20 | 62 | CGG GGT RTG GAG TAA AGT GG | Desulfomonile spp. | This study |
| DSMON1419R | S-G-Dsmon-1419-a-A-20 | 62 | CGA CTT CTG GTG CAG TCA RC | Desulfomonile spp. | This study |
| SYBAC+282F | S-*-Sybac-0282-a-S-18 | 60 | ACG GGT AGC TGG TCT GAG | “Syntrophobacteraceae” and some other Bacteria | This study |
| SYBAC1427R | S-*-Sybac-1427-a-A-18 | 60 | GCC CAC GCA CTT CTG GTA | “Syntrophobacteraceae” | This study |
| DBACCA65F | S-S-Dbacca-0065-a-S-18 | 58 | TAC GAG AAA GCC CGG CTT | Desulfobacca acetoxidans | This study |
| DBACCA1430R | S-S-Dbacca-1430-a-A-18 | 58 | TTA GGC CAG CGA CAT CTG | Desulfobacca acetoxidans | This study |
Short name used in the reference or in this study.
Name of 16S rRNA gene-targeted oligonucleotide primer based on established nomenclature (6).
The annealing temperature was 52°C when the primer was used with forward primer 616V or ARGLO36F, and the annealing temperature was 60°C when the primer was used with forward primer DSBAC355F.
TABLE 2.
Dissimilatory (bi)sulfite reductase gene (dsrAB)-targeted primersa
| Primer | Sequence (5′-3′) | Specificity | Reference |
|---|---|---|---|
| DSR1Fb | ACS CAC TGG AAG CAC G | Archaeoglobus fulgidus, Archaeoglobus profundus, Desulfovibrio vulgaris | 74 |
| DSR1Fab | ACC CAY TGG AAA CAC G | Desulfotomaculum thermocisternum, Desulfobulbus rhabdoformis, Desulfobacter vibrioformis | This study |
| DSR1Fbb | GGC CAC TGG AAG CAC G | Thermodesulforhabdus norvegica | This study |
| DSR4Rb | GTG TAG CAG TTA CCG CA | Archaeoglobus fulgidus, Desulfovibrio vulgaris, Desulfobulbus rhabdoformis | 74 |
| DSR4Rab | GTG TAA CAG TTT CCA CA | Archaeoglobus profundus | This study |
| DSR4Rbb | GTG TAA CAG TTA CCG CA | Desulfobacter vibrioformis | This study |
| DSR4Rcb | GTG TAG CAG TTK CCG CA | Thermodesulforhabdus norvegica, Desulfotomaculum thermocisternum | This study |
| DSR978Fac | GGT CAT CGA CCT TTG TCC | Schlöppnerbrunnen I soil OTU 5 | This study |
| DSR978Fbc | CGT CGT CGG GAA GTG CCC | Schlöppnerbrunnen I soil OTU 8 | This study |
| DSR978Fcc | AGT AGT CGA CCT TTG CCC | Schlöppnerbrunnen I and II soil OTU 6 | This study |
| DSR978Fdc | TGT CAC CGA TCT CTG CCC | Schlöppnerbrunnen I soil OTU 1 | This study |
| DSR978Fec | TGT TAC CGA CCT CTG CCC | Schlöppnerbrunnen II soil OTU 1 (dsrSbII-20) | This study |
| DSR978Ffc | TGT CAC CGA TCT TTG CCC | Schlöppnerbrunnen II soil OTU 4 (dsrSbII-15) | This study |
| DSR978Fgc | CGT CAC CAT TCT CTG CCC | Schlöppnerbrunnen II soil OTU 4 (dsrSbII-9) | This study |
| DSR978Fhc | GGT CGT TGA CAT GTG TCC | Schlöppnerbrunnen II soil OTU 11 | This study |
| DSR978Fic | GGT CTG CAA TCT CTG YCC | Schlöppnerbrunnen I and II soil OTU 2 and 3 | This study |
| DSR978Fjc | GGT TGT TGA CCT TTG CCC | Schlöppnerbrunnen I soil OTU 9 | This study |
| DSR978Fkc | CGT TTG CGA TCT CTG CCC | Schlöppnerbrunnen II soil OTU 7 | This study |
| DSR860Fc | AGA TCC GGC GGG ACG ATG | Schlöppnerbrunnen I soil OTU 10 | This study |
The target sites of all DSR1 and DSR4 primers were analyzed for the SRPs (n = 7) for which complete dsrAB operons were available in the GenBank database (9). SRPs with a target sites fully complementary to the primers are indicated.
The primer was used under nonstringent conditions by using an annealing temperature of 48°C for PCR in order to target a wide diversity of SRPs.
Internal sequencing primer used to complete dsrAB sequences retrieved from acidic fen sites at Schlöppnerbrunnen I and II.
Both positive controls (purified DNA from suitable reference organisms) and negative controls (no DNA) were included in all PCR amplification experiments. For 16S rRNA gene and dsrAB amplifications, reaction mixtures (total volume, 50 μl) containing each primer at a concentration of 25 pM were prepared by using 10× Ex Taq reaction buffer and 2.5 U of Ex Taq polymerase (Takara Biomedicals, Otsu, Shiga, Japan). Additionally, 20 mM tetramethylammonium chloride (Sigma, Deisenhofen, Germany) was added to each amplification mixture to enhance the specificity of the PCR (35). Thermal cycling was carried out by using an initial denaturation step at 94°C for 1 min, followed by 30 (16S rRNA genes) or 35 cycles (dsrAB) of denaturation at 94°C for 40 s, annealing at temperatures from 48 to 62°C (depending on the primer pair [Tables 1 and 2]) for 40 s, and elongation at 72°C for 1.5 min. The cycling was completed by a final elongation step at 72°C for 10 min. The presence and sizes of the amplification products were determined by agarose (1%) gel electrophoresis. Ethidium bromide-stained bands were digitally recorded with a video documentation system (Cybertech, Hamburg, Germany).
DNA microarray analyses.
Fluorescence labeling of PCR products, manufacturing and processing of SRP-PhyloChips, reverse hybridization on microarrays, and scanning and image analyses of microarrays were performed as previously described (44). Spots that had signal-to-noise ratios equal to or greater than 2.0 were considered positive. Oligonucleotides used for printing of the SRP-PhyloChips were obtained from MWG Biotech (Ebersberg, Germany). For each site and soil depth, two separate microarrays with duplicate spots for each probe were hybridized with labeled PCR products. The sequences and specificities of all probes are listed in the DNA microarray section of the probeBase website (43) http://www.microbial-ecology.net/probebase/.
Microarray hybridization patterns for the different depths of the samples from the two fens were used to infer binary similarities in order to provide a quantitative measure for comparison of hybridization data. The Jaccard coefficient (CJ) and the Sorenson coefficient (CS) for two samples were calculated by using the following formulas: CJ = 100 × c × (a + b − c)−1 and CS = 100 × 2c × (a + b)−1, where a is the number of positive SRP-PhyloChip probes in the first sample, b is the number of positive SRP-PhyloChip probes in the second sample, and c is the number of SRP-PhyloChip probes positive in both samples. These coefficients are usually calculated based on presence-absence data for species in the ecosystems compared. In this context, it should be noted that due to the multiple-probe design strategy of the SRP-PhyloChip, the number of positive probe signals on the microarray is generally much higher than the number of species actually detected in the sample analyzed. Therefore, the calculated coefficients can only be interpreted as indications of the similarity of hybridization patterns.
Cloning and sequencing.
Prior to cloning, the PCR amplification products were purified by low-melting-point agarose (1.5%) gel electrophoresis (NuSieve 3:1; FMC Bioproducts, Biozym Diagnostics GmbH, Oldendorf, Germany) and stained in a SYBR Green I solution (10 μl of SYBR Green I [Biozym Diagnostics GmbH] in 100 ml of Tris-acetate-EDTA buffer [40 mM Tris, 10 mM sodium acetate, 1 mM EDTA; pH 8.0]) for 45 min. Bands of the expected size were excised from the agarose gel with a glass capillary and melted with 80 μl of double-distilled water for 10 min at 80°C. Four microliters of each solution was ligated as recommended by the manufacturer (Invitrogen Corp.) either into the cloning vector pCR2.1 of a TOPO TA cloning kit (16S rRNA gene PCR products) or into the cloning vector pCR-XL-TOPO of a TOPO XL cloning kit (dsrAB PCR products). Nucleotide sequences were determined by a modification of the dideoxynucleotide method (58) as described previously (54). In addition, internal dsrAB-targeted sequencing primers (Table 2) were used to complete the dsrAB sequences.
Phylogeny.
Phylogenetic analyses were performed by using the alignment and treeing tools implemented in the ARB program package (46). New 16S rRNA gene sequences were added to an ARB alignment of about 20,000 small-subunit rRNA gene sequences (which included sequences from all recognized SRPs and clone sequences from uncultured prokaryotes from sulfate-reducing environments) by using the alignment tool ARB_EDIT. Alignments were refined by visual inspection. 16S rRNA gene phylogenetic analyses were performed exclusively with sequences having more than 1,150 bases by using distance matrix, maximum-parsimony, and maximum-likelihood methods (45). The composition of the 16S rRNA gene sequence data sets varied with respect to the reference sequences and alignment positions. The variability of the individual alignment positions was determined by using the ARB_SAI tools and was used as a criterion to remove or include variable positions for phylogenetic analyses.
New dsrAB sequences were added to an ARB alignment that contained all dsrAB sequences of recognized SRPs (22, 34, 74) and uncultured SRPs available in the GenBank database (9). Deduced amino acid sequences were manually aligned by using the editor GDE 2.2 (64). Nucleic acid sequences were aligned according to the alignment of amino acids. For phylogenetic inference of DsrAB amino acid sequences, insertions and deletions were removed from the data set by using a suitable alignment mask (indel filter), which left a total of 543 amino acid positions (alpha subunit, 327 positions; beta subunit, 216 positions) for comparative analyses. Distance matrix (using FITCH with global rearrangements and randomized input order of species) and maximum-parsimony trees were calculated with the Phylogeny Inference Package (PHYLIP) (21). In addition, the programs MOLPHY (2) and TREE-PUZZLE (67) were used to infer maximum-likelihood trees with JTT-f as the amino acid replacement model. To determine the level of amino acid identity between two DsrAB sequences, ambiguous amino acid positions and the alignment regions of insertions and deletions (indel filter) were omitted.
Parsimony bootstrap analyses for nucleic acid (16S rRNA gene) and protein (DsrAB) trees were performed with PHYLIP. One hundred bootstrap resamplings were analyzed for each calculation. All phylogenetic consensus trees were drawn by using established protocols (45).
Bacterial nomenclature.
The names of bacterial taxa used here are in accordance with the prokaryotic nomenclature proposed in the taxonomic outline of Bergey's Manual of Systematic Bacteriology, 2nd ed. (23; http://dx.doi.org/10.1007/bergeysoutline200210).
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession numbers AY167444 to AY167462 (16S rRNA gene clones) and AY167464 to AY167483 (dsrAB clones).
RESULTS
Oxidized and reduced inorganic sulfur in fen soils.
The concentrations of sulfate in the soil solutions from Schlöppnerbrunnen I and Schlöppnerbrunnen II varied over the year; the minimum concentration was 20 μM in late autumn, and the maximum concentration was 200 μM after the snow melt in February. At Schlöppnerbrunnen I, the average concentrations of TRIS in triplicate soil samples obtained from depths of 0 to 10, 10 to 20, and 20 to 30 cm in December were approximately 0.05 μmol g (fresh weight) of soil−1 at each depth. In contrast, the average concentration of AVS increased from 0.01 to 0.05 μmol g (fresh weight) of soil−1 as the soil depth increased. At Schlöppnerbrunnen II, the average concentrations of TRIS were approximately 0.29, 0.47, and 0.63 μmol g (fresh weight) of soil−1 at depths of 0 to 10, 10 to 20, and 20 to 30 cm, respectively; in contrast, the average concentrations of AVS were more uniform and were approximately 0.05, 0.06, and 0.05 μmol g (fresh weight) of soil−1 at the three depths, respectively.
Capacity of fen soils to consume sulfate.
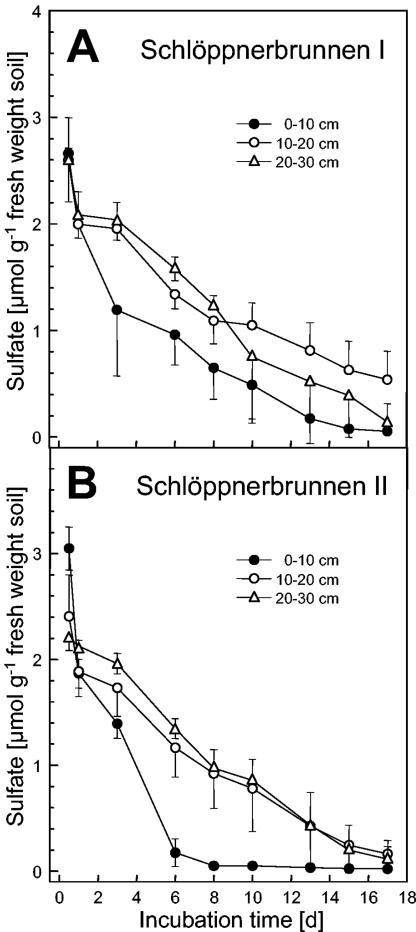
Supplemental sulfate was rapidly consumed without an apparent delay in anoxic microcosms that contained soil from the upper peat layers (Fig. 1). Soils from the three depths yielded similar concentrations of TRIS and AVS (Table 3). The average concentrations of TRIS and AVS were 0.67 and 0.12 μmol g (fresh weight) of soil−1, respectively, in sulfate-supplemented Schlöppnerbrunnen I microcosms at the end of incubation. The average concentrations of TRIS and AVS in unsupplemented controls were 0.16 and 0.03 μmol g (fresh weight) of soil−1, respectively. The Schlöppnerbrunnen II soils yielded results similar to those obtained with the Schlöppnerbrunnen I soils, although the values were slightly higher. However, the average amount of reduced sulfur recovered was only approximately 18% of the amount of sulfate-derived sulfur that was consumed. Part of the reduced sulfur might have been lost in the headspace as H2S due to the low soil pH. Despite this discrepancy, the concentrations of TRIS and AVS in soils were higher at the end of incubation in sulfate-supplemented microcosms than at the end of incubation in unsupplemented controls, indicating that the consumption of supplemental sulfate was linked to the dissimilatory reduction of sulfate.
FIG. 1.
Consumption of supplemental sulfate (500 μM) in anoxic microcosms of soil obtained from Schlöppnerbrunnen I (A) and II (B). The values are averages ± standard deviations for triplicate determinations.
TABLE 3.
Effect of the consumption of supplemental sulfate on the formation of reduced sulfur compounds in anoxic microcosms of soil obtained from Schlöppnerbrunnen I and IIa
| Sampling site | Depth (cm) | Concn (μmol g [fresh wt]−1)
|
|||
|---|---|---|---|---|---|
| AVSb | AVS control (no sulfate added) | TRISc | TRIS control (no sulfate added) | ||
| Schlöppnerbrunnen I | 0-10 | 0.057 ± 0.023 | 0.031 ± 0.004 | 0.648 ± 0.493 | 0.101 ± 0.020 |
| 10-20 | 0.127 ± 0.051 | 0.023 ± 0.003 | 0.646 ± 0.192 | 0.146 ± 0.168 | |
| 20-30 | 0.167 ± 0.100 | 0.040 ± 0.024 | 0.716 ± 0.389 | 0.234 ± 0.051 | |
| Schlöppnerbrunnen II | 0-10 | 0.389 ± 0.011 | 0.046 ± 0.006 | 0.586 ± 0.158 | 0.174 ± 0.037 |
| 10-20 | 0.147 ± 0.020 | 0.058 ± 0.017 | 0.786 ± 0.066 | 0.477 ± 0.164 | |
| 20-30 | 0.295 ± 0.089 | 0.052 ± 0.015 | 1.202 ± 0.353 | 0.530 ± 0.234 | |
The data are averages ± standard deviations for triplicate soil samples obtained at the end of incubation.
AVS is amorphous FeS.
TRIS is amorphous FeS, S0, and FeS2.
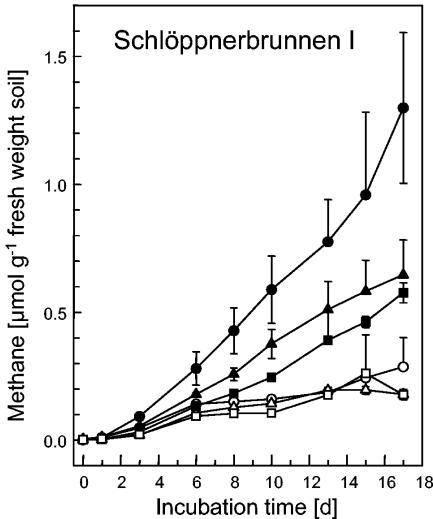
The maximum rates at which supplemental sulfate was consumed in the soil microcosms prepared from soils obtained at depths of 0 to 10, 10 to 20, and 20 to 30 cm from Schlöppnerbrunnen I were approximately 0.14, 0.11, and 0.14 μmol g (fresh weight) of soil−1 day−1, respectively, and the corresponding rates for soils from Schlöppnerbrunnen II were 0.41, 0.13, and 0.13 μmol g (fresh weight) of soil−1 day−1, respectively. In the sulfate-supplemented Schlöppnerbrunnen I soil microcosms the rates of CH4 production were 0.02, 0.01, and 0.01 μmol g (fresh weight) of soil−1 day−1, respectively, and the rates in the Schlöppnerbrunnen II soil microcosms were 0.06, 0.03, and 0.02 μmol g (fresh weight) of soil−1 day−1, respectively; the rates of production of CH4 in control Schlöppnerbrunnen I microcosms not supplemented with sulfate were 0.07, 0.04, and 0.04 μmol g (fresh weight) of soil−1 day−1, respectively, and the rates in the corresponding Schlöppnerbrunnen II microcosms were 0.19, 0.09, and 0.09 μmol g (fresh weight) of soil−1 day−1, respectively (Fig. 2 and data not shown). Thus, supplemental sulfate caused a 71% decrease in the average rate at which CH4 was produced.
FIG. 2.
Effect of the consumption of supplemental sulfate (500 μM) on the production of methane in anoxic microcosms of soil obtained from Schlöppnerbrunnen I. The values are averages ± standard deviations for triplicate determinations. Open symbols, methane production with supplemental sulfate; solid symbols, controls (no sulfate added); circles, 0 to 10 cm, triangles, 10 to 20 cm; squares, 20 to 30 cm.
16S rRNA-based phylogenetic profiles of SRPs at different fen soil depths.
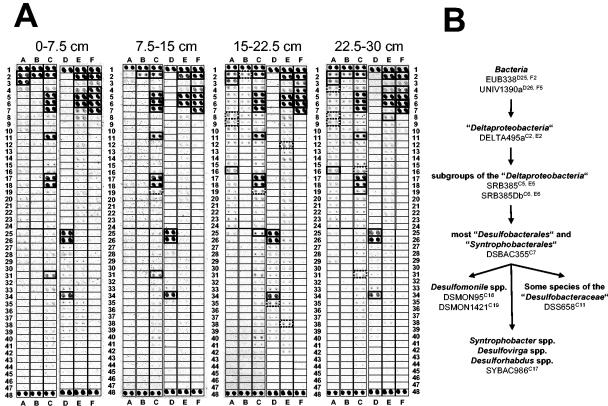
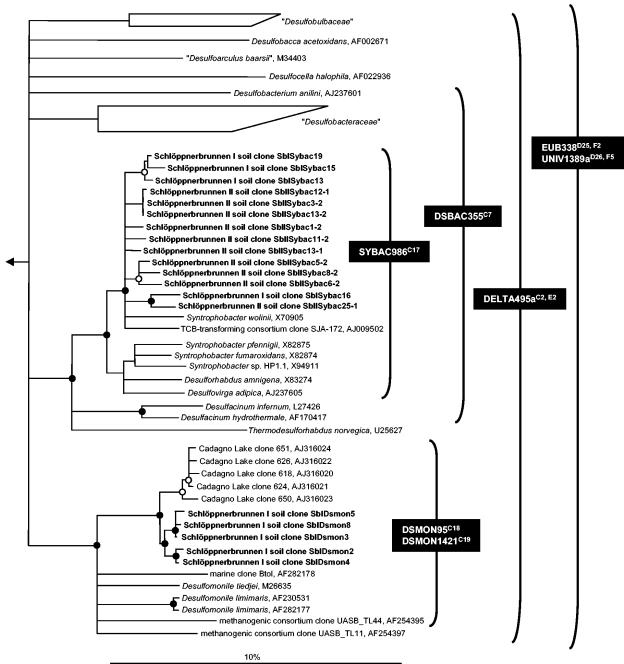
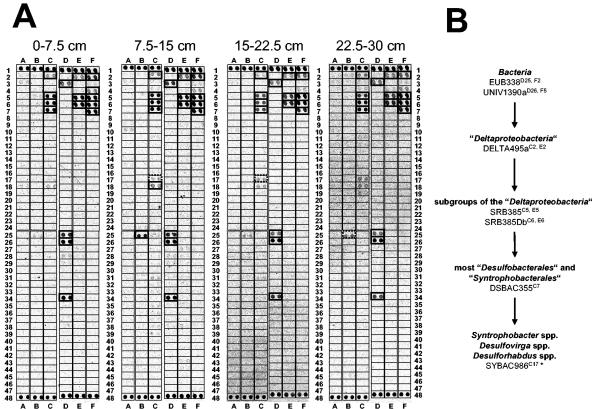
All soil depths at Schlöppnerbrunnen I yielded similar microarray hybridization patterns (Fig. 3A), indicating that there were minimal depth-dependent changes in the richness of recognized SRP phylotypes. Consistent with this observation, high Jaccard and Sorensen coefficient values (56 to 83% and 71 to 91%, respectively) were inferred by comparing the hybridization patterns for the different soil depths (Table 4). The microarray results indicated the presence of (i) Desulfomonile spp., (ii) some species of the family “Desulfobacteraceae,” and (iii) bacteria belonging to the Syntrophobacter-Desulfovirga-Desulforhabdus line of descent of the family “Syntrophobacteraceae” (order “Syntrophobacterales”) (Fig. 3B). Archaeoglobus 16S rRNA genes were not detected by PCR with genus-specific primers (Table 1). Consistent with the microarray patterns, direct PCR of Schlöppnerbrunnen I soil DNA with Desulfomonile- and “Syntrophobacteraceae”-specific primer pairs (Table 1) yielded increasing amounts of PCR products of the expected size with increasing soil depth (data not shown). Cloning and sequencing of the PCR products from soil at a depth of 22.5 to 30 cm confirmed that Desulfomonile spp. and Syntrophobacter wolinii-related bacteria were present at Schlöppnerbrunnen I (Fig. 4).
FIG. 3.
(A) Use of SRP-PhyloChip for surveys of SRP diversity at four different depths at Schlöppnerbrunnen I. Each probe was spotted in duplicate. The specificity and microarray position of each probe have been described previously (44). Probe spots having a signal-to-noise ratios equal to or greater than 2.0 are indicated by boldface boxes and were considered to be positive. The dotted boldface boxes indicate that only one of the duplicate spots had a signal-to-noise ratio equal to or greater than 2.0. (B) Flow chart illustrating the presence of distinct SRP groups in Schlöppnerbrunnen I as inferred from positive signals for sets of probes with nested and/or parallel specificity. For each probe the position on the microarray is indicated by a superscript.
TABLE 4.
Similarity matrix for SRP communities in the fen samples based on the presence or absence of SRP-PhyloChip probe signals for soil samples taken from four depths at Schlöppnerbrunnen I and II
| Sample
|
Jaccard coefficient (%)/Sorenson coefficient (%)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Sitea | Depth | SbI, 0-7.5 cm | SbI, 7.5-15 cm | SbI, 15-22.5 cm | SbI, 22.5-30 cm | SbII, 0-7.5 cm | SbII, 7.5-15 cm | SbII, 22.5-30 cm |
| SbI | 0-7.5 | |||||||
| 7.5-15 | 83/91 | |||||||
| 15-22.5 | 59/74 | 56/71 | ||||||
| 22.5-30 | 65/79 | 62/76 | 68/81 | |||||
| SbII | 0-7.5 | 53/69 | 56/72 | 35/51 | 35/51 | |||
| 7.5-15 | 56/71 | 59/74 | 37/54 | 37/54 | 67/80 | |||
| 15-22.5 | 53/69 | 56/72 | 35/51 | 35/51 | 80/89 | 82/90 | ||
| 22.5-30 | 41/58 | 44/61 | 27/42 | 27/42 | 78/88 | 64/78 | 78/88 | |
SbI, Schlöppnerbrunnen I; SbII, Schlöppnerbrunnen II.
FIG. 4.
16S rRNA gene phylogenetic consensus tree based on neighbor-joining analysis performed with a 50% conservation filter for the Deltaproteobacteria. The tree shows the affiliations of clone sequences from Schlöppnerbrunnen I and II soils (indicated by boldface type). Bar = 10% estimated sequence divergence. Polytomic nodes connect branches for which a relative order could not be determined unambiguously by applying distance matrix, maximum-parsimony, and maximum-likelihood treeing methods. Parsimony bootstrap values for branches are indicated by solid circles (>90%) and open circles (75 to 90%). Branches without circles had bootstrap values of less than 75%. Brackets indicate the perfect-match target organisms of the probes. The microarray position is indicated by a superscript after each probe designation. Cadagno Lake clones were not sequenced at the target site for probe DSMON1421. TCB, trichlorobenzene.
The microarray hybridization patterns for soils from different depths at Schlöppnerbrunnen II did not vary a great deal (Fig. 5A and Table 4), but they were less complex than those of the Schlöppnerbrunnen I soils. The binary similarities of the hybridization patterns obtained for the different depths yielded high Jaccard and Sorensen coefficient values, which were comparable to the values obtained for the samples from different depths at Schlöppnerbrunnen I (Table 4). Only probes targeting SRPs at higher taxonomic levels (e.g., DELTA495a [microarray positions C2 and E2] and DSBAC355 [microarray position C7]) were unambiguously positive. However, the mean signal-to-noise ratios of the probe spots that were specific for members of the Syntrophobacter-Desulfovirga-Desulforhabdus lineage were just below the threshold value (Fig. 5B). Direct PCR of Schlöppnerbrunnen II soil DNA with “Syntrophobacteraceae”-specific primer pairs (Table 1) yielded increasing amounts of PCR products of the expected size with increasing soil depth (data not shown) and verified the microarray patterns (data not shown). Cloning and sequencing of the PCR products from soil obtained at a depth of 22.5 to 30 cm confirmed that S. wolinii-related bacteria were present at Schlöppnerbrunnen II (Fig. 4).
FIG. 5.
(A) Use of SRP-PhyloChip for surveys of SRP diversity at four different depths at Schlöppnerbrunnen II. See the legend to Fig. 3 for additional details. (B) Flow chart illustrating the presence of distinct SRP groups in Schlöppnerbrunnen II soil as inferred from positive signals for sets of probes with nested specificity. For each probe the position on the microarray is indicated by a superscript. The asterisk indicates that the mean signal-to-noise ratios of the duplicate SYBAC986 spots for 7.5 to 15, 15 to 22.5, and 22.5 to 30 cm were just below the threshold value of 2.0 (1.88, 1.95, and 1.70, respectively).
The primer pair DSBAC355F-1492R is specific for most members of the orders “Desulfobacterales” and “Syntrophobacterales” (Table 1). This primer pair was used to screen for SRPs belonging to these orders which were not covered by the specific primer pairs described above. Twelve DSBAC355F-1492R-dependent clone sequences were determined for the deepest soil samples from each field site, but none of these sequences were closely related to recognized SRP 16S rRNA gene sequences (data not shown).
dsrAB diversity in fen soils.
As observed for SRP 16S rRNA gene PCR products (see above), the amount of dsrAB PCR products increased as the soil depth increased (data not shown). The dsrAB PCR products that were retrieved from the deepest soils (22.5 to 30 cm) from Schlöppnerbrunnen I and II were used to construct dsrAB clone libraries; the clone libraries for Schlöppnerbrunnen I and II were designated dsrSbI and dsrSbII, respectively; 41 of 42 randomly picked dsrSbI clones and 35 of 48 randomly picked dsrSbII clones had an insert of the expected size (1.9 kb). However, partial sequencing followed by BLAST analyses (7) demonstrated that only 29 clones from the dsrSbI library and 24 clones from the dsrSbII library contained dsrAB sequences. Comparative sequence analysis of the partial DsrAB sequences grouped the 53 Schlöppnerbrunnen dsrAB clones in 11 clusters. The complete sequence of at least one dsrAB clone per cluster was subsequently determined. All dsrAB clones with deduced DsrAB amino acid sequence identities equal to or greater than 90% with each other were grouped into an operational taxonomic unit (OTU). This grouping yielded 11 OTUs for both libraries (Table 5). Three OTUs (OTUs 1, 3, and 6) were present at both fen sites; in contrast, eight OTUs contained dsrAB clones that were found only at either Schlöppnerbrunnen I or Schlöppnerbrunnen II (four OTUs each).
TABLE 5.
OTUs of sulfate-reducing prokaryotes based on comparative sequence analyses of dsrAB retrieved from acidic fen soil at the Schlöppnerbrunnen I and II sampling sites
| OTUa | No. of clonesb
|
dsrAB clonesc | Most similar dsrAB sequence in GenBank as determined by BLAST search (accession no./% amino acid identity) | Inferred phylogenyd | |
|---|---|---|---|---|---|
| dsrSbI | dsrSbII | ||||
| 1 | 21 | 1 | dsrSbI-56, -57, -58, -59, -60, 61-, -62, -65, -66, -67, -69, -72, -73, -74, -78, -79, -81, -83, -84, -86, and -87; dsrSbII-20 | Uranium mill tailings clone UMTRAdsr828-17 (AY015508, AY015597/86-89) | Desulfobacca acetoxidans related, Deltaproteobacteria |
| 2 | 9 | dsrSbII-3, -18, -21, -22, -23, -28, -34, -42, and -47 | Everglades clone FISU-12 (AY096051/83-84) | Unaffiliated with known SRPs | |
| 3 | 1 | 6 | dsrSbI-71; dsrSbII-4, -5, -8, -12, -25, and -36 | Uranium mill tailings clone UMTRA826-5 (AY015548, AY015614/88) | Unaffiliated with known SRPs |
| 4 | 4 | dsrSbII-9, -11, -15, and -33 | Uranium mill tailings clone UMTRAdsr828-17 (AY015508, AY015597/87-89) | Desulfobacca acetoxidans related, Deltaproteobacteria | |
| 5 | 2 | dsrSbI-82 and -50 | “Desulfobacterium oleovorans” (AF418201/80) | Deltaproteobacteria | |
| 6 | 1 | 1 | dsrSbI-54; dsrSbII-40 | Syntrophobacter wolinii (AF418192/87-88) | Syntrophobacter wolinii related, Deltaproteobacteria |
| 7 | 2 | dsrSbII-2 and -16 | Everglades clone F1SU-12 (AY096051/83) | Unaffiliated with known SRPs | |
| 8 | 2 | dsrSbI-75 and -85 | Only distantly related sequences in GenBank | Unaffiliated with known SRPs | |
| 9 | 1 | dsrSbI-88 | Desulfomonile tiedjei (AF334595/85) | Desulfomonile-related, Deltaproteobacteria | |
| 10 | 1 | dsrSbI-64 | Uranium mill tailings clone UMTRAdsr626-20 (AY015569, AY015611/90) | Unaffiliated with known SRPs | |
| 11 | 1 | dsrSbII-39 | Uranium mill tailings clone UMTRAdsr624-8 (AY015519, AY015596/92) | Unaffiliated with known SRPs | |
dsrAB clones exhibiting deduced DsrAB sequence identity equal to or greater than 90% were grouped in an OTU. The OTUs are listed and sequentially numbered according to the total number of clones retrieved.
The dsrAB clone libraries for Schlöppnerbrunnen I and II were designated dsrSbI and dsrSbII, respectively. Homologous coverage (C) was calculated as follows: C = [1 − (n1 × N−1)]× 100%, where n1 is the number of OTUs containing only one sequence and N is the total number of dsrAB clones analyzed (24, 31, 62). The homologous coverage for the 29 clones in dsrSbI was 86%, and the homologous coverage for the 24 clones in dsrSbII was 88%.
Completely sequenced dsrAB clones (>1,750 bases) are indicated by boldface type.
Phylogeny of dsrAB clones as inferred from Fig. 6.
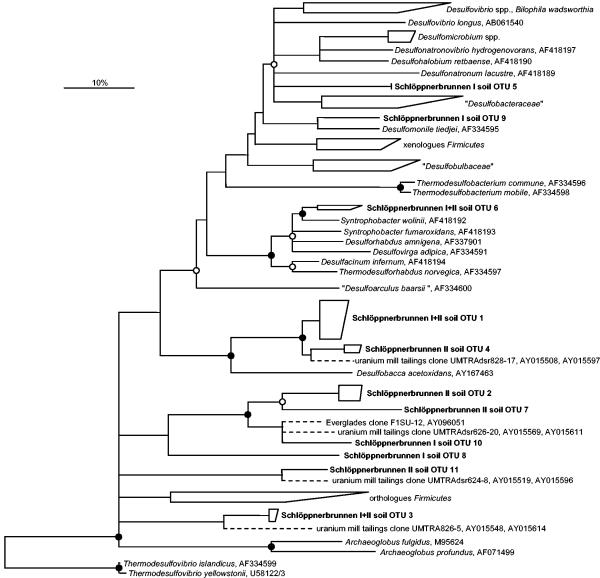
The phylogenetic affiliations of deduced DsrAB sequences are shown in Fig. 6. OTU 1, which comprised most of the dsrSbI clones, and the dsrSbII-specific OTU 4 displayed high sequence identity to the OTU of a groundwater clone derived from a uranium mill tailings site (Table 5). Clones in OTUs 1 and 4 formed a stable, monophyletic group with the deltaproteobacterial SRP Desulfobacca acetoxidans. One dsrAB clone derived from each fen site formed OTU 6, which was closely related to S. wolinii. OTU 9, which consisted of a single clone derived from Schlöppnerbrunnen I and was affiliated with Desulfomonile tiedjei, constituted an additional deltaproteobacterial lineage. OTU 5, which was composed of two clones derived from Schlöppnerbrunnen I, formed an independent branch within a monophyletic deltaproteobacterial cluster consisting of the family “Desulfobacteraceae” and different groups belonging to the order “Desulfovibrionales.” The remaining six OTUs formed three deeply branching evolutionary lines of descent that were different from any cultured SRP lineage (Fig. 6). One of these three deeply branching lines of descent contained OTUs 2, 7, 8, and 10; the other two lines of descent contained either OTU 3 or OTU 11. OTUs 2 and 7 consisted exclusively of dsrSbII clones. In contrast, only dsrSbI clones were present in OTUs 8 and 10. Interestingly, each of these three deeply branching lines of descent also contained at least one dsrAB clone that was derived from uranium mill tailings groundwater (16). It is also noteworthy that a dsrAB clone related to OTU 10 was recently retrieved from Everglades soil (14).
FIG. 6.
Phylogenetic consensus tree (based on FITCH analysis) for DsrAB amino acid sequences deduced from dsrAB sequences longer than 1,750 bases, showing the affiliation of OTUs from Schlöppnerbrunnen fen soils (indicated by boldface type). DsrAB sequences deduced from dsrAB sequences shorter than 1,750 bases (indicated by dashed branches) were individually added to the distance matrix tree without changing the overall tree topology by using the ARB treeing tool PARSIMONY_INTERAKTIV. Bar = 10% estimated sequence divergence. See the legend to Fig. 4 for additional details.
Molecular assessment of D. acetoxidans-related SRPs.
The DsrAB analyses indicated that fen soils at both Schlöppnerbrunnen I and II harbored D. acetoxidans-related SRPs. Because no species-specific probe for D. acetoxidans was included on the microarray, a 16S rRNA-based PCR assay with the D. acetoxidans-specific primer pair DBACCA65F-DBACCA1430R (Table 1) was used to retrieve 16S rRNA gene sequences of D. acetoxidans-related bacteria. PCRs were carried out at low stringency to allow amplification of 16S rRNA genes of D. acetoxidans-related SRPs even if they had mismatches in the primer target sites. Although this primer pair produced a PCR product of the expected size (1.4-kb) when D. acetoxidans was used as a positive control, it did not yield any D. acetoxidans-specific PCR product from Schlöppnerbrunnen I or II soils (data not shown).
DISCUSSION
SRP activities in fen soils.
The occurrence of TRIS and AVS in the upper peat layers and the capacity of the soils to consume sulfate and form TRIS and AVS under anoxic conditions corroborated previous results (3, 5) that indicated that the dissimilatory reduction of sulfate is an on-going process in the low-sulfate, acidic fens of the Lehstenbach catchment. However, the maximum concentrations of sulfate detected in the soil solution never exceeded 200 μM, and the low concentrations (20 μM) of sulfate in the soil solution obtained in autumn might be insufficient for the dissimilatory reduction of sulfate (42). It can be projected that the low concentration of sulfate is a limiting seasonal factor for the sulfate-reducing activity of SRPs in these fens.
Fens of the Lehstenbach catchment emit 0.02 to 15 mmol of CH4 m−2 day−1 (28). Methanogenesis was inhibited in anoxic microcosms supplemented with sulfate; this result supports previous findings suggesting that SRPs outcompete methanogens in peatlands under certain in situ conditions (11, 79). The rates of consumption of supplemental sulfate in soil microcosms were twice as high as the rates of formation of methane in microcosms lacking sulfate and were comparable to the rates for eutrophic waterlogged Everglades soils (14). These collective observations indicate that sulfate, when available, is subject to rapid reduction in these fens.
SRP diversity in fen soils.
Both the 16S rRNA-based (microarray and clone library) and dsrAB-based analyses indicated that the SRP community compositions of the two fen soils were not identical. Soil from Schlöppnerbrunnen I contained members of the genus Desulfomonile and the family “Syntrophobacteraceae,” while only the latter group was detected in soil from Schlöppnerbrunnen II. Only 7 and 4% of the dsrAB clones from Schlöppnerbrunnen I and II, respectively, were affiliated with these organisms. A large fraction of the remaining dsrAB clones (72 and 21% of the clones derived from Schlöppnerbrunnen I and II, respectively) formed a monophyletic group with D. acetoxidans, a result that is paradoxical in that this organism was not detected by nonstringent 16S rRNA gene amplification with D. acetoxidans-specific primers. The failure to detect the D. acetoxidans-like organisms via their 16S rRNA genes possibly reflects their moderate phylogenetic relatedness to D. acetoxidans (73 to 75% DsrAB amino acid identity). Consequently, the D. acetoxidans-like organisms of the fens might have 16S rRNA genes that were not targeted by the D. acetoxidans-specific primers. An encompassing 16S rRNA gene library from the Schlöppnerbrunnen sites obtained with general bacterial primers could help determine whether such organisms actually occur in the acidic fens. Alternatively, the D. acetoxidans-related dsrAB sequences might originate from SRPs which are phylogenetically not closely related to D. acetoxidans but received their dsrAB genes via lateral gene transfer from a D. acetoxidans-related donor. The fact that several horizontal dsrAB gene transfer events have been described (34; V. Zverlov, unpublished data) lends weight to this hypothesis.
The molecular analyses indicated that S. wolinii-related bacteria are present in these acidic fen soils, a result that is consistent with detection of this species in an acid-tolerant methanogenic consortium derived from a Sphagnum peat bog (63). 16S rRNA gene sequences obtained from rice paddy soil indicate that S. wolinii-like bacteria also occur in other types of soils (61). S. wolinii was first isolated from an anaerobic municipal sewage digestor in syntrophic cocultures with methanogens or SRPs (12) and oxidizes propionate to acetate and CO2. The electrons that are obtained from the oxidation of propionate are transferred via hydrogen and/or formate to methanogens that reduce CO2 to methane. The oxidation of propionate is exergonic only when hydrogen and/or formate is continuously removed by the syntrophic, methanogenic partner. The discovery that S. wolinii is capable of dissimilating sulfate led to its isolation (75) and proved that it was not an obligate syntroph (12). Due to the low concentration of sulfate in the Schlöppnerbrunnen fens and the likelihood that H2 is an important substrate for moderately acid-tolerant methanogens in this ecosystem (28), it is tempting to speculate that the S. wolinii-like bacteria of these fens also have a syntrophic lifestyle and interact with methanogens. However, the S. wolinii-related 16S rRNA gene sequences that were retrieved from the fen soils were 3.6 to 5.5% dissimilar to the 16S rRNA gene sequence of S. wolinii; likewise, the levels of sequence dissimilarity among the S. wolinii-like fen clones ranged from 0.1 to 5.2%. Thus, the cloned sequences represent several novel genomospecies (65) that might have physiologies that are at least partially dissimilar to that of S. wolinii.
Desulfomonile-related organisms appear to be members of the soil community at Schlöppnerbrunnen I. Members of this genus occur in other low-sulfate systems, such as an alpine lake (8), a forested wetland (13), deep crystalline bedrock (52), and a uranium mill tailings site (16), but they also occur in a hypersaline, sulfide-rich microbial mat (44). An environmentally important feature of the two cultured species of this genus, Desulfomonile limimaris and Desulfomonile tiedjei, is their ability to reductively dehalogenate anthropogenic compounds, such as polychlorinated biphenyls, perchloroethene, and chlorobenzenes (47, 48, 69). These species can also use H2 as an electron donor (55, 69). Thus, Desulfomonile-related organisms might compete for H2 and limit hydrogenotrophic methanogenesis when sulfate becomes available in the Schlöppnerbrunnen I fen. However, the 16S rRNA gene sequences of Desulfomonile-related clones were 5.2 to 7.5% dissimilar to those of the cultured members of this genus, suggesting that the ecophysiological capacities of the organisms represented by the cloned sequences might not be identical to those of known Desulfomonile species.
D. acetoxidans-like organisms appear to be present in both fens and have previously been detected in other low-sulfate habitats (13, 14, 16). The only cultured member of the genus Desulfobacca, D. acetoxidans, was isolated from a laboratory-scale upflow anaerobic sludge bed reactor fed with acetate and sulfate. This organism is specialized in acetate consumption and can outcompete acetoclastic methanogens (51). Because the D. acetoxidans dsrAB clone group was numerically the largest clone group obtained, we hypothesized that D. acetoxidans-like bacteria are abundant in these fens and are at least partially responsible for the lack of acetate accumulation and low abundance of acetoclastic methanogens in the fen soils (28).
SRP novelties in fen soils.
In addition to molecular signatures of well-known SRP lineages, novel types of dissimilatory (bi)sulfite reductases were detected in both fens. The deduced DsrAB amino acid sequences were remarkably dissimilar to those of cultured SRPs (the maximum deduced level of DsrAB amino acid sequence identity between one of the clones [dsrSbII-36] and a cultured SRP [Desulfotomaculum alkaliphilum DSM 12247] was 68%) and formed six deeply branching OTUs in the DsrAB tree (Fig. 6). Only one of these OTUs was found in both fens. The estimated diversity coverage of the established dsrAB libraries was almost 90% (Table 5), which suggests that the actual diversity of dsrAB sequences (detectable by the PCR approach used) present in the fens was largely recovered. Thus, the fact that five of the six deeply branching dsrAB OTUs were not detected at both fen sites is indicative of site-specific occurrence of the organisms. On the other hand, three of the retrieved lineages displayed affiliations with dsrAB sequences that were derived from a uranium mill tailings soil (16) or an Everglades soil (14), indicating that some of the detected lineages are widely distributed in low-sulfate ecosystems.
The novel types of dissimilatory (bi)sulfite reductases detected in the present study originated from SRPs that belong to either previously undescribed phylogenetic groups or phyla not yet known to contain organisms capable of dissimilating sulfate or sulfite. Certain type II methanotrophs harbor two different genes for subunit A of particulate methane monooxygenase (20, 70), and the novel dsrAB types could likewise represent additional dissimilatory (bi)sulfite reductases in well-known SRPs. However, SRPs that possess multiple dsr operons with significantly different sequences have not been reported. Theoretically, the novel dsrAB types from the Schlöppnerbrunnen fens could be pseudogenes whose sequences differ from recognized dsrAB sequences due to a higher mutation frequency caused by lack of selective pressure. However, there are three lines of evidence that argue against this possibility. (i) The dsrAB sequences did not contain any unexpected stop codons. (ii) Both subunits of the deduced DsrAB amino acid sequences contained the Cys motif consensus sequences Cys-X5-Cys and Cys-X3-Cys that are essential for binding the [Fe4S4]-siroheme cofactor of (bi)sulfite reductase (17); as in other (bi)sulfite reductases (26, 34), the Cys-X5-Cys motif of some of the deduced DsrAB amino acid sequences (OTUs 2, 7, 10, and 11) was truncated in the beta subunit. (iii) The nonsynonymous/synonymous substitution rate ratios of branches leading to the new dsrAB sequence clusters are low (<0.1), suggesting that there was strong purifying (negative) selection indicative of expressed and functionally active proteins (78).
Conclusions
In this study we assessed the potential activities and molecular signatures of SRPs in two acidic fens within the Lehstenbach catchment. Although some of the geochemical features (e.g., pH, temperature, and low concentration of sulfate) of these fens are similar, the types of vegetation in the two fens are different. In addition, only Schlöppnerbrunnen II is enriched both with dithionite-extractable pedogenic iron oxides and oxalate-extractable poorly crystallized iron oxides (36). Poorly crystallized iron oxides are the favored reducible forms of Fe3+ for microbial reduction, and Fe3+ might be an alternative electron acceptor for the anaerobic oxidation of organic matter at Schlöppnerbrunnen II. The collective data obtained in this study revealed stable diversity profiles for SRPs in the upper peat layers of the fen soils but also revealed site-specific novel SRP lineages. Thus, one might speculate that the different types of vegetation and different bioavailabilities of iron of the two fens are important factors in determining which SRPs become established in either one or both fens. Likewise, the seasonal variability in the water content of the two fens might be a contributing factor. Resolving the genomic and ecophysiological characteristics of the SRP community members of these fens is a major challenge for future research and could yield new insights into the novel physiologies and structure-function relationships of resident SRPs that enable them to compete in low-sulfate, acidic habitats.
Acknowledgments
We thank Helga Gaenge, Sibylle Schadhauser, Silvia Weber, Sandra Praßl, and Sonja Trenz for their excellent technical assistance. Michael Friedrich is acknowledged for providing the dsrAB sequence of D. acetoxidans DSM 11109T (GenBank accession number AY167463). We also thank Matthias Horn for critical reading of the manuscript and for helpful discussions and Michael Klein for help with dsrAB primer design. The tedious work of Michael Klein and Natuschka Lee required for establishing and maintaining the dsrAB database is highly appreciated.
This research was supported by grants from the bmb+f (01 LC 0021 subproject 2 in the framework of the BIOLOG program to M.W. and PT BEO 51-0339476 D) and the Bayerische Forschungsstiftung (Development of Oligonucleotide DNA Chips in cooperation with MWG Biotech; project 368/99 to M.W.).
REFERENCES
- 1.Abdelouas, A., W. Lutze, W. Gong, E. H. Nuttall, B. A. Strietelmeier, and B. J. Travis. 2000. Biological reduction of uranium in groundwater and subsurface soil. Sci. Total Environ. 250:21-35. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, J., and M. Hasegawa. 1996. MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr. 28:1-150. [Google Scholar]
- 3.Alewell, C., and M. Gehre. 1999. Patterns of stable S isotopes in a forested catchment as indicators for biological S turnover. Biogeochemistry 47:319-333. [Google Scholar]
- 4.Alewell, C., and A. Giesemann. 1996. Sulfate reduction in a forested catchment as indicated by d34S values of sulfate in soil solutions and runoff. Isot. Environ. Health Stud. 32:203-210. [DOI] [PubMed] [Google Scholar]
- 5.Alewell, C., and M. Novak. 2001. Spotting zones of dissimilatory sulfate reduction in a forested catchment: the 34S-35S approach. Environ. Pollut. 112:369-377. [DOI] [PubMed] [Google Scholar]
- 6.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 8.Bensadoun, J., M. Tonolla, A. Demarta, F. Barja, and R. Peduzzi. 1998. Vertical distribution and microscopic characterization of a non-culturable microorganism (morphotype R) of Lake Cadagno. Doc. Ist. Ital. Idrobiol. Dott. Marco Marchi 63:45-51. [Google Scholar]
- 9.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2002. GenBank. Nucleic Acids Res. 30:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berge, E., J. Bartnicki, K. Olendrzynski, and S. G. Tsyro. 1999. Long-term trends in emissions and transboundary transport of acidifying air pollution in Europe. J. Environ. Manag. 57:31-50. [Google Scholar]
- 11.Blodau, C., C. L. Roehm, and T. R. Moore. 2002. Iron, sulfur, and dissolved carbon dynamics in a northern peatland. Arch. Hydrobiol. 154:561-583. [Google Scholar]
- 12.Boone, D. R., and M. P. Bryant. 1980. Propionate-degrading bacterium, Syntrophobacter wolinii sp. nov., gen. nov., from methanogenic ecosystems. Appl. Environ. Microbiol. 40:626-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 14.Castro, H., K. R. Reddy, and A. Ogram. 2002. Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68:6129-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, B. V., L. C. Shiung, and S. Y. Yuan. 2002. Anaerobic biodegradation of polycyclic aromatic hydrocarbon in soil. Chemosphere 48:717-724. [DOI] [PubMed] [Google Scholar]
- 16.Chang, Y. J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crane, B. R., L. M. Siegel, and E. D. Getzoff. 1995. Sulfite reductase structure at 1.6 A: evolution and catalysis for reduction of inorganic anions. Science 270:59-67. [DOI] [PubMed] [Google Scholar]
- 18.Devereux, R., and G. W. Mundfrom. 1994. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl. Environ. Microbiol. 60:3437-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunfield, P. F., M. Tchawa Yimga, S. N. Dedysh, U. Berger, W. Liesack, and J. Heyer. 2002. Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol. Ecol. 41:17-26. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein, J. 1995. PHYLIP: phylogeny inference package, version 3.57c. Department of Genetics, University of Washington, Seattle.
- 22.Friedrich, M. W. 2002. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J. Bacteriol. 184:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrity, G. M., and J. G. Holt. 2001. The road map to the manual, p. 119-166. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y. [Google Scholar]
- 24.Giovannoni, S. J., T. D. Mullins, and K. G. Field. 1995. Microbial diversity in oceanic systems: rRNA approaches to the study of unculturable microbes, p. 217-248. In J. Joint (ed.), Molecular ecology of aquatic microbes, vol. G38. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 25.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hipp, W. M., A. S. Pott, N. Thum-Schmitz, I. Faath, C. Dahl, and H. G. Trüper. 1997. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology 143:2891-2902. [DOI] [PubMed] [Google Scholar]
- 27.Höpner, T. 1981. Design and use of a diffusion sampler for interstitial water from fine grained sample. Environ. Technol. Lett. 2:187-196. [Google Scholar]
- 28.Horn, M. A., C. Matthies, K. Küsel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jørgensen, B. B. 1982. Mineralization of organic matter in the sea-bed—the role of sulphate reduction. Nature 296:643-645. [Google Scholar]
- 30.Jørgensen, B. B. 1977. The sulfur cycle of a coastal marine sediment (Limfjorden, Denmark). Limnol. Oceanogr. 22:814-832. [Google Scholar]
- 31.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 32.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Roser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovárová, M., and P. Dráber. 2000. New specificity and yield enhancer of polymerase chain reactions. Nucleic Acids Res. 28:E70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Küsel, K., and C. Alewell. 2004. Riparian zones in a forested catchment: hot spots for microbial reductive processes. Ecol. Stud. 172:377-395. [Google Scholar]
- 37.Küsel, K., and H. L. Drake. 1995. Effect of environmental parameters on the formation and turnover of acetate in forest soils. Appl. Environ. Microbiol. 61:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Küsel, K., H. C. Pinkart, H. L. Drake, and R. Devereux. 1999. Acetogenic and sulfate-reducing bacteria inhabiting the rhizoplane and deep cortex cells of the sea grass Halodule wrightii. Appl. Environ. Microbiol. 65:5117-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Küsel, K., U. Roth, T. Trinkwalter, and S. Peiffer. 2001. Effect of pH on the anaerobic microbial cycling of sulfur in mining-impacted freshwater lake sediments. Environ. Exp. Bot. 46:213-223. [Google Scholar]
- 40.Li, J.-H., K. J. Purdy, S. Takii, and H. Hayashi. 1999. Seasonal changes in ribosomal RNA of sulfate-reducing bacteria and sulfate reducing activity in a freshwater lake sediment. FEMS Microbiol. Ecol. 28:31-39. [Google Scholar]
- 41.Llobet-Brossa, E., R. Rossello-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lovley, D. R., and M. J. Klug. 1983. Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl. Environ. Microbiol. 45:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 46.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohn, W. W., and K. J. Kennedy. 1992. Reductive dehalogenation of chlorophenols by Desulfomonile tiedjei DCB-1. Appl. Environ. Microbiol. 58:1367-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohn, W. W., and J. M. Tiedje. 1992. Microbial reductive dehalogenation. Microbiol. Rev. 56:482-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noh, S. L., J. M. Choi, Y. J. An, S. S. Park, and K. S. Cho. 2003. Anaerobic biodegradation of toluene coupled to sulfate reduction in oil-contaminated soils: optimum environmental conditions for field applications. J. Environ. Sci. Health Part A 38:1087-1097. [DOI] [PubMed] [Google Scholar]
- 50.Novak, M., R. K. Wieder, and W. R. Schell. 1994. Sulfur during early diagenesis in sphagnum peat: insights from delta34S ratio profiles in 210Pb-dated peat cores. Limnol. Oceanogr. 39:1172-1185. [Google Scholar]
- 51.Oude Elferink, S. J., W. M. Akkermans-van Vliet, J. J. Bogte, and A. J. Stams. 1999. Desulfobacca acetoxidans gen. nov., sp. nov., a novel acetate-degrading sulfate reducer isolated from sulfidogenic granular sludge. Int. J. Syst. Bacteriol. 49:345-350. [DOI] [PubMed] [Google Scholar]
- 52.Pedersen, K., J. Arlinger, L. Hallbeck, and C. Pettersson. 1996. Investigations of subterranean bacteria in deep crystalline bedrock and their importance for the disposal of nuclear waste. Can. J. Microbiol. 42:382-391. [Google Scholar]
- 53.Purdy, K. J., D. B. Nedwell, and T. M. Embley. 2003. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 69:3181-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purkhold, U., A. Pommering-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabus, R., T. Hansen, and F. Widdel. 2000. Dissimilatory sulfate- and sulfur-reducing prokaryotes. .In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. [Online.] Springer-Verlag, New York, N.Y.
- 56.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sahm, K., C. Knoblauch, and R. Amann. 1999. Phylogenetic affiliation and quantification of psychrophilic sulfate-reducing isolates in marine arctic sediments. Appl. Environ. Microbiol. 65:3976-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sass, H., H. Cypionka, and H.-D. Babenzien. 1997. Vertical distribution of sulfate-reducing bacteria at the oxic-anoxic interface in sediments of the oligotrophic Lake Stechlin. FEMS Microbiol. Ecol. 22:245-255. [Google Scholar]
- 60.Sass, H., E. Wieringa, H. Cypionka, H.-D. Babenzien, and J. Overmann. 1998. High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch. Microbiol. 170:243-251. [DOI] [PubMed] [Google Scholar]
- 61.Scheid, D., and S. Stubner. 2001. Structure and diversity of Gram-negative sulfate-reducing bacteria on rice roots. FEMS Microbiol. Ecol. 36:175-183. [DOI] [PubMed] [Google Scholar]
- 62.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sizova, M. V., N. S. Panikov, T. P. Tourova, and P. W. Flanagan. 2003. Isolation and characterization of oligotrophic acido-tolerant methanogenic consortia from a Sphagnum peat bog. FEMS Microbiol. Ecol. 45:301-315. [DOI] [PubMed] [Google Scholar]
- 64.Smith, S. W., R. Overbeek, C. R. Woese, W. Gilbert, and P. M. Gillevet. 1994. The Genetic Data Environment (GDE): an expandable graphic interface for manipulating molecular information. Comput. Appl. Biol. Sci. 10:671-675. [DOI] [PubMed] [Google Scholar]
- 65.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 66.Stahl, D. A., S. Fishbain, M. Klein, B. J. Baker, and M. Wagner. 2002. Origins and diversification of sulfate-respiring microorganisms. Antonie Leeuwenhoek 81:189-195. [DOI] [PubMed] [Google Scholar]
- 67.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 68.Stubner, S., and K. Meuser. 2000. Detection of Desulfotomaculum in an Italian rice paddy soil by 16S ribosomal nucleic acid analyses. FEMS Microbiol. Ecol. 34:73-80. [DOI] [PubMed] [Google Scholar]
- 69.Sun, B., J. R. Cole, and J. M. Tiedje. 2001. Desulfomonile limimaris sp. nov., an anaerobic dehalogenating bacterium from marine sediments. Int. J. Syst. Evol. Microbiol. 51:365-371. [DOI] [PubMed] [Google Scholar]
- 70.Tchawa Yimga, M., P. F. Dunfield, P. Ricke, J. Heyer, and W. Liesack. 2003. Wide distribution of a novel pmoA-like gene copy among type II methanotrophs, and its expression in Methylocystis strain SC2. Appl. Environ. Microbiol. 69:5593-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Telang, A. J., G. Voordouw, S. Ebert, N. Sifeldeen, J. M. Foght, P. M. Fedorak, and D. W. Westlake. 1994. Characterization of the diversity of sulfate-reducing bacteria in soil and mining waste water environments by nucleic acid hybridization techniques. Can. J. Microbiol. 40:955-964. [DOI] [PubMed] [Google Scholar]
- 72.Teske, A., C. Wawer, G. Muyzer, and N. B. Ramsing. 1996. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ulrich, G. A., L. R. Krumholz, and J. M. Suflita. 1997. A rapid and simple method for estimating sulfate reduction activity and quantifying inorganic sulfides. Appl. Environ. Microbiol. 63:1627-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wallrabenstein, C., E. Hauschild, and B. Schink. 1994. Pure culture and cytological properties of Syntrophobacter wolinii. FEMS Microbiol. Lett. 123:249-254. [Google Scholar]
- 76.Wind, T., and R. Conrad. 1995. Sulfur compounds, potential turnover of sulfate and thiosulfate, and numbers of sulfate-reducing bacteria in planted and unplanted paddy soil. FEMS Microbiol. Ecol. 18:257-266. [Google Scholar]
- 77.Wind, T., S. Stubner, and R. Conrad. 1999. Sulfate-reducing bacteria in rice field soil and on rice roots. Syst. Appl. Microbiol. 22:269-279. [DOI] [PubMed] [Google Scholar]
- 78.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yavitt, J. B., G. E. Lang, and R. K. Wieder. 1987. Control of carbon mineralization to CH4 and CO2 in anaerobic, sphagnum derived peat from Big Run, West Virginia. Biogeochemistry 4:141-157. [Google Scholar]