Abstract
Breast carcinoma comprises a group of diseases with specific clinical, histopathologic and molecular properties. Traditional classification use morphology to divide tumors into separate categories with differing behavior and prognosis. However, there are limitations of traditional classification systems, and new molecular methods are expected to improve classification systems. Molecular subtypes of breast carcinomas have been characterized in the last 11 years, and have been studied extensively. Much of the information accumulated in recent years, and molecular taxonomy seems to be still developing and undergoing change. The main question is whether new molecular techniques such as gene expression profiling will be accepted as gold standard in determining breast cancer subtypes, and whether molecular classification is useful in specific subtypes of breast cancer as it is in ductal carcinoma (nonspecific type). In addition, critical review of the literature reveals major problems such as poor definition, lack of reproducibility and lack of quality control in current molecular techniques and classifications. Therefore, current molecular approaches are not yet used in routine clinical practice and treatment guidance since they are immature and can even lead to incorrect assessment.
Keywords: Breast cancer, the molecular classification, immunohistochemistry, microarray gene expression
Introduction
Breast cancer is a significant and common disease that has a negative effect on women health, and is one of the leading causes of cancer related deaths. It constitutes 23% of all cancer patients and account for 14% of cancer related deaths (1). Egyptian physicians have accepted it as a fatal disease, and extensive surgical treatment was applied for many years until the end of the 19th century (2). At the beginning of the last century, it was enough to know that the patient had breast malignancy and all patients were administered a uniform treatment. Over time, the observation that patients with the same type of cancer show varying prognosis, and identification of increasing form of different morphological variants by pathologists during the last 50 years has led to discussion on breast cancer classification. Currently there are 20 major types and 18 minor subtypes of breast cancer that have been defined and included in the recently published WHO classification (3). However, there are doubts as to whether these variants are biologically significant or not. In addition, definition of so many variants is suggested to result from the pathologist’s own design. On the other hand, pathologists have been stating that breast cancer is a heterogeneous disease rather than a single disease for quite a long time. Now, it is obvious that breast cancer is a heterogeneous disease with different histological and biological properties due to genetic, epigenetic and transcriptome changes, with varying clinical findings and treatment responses, and with multiple entities. This phenotypic difference influences breast cancer diagnosis, treatment and thus prognosis. The basis of all this chaos seems to be based on absence of specific markers, and not fully understanding epithelial cellular development of breast tissue (4, 5). With the advancement of molecular techniques such as gene expression profiling, “heterogeneity in breast cancer concept” has now become generally accepted. Thus, a new “taxonomy” began to develop in the classification of breast cancer. This development has led to a concern between surgeons and oncologists that standard histopathological analysis reports prepared by pathologists will not contain some important data in the regulation of patient care and consequently patient treatment may not be regulated properly. Thus, pathologists were introduced to the so-called new era “Molecular Classification” that is developed from the traditional old fashioned “morphological” classification new age classification. Targeted therapies and more importantly, individualized treatment programs have become possible with the implementation of this classification.
Traditional, Old Fashioned Practice
Invasive breast cancer is currently classified as non-specific ductal carcinoma and specific subtypes. Special subtypes of breast cancer have specific definitions, while the non-specific type is like a dumpster containing all carcinomas other than specific subtypes. Non-specific invasive ductal carcinomas constitute about 60–75% of all breast cancers. Specific types constitute 20–25% of all, and the most common types within this group are lobular, tubular, papillary, and mucinous tumors (3, 4). Heterogeneity within a single tumor (intratumoral) or between morphologically similar same type of tumors (intertumoral) is currently well-known and accepted (Figure 1). Therefore, pathologists have attempted to produce new systems to allow clinicians to monitor their patients better. An absolute necessary component of pathology reports is “histological grade”, and is determined by the evaluation of the degree of tumor differentiation (tubule formation), nuclear pleomorphism / degree and proliferation (mitosis rate).
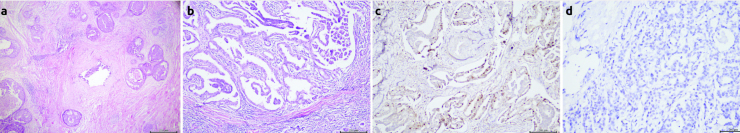
Figure 1. a–d.
Intratumoral heterogenity with H&E, IHC and CISH. (a) Comedo-type ductal carcinoma insitu morphology containing focal invasive ductal carcinoma (b) Heterogenity in breast carcinoma lymph node metastasis (papillary, micropapillary and ductal morphology) (c) ER positivity and heterogeneity in breast carcinoma (d) Intratumoral heterogeneity with CISH
Microscopic Grading in Breast Carcinoma (Nottingham Modification of the Bloom-Richardson system)
Tubule formation
point: Tubule formation constitutes more than 75% of the tumor,
points: Tubule formation constitutes 10–75% of the tumor,
points: Tubule formation constitutes less than 10% of the tumor.
Note: Tubule formation evaluation should take into account the entire tumor.
Nuclear pleomorphism
point: Nucleus shape and size difference mild,
points: Nucleus shape and size difference moderate,
points: Nucleus shape and size difference significant.
Note: The area containing cells with most prominent nuclear pleomorphism should be evaluated.
Mitotic Count
Mitotic counting process should only be done at the periphery of the tumor and should be started from the most mitotic active areas. The suggested application is counting within the same field, but it is not necessary to use subsequent fields. Areas rich in tumor that are free of normal breast tissue are preferred as much as possible. Prophase cells should not be counted. Due to differences in image area due to varying brands of microscopes, there are determined and accepted values for the number of mitotic count. Based on these values mitotic count are scored as 1, 2 and 3.
A total score is obtained by scores on tubule formation, nuclear pleomorphism and mitotic count. The histological grade is determined based on the obtained total score, as shown in Table 1.
Table 1.
Histologic type scoring
| Total Score | Histologic Grade |
|---|---|
| 3–5 | I |
| 6–7 | II |
| 8–9 | III |
Histological evaluation by this method is semi-quantitative, but provides very strong prediction for determining patient prognosis (6). In addition, it is known that histological grading is associated with histological type as well as with molecular changes such as estrogen-progesterone receptor expression and HER-2 amplification (7). In the traditional approach, a number of powerful parameters such as tumor size and extension pattern (particularly lymph node involvement status) determine the stage of the disease, and these are important prognostic factors. The principles of a staging system that can be applied to all types of cancer and parameters of tumor, node, metastasis (TNM) were defined by Pierre Denoix, and has gained wide acceptance shortly afterwards (8, 9). The TNM system is used as a common language among treatment centers widely all over the world, to guide treatment planning, provide a possibility to demonstrate the effectiveness of the treatment during follow-up and predict prognosis. However, with advances in diagnosis and treatment of breast cancer, improving technology and increased knowledge, initiatives to evaluate tumor biology in detail, accumulation of new data showing that most prognostic factors are related to biological features of the tumor, and most importantly the observation of very different survival rates within tumors with the same TNM group and same histological type have led to the search for alternative solutions.
The TNMEIO system was suggested by the European Institute of Oncology (EIO) in an effort to include tumor characteristics affecting treatment decisions in the TNM system (10). In this system, all anatomical and biological properties such as ER, PR, HER-2 of the tumor are included. It was suggested that if the diameter of the breast carcinoma is 1.3 cm it can be defined as T1.3, similar application can be used for lymph node invasion, the number of all examined lymph nodes including sentinel lymph and the number of lymph nodes with invasion (e.g. N0/1, N3/9, Ns0/9, s: sentinel lymph node), and the site of metastasis is presented with a suffix to M (MH: Hepatic metastases, ML: Lung metastasis). According to this system, a tumor with a maximum size of 1.8 cm, ER positive, PR negative, HER2 positive, with liver metastases, and invasion in 2 out of 26 lymph nodes is coded as T1.8, ER +, PR−, HER2 +, N2/26, MH. Some centers prefer more aggressive treatment modality in tumors with lympho-vascular invasion.
Indexes such as Nottingham Prognostic Index (11), Adjuvant! Online (AO) (12) and St. Gallen criteria (13) are widely used when deciding on treatment, in order to increase the success of predicting survival and development of metastatic disease. Histopathologic evaluation is very effective in directing clinical treatment of breast cancer patients. However, the significant differences detected among patients with the same histological subtype (eg, tubular carcinoma) and the same histological grade-the same stage (eg, node-negative disease) in response to treatment and long-term survival, as well as benefits of tamoxifen treatment in ER positive patients, and of trastuzumab treatment in patients with HER-2 amplification, all support the belief that breast cancer is a heterogeneous group of diseases, thus pointing out the importance of biological properties of the tumor in its management. Although the value of clinical data and algorithms are limited with results from undersized clinical studies, it is promising that new generation molecular methods can provide a more precise classification, thus more targeted and perhaps individualized treatment options.
New Era, New Beginning
All kinds of molecular analytical methods applied to cancer tissue help us determine the prognostic and predictive factors of the cancer. Together with the introduction of microarray-based technological applications, which is one of these beneficial molecular analytical methods, the development and use of genomic and expression profiling studies has led to development of a breast cancer classification system based on tumor biology rather than morphology. Studies conducted with this method also support the idea that breast cancer is a molecularly heterogeneous disease with different clinics, and that it is a complex disease containing different gene expression patterns that influence prognosis (14–17). The results of these studies are believed to be more objective than the currently used relatively subjective histopathological evaluation. New era, new methods applied with new technology provide definition of various aspects of breast cancer again but in a different way, and allows us correlating these with morphological appearance of breast cancer. Nevertheless, it must be remembered that new molecular technological evaluation methods are not completely independent, and in fact, the data obtained incorporate many assumptions.
Perou and Sorlie proposed “Molecular Classification” terminology in breast cancer for the first time with a comprehensive study showing the differences in gene expression in 2000 (14). In this study breast cancer was divided into different sub-groups according to various gene expression: “Luminal” (often differentiated in two or three subgroups; reflecting ER, ER regulatory genes and the expression of genes expressed in normal luminal epithelial cells), “HER-2 positive “(reflecting ErbB2 / HER-2 amplification and overexpression),” basal “(reflecting ER, PR, and HER-2 negative and the expression of genes expressed in normal breast basal and / myoepitelial cells). A normal-like subgroup has been described, but the importance of identifying this subgroup and its consequences are not clear, because it seems to represent samples with low tumor cell content and more normal tissue components. Such molecular subtypes were formed by differentiation of numerous intrinsic genes (showing very little difference in the repeated samples of the same tumor but high rates of difference in different tumors) and clustering of patients in a hierarchical order to separate into different groups in terms of transcription (14). Since only samples belonging to a retrospective evaluation of numerous cases can be classified with this method, the “Single Sample Predictor (SSP)” has been defined. With the implementation of SSP, inclusion of a single tumor to a specific subtype was provided by using the closest main class of the tumor (16–18). SSP2003, the first described SSP, includes 500-gene expression (16). SSP2003 is further differentiated with repetitions of intrinsic gene lists, yielding two new SSPs: SSP2006 and PAM50 (17, 18). There are certain limitations of SSPs defined in this way. Pusztai et al. (19) emphasized such limitations in detail, and showed that even small changes in the initial set of defined SSPs can lead to significant changes in hierarchical clustering that is used defining subgroups. Thus, the stability of the method has been questioned (19, 20).
It was shown that three main subtypes can be identified in a stable manner by only using genes related to ER and HER-2 phenotypes instead of using hundreds of intrinsic genes (21). These subtypes are ER− / HER2− (basal-like), HER2+(HER2-Enriched), and ER+/ HER2− (luminal A and B combined). Mackay et al. (22) pointed out that there was no inter-observer harmony in defining subtypes from dendrograms generated by hierarchical clustering. Such complex studies and assessments have led to new, alternative classification approaches (23): In this approach model, an mRNA expression predictor that classifies breast tumors in four molecular entities, by quantitative measurement of three genes, namely ESR1, ERBB2 and AURKA. The AURKA mentioned in this context is a proliferation module providing separation between low and high-proliferative tumors (aurora kinase A [AURKA]). The four entities defined by this model are as follows: ER +/ HER2−/ low proliferative, ER +/ HER2−/ high proliferative, HER2+and ER−/ HER2−.
The defined model is a simplified subtype classification model (Subtype Classification Model-SCM) and contains only ESR1, ERBB2 and AURKA genes, and is simply known as SCMGENE. It was reported that major breast cancer intrinsic subtypes can be defined by SCMGENE determinants (ESR1, ERBB2 and AURKA), and that it provides a firm distinction for clinical use similar to 50 genes predictor (PAM50). A recent study compared results from PAM50 and SCMGENE in terms of different factors (such as patient prognosis, pathologic complete response, biological differences) (24). This study concluded that classification into major molecular subtypes that are clinically relevant was the best model of those including wider gene panels.
Despite the ongoing debate on advantages of molecular subtyping methods of breast cancer over each other, basically luminal A, luminal B, HER2, basal and normal-like molecular subgroups represent different prognostic subgroups, has led to rapid acceptance of the proposed classification system into clinical practice. Efforts have been made to ensure widespread use of molecular classification system as a diagnostic tool and prove its validity. It was proved that various data from different patient study groups and different array platforms overlap with the classification system quite tightly (15–18).
Detection of difference in response to treatment and metastatic pattern according to molecular subtypes further increased the value of molecular classification (25, 26).
Ultimately, the idea that a patient with breast cancer can be classified according to the molecular subtype of the tumor and thus directed to appropriate, specific, targeted therapies has become very attractive. Nowadays the search for specific, targeted, personalized treatment programs are ongoing in all types of cancer. There are controversies if data obtained from the assessment of histological grade of the tumor, ER and HER-2 status can provide appropriate treatment that the molecular classification will bring.
What are the molecular subgroups?
The current molecular classification divides breast cancer into five groups as luminal A, luminal B, HER-2, basal and normal breast like. Further grouping of these subgroups seem possible and necessary. Recently, HER-2 subgroup is divided into three groups that clinically behave differently, one of them reported to have a highly aggressive behavior. Because of these differences, determinants that can explain difference in prognosis of patients with HER-2 have been tried to develop (27). Based on these studies, it is expected that differences in prognosis despite treatment programs directed with molecular indicators that provide data on tumor molecular identity, prognosis, and individualized treatment will be an even more pronounced subject in the near future. For example, based on HER-2 patients who were resistant to treatment with monoclonal antibodies (Herceptin) targeting the extracellular domain or those who relapsed after treatment, possible causative mechanisms were investigated (28–30). It was reported that a portion of HER-2 positive breast cancer patients with poor prognosis express a heterogeneous group of HER2 carboxy-terminal fragments known as p95HER2 (29). One of these fragments, 611-CTF, is the oncogene of HER2. Thus, it was concluded that 611-CTF gene status was probably effective in the progression of p95HER2 positive tumors. In addition, the expression of HER2 isoform that encodes a receptor without exon 16 is known as delta 16HER2, and is recognized today as one of the trastuzumab resistance mechanisms (31).
Molecular Subgrouping Valid in Clinical Use
Most of molecular subgrouping studies were performed on non-specific types of ductal carcinoma and is well known, this histological group contains non-specific tumor types. Therefore, efforts aiming to separate this group of heterogeneous group into subgroups seem to be meaningful. Molecular subgrouping can be provided by using a few immunohistochemical markers. A panel including ER, PR, HER2, Ki-67, epidermal growth factor receptor (EGFR) and basal cytokeratins (CK14 and CK5 / 6 etc) can be used to distinguish between “luminal”, HER2 and triple negative tumors. In fact, there is no consensus on the determinants defining “basal” tumors, nevertheless it is considered that the use of EGFR and CK5 / 6 can aid in identification of this subgroup and predict prognosis (32).
Proliferation markers are very important in molecular subgrouping besides ER and HER-2, especially in ER-positive tumors. However, the appropriateness of using Ki-67 or more detailed mitotic index scoring system as a proliferation marker has been questioned. The application of a Ki67 scoring as positive / negative or high / low in patient follow-up and treatment is controversial and there is no consensus on this issue today. Separation of luminal tumors into subgroups is mainly based on proliferation intensity. Therefore, a gene expression profiling study combined with IHC was conducted in order to determine the Ki67 limit value that can be used routinely to distinguish luminal a tumors from luminal B tumors (33). The limit value in this study was determined as 13.25%. It is obvious that application of such a precise limit in clinical practice is not very realistic. It seems that there is need for more practical prognostic tests that can be evaluated in routine diagnostic pathology laboratories and used in clinical practice. Similar to histological evaluation, molecular tests that show the average gene expression level show that breast cancer is a heterogeneous tumor. This situation could partly explain the mismatch in tumor distinction into molecular subtypes. Various algorithms have been developed to place each tumor into one of the five basic molecular subtypes (luminal or luminal B, normal breast-like, HER2 and basal) (16–18). These algorithms are defined as “Single Sample predictors” (SSP), as previously referred to. Since each new diagnosed breast cancer patient should be separated into a specific molecular subgroup in order to determine the prognosis and decide on specific treatment, application of such algorithms will be required.
Major molecular subtypes According to gene expression profiles in breast cancer are summarized in Table 2 (34, 35).
Table 2.
Major molecular subtypes of breast cancer
| Molecular Subtype | ||||
|---|---|---|---|---|
| Luminal A | Luminal B | HER2/neu | Basal likea | |
| Gene expression pattern | Expression of luminal (low molecular weight) cytokeratins, high expression of hormone receptors and related genes | Expression of luminal (low molecular weight) cytokeratins, moderate-low expression of hormone receptors and related genes | High expression of HER2/neu, low expression of ER and related genes | High expression of basal epithelial genes and basal cytokeratins, low expression of ER and related genes, low expression of HER2/neu |
| Clinical and biologic properties | 50% of invasive bresat cancer, ER/PR positive, HER2/neu negative | 20% of invasive breast cancer, ER/PR positive, HER2/neu expression variable, higher proliferation than Luminal A, higher histologic grade than Luminal A | 15% of invasive breast cancer, ER/PR negative, HER2/neu positive, high proliferation, diffuse TP53 mutation, high histologic grade and nodal positivity | ~15% of invasive breast cancer, most ER/PR/HER2/neu negative (triple negative), high proliferation, diffuse TP53 mutation, BRCA1 dysfunction (germline, sporadic) |
| Histologic correlation | Tubular carcinoma, Cribriform carcinoma, Low grade invasive ductal carcinoma, NOS, Classic lobular carcinomab | Invasive ductal carcinoma, NOS Micropapillary carcinoma | High grade invasive ductal carcinoma, NOS | High grade invasive ductal carcinoma, NOS Metaplastic carcinoma, Medullary carcinoma |
| Response to treatment and prognosis | Response to endocrine therapy | Response to endocrine therapy (tamoxifene and aromatase inhibitors) not as good as Luminal A | Response to trastuzumab (Herceptin) | No response to endocrine therapy or trastuzumab |
| Variable response to chemotherapy | Variable response to chemotherapy (better than Luminal A) | Response to chemotherapy with antracyclins | Sensitive to platinum group chemotherapy and PARP inhibitors | |
| Good prognosis | Prognosis not as good as Luminal A | Usually unfavorable prognosis | Not all, but usually worse prognosis | |
PARP poly-adenosinediphosphate ribose polymerase
Basal like tumor group includes a low-grade group with low proliferation but expression of basal type (high molecular weight) cytokeratin and triple negative phenotype ( like adenoid cystic carcinoma, secretuar carcinoma).
Classical lobuler carcinoma generally exhibits luminal A properties, while pleomorphic lobular carcinoma usually shows features of other molecular subtypes.
Special Type Tumors
Histopathological special types of breast cancer constitute approximately 25%, have different architectural patterns, are less associated with clinical features, and have a better prognosis than non-specific type ductal carcinoma. Most of the gene expression profiling studies are related to non-specific type ductal carcinoma. Evaluation of molecular properties for specific types of breast cancer has not been systematically studied as much as in non-specific ductal carcinoma. Genomic and gene expression studies have also shown that special histological types are much more homogenous than the non-specific ductal carcinoma. This emphasizes the importance of morphological assessment made by histopathologists (Figure 2). Special types of breast cancer can be explained by specific somatic re-organization, resulting in specific development patterns that pathologists have been defining morphologically for years. Indeed, some special types are associated with specific genetic alterations that describe both their special developmental pattern and their specific behavior (36). The most descriptive molecular feature of lobular carcinoma is the loss of E-cadherin. In addition, translocations observed in various malignancies other than the breast were observed in secretory carcinoma, and adenoid cystic carcinomas (37, 38). Weigelt et al. (39) demonstrated that each specific subtype in 113 breast tumors containing 11 different special types (other than lobular and apocrine special type) actually belonged to only one molecular subtype (luminal, HER2, have proven that normal breast-like and basal) by gene expression profiling study. According to this study, neuroendocrine and mucinous carcinomas have very similar genetic profiles, which was an expected finding due to the histopathologic observation of these two components within the same tumor. Adenoid cystic, medullary, and metaplastic carcinomas were within basal group, as expected. However, it was interesting that despite these carcinoma’s good prognosis, they were included in poor prognosis category with other basal tumors (39). This situation is defined in basal phenotype non-specific type ductal carcinoma and reflects the significant heterogeneity of basal-type. In addition, it should be emphasized that “histopathological evaluation” is much more important than molecular subtyping in diagnostic application for special types such as these.
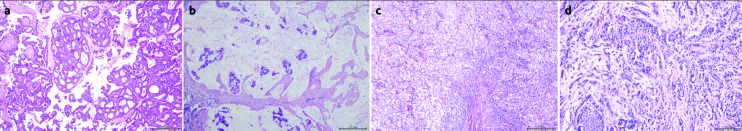
Figure 2. a–d.
Specific subtype examples (a) Invasive cribriform carcinoma (b) Mucinous carcinoma (c) Medullary carcinoma (d) Invasive lobular carcinoma, containing lobular carcinoma insitu foci
Histological and molecular characteristics of specific types of breast cancer are shown in Table 3.
Table 3.
Histologic and molecular properties of specific type breast cancer
| Molecular subtype | Common histologic types | HG | ER status (by IHC) | HER2 status (by ISH/IHC) | Ki67 (by IHC) | Specific IHC/ molecular properties |
|---|---|---|---|---|---|---|
| Luminal A | Classical, lobular, tubular, cribriform | 1 or 2 | + | − | Low | Luminal CK +, E−cadherin +/− |
| Luminal B | Micropapillary | 2 or 3 | +/− | −/+ | High | Luminal CK +, p53 mutations |
| Basal like | Medullary, metaplastic, adenoid cystic, secretory | 3 | − | − | High | Basal CK+, p53 mDNA repair loss, EGFR+/− mutations |
| Mol. apocrine | Apocrine, plemorphic lobular | 2 or 3 | − | −/+ | High | Androgene receptor+ |
| Claudin-low | Metaplastic | 3 | − | − | High | Cancer like stem cell, EMT like, low E-cadherin level |
As depicted in the table, most specific type breast cancer are homogenous entities and are included in only one molecular subtype (such as adenoid cystic carcinoma basal like, micropapillary carcinoma luminal type). However, specific types such as classical and pleomorphic lobular carcinoma and apocrine carcinoma are heterogenous. The table depicts probable histologic grade, ER, HER2, Ki67 status for each molecular classification.
Exclusive Subtype: Lobular Carcinoma
Five to 15% of all breast cancers are invasive lobular carcinomas (ILC), and it is the most common “special type”. The very special biological and clinical behavior patterns of ILC are more important than the fact that they are histopathologically different forms from non-specific ductal carcinoma. Although ILC has better prognostic features as compared to IDC, they exhibit a similar to or worse outcome than ductal carcinoma in long-term follow-up (40–43). The slow progress and diffuse growth pattern that results in diagnostic delay, difficulty in detection by routine screening programs, and difficulty in obtaining reliable and safe surgical margins may be suggested as reasons for this situation. Thus, ILC presents with distant metastases. The ILC metastasis pattern is also very interesting by its being quite different from IDC, with metastasis to bone, gastrointestinal tract, gynecological organs and peritoneal cavity (44–46). This complex structure of ILC may also partly be explained by a large number of variants. These variants can be listed as solid, alveolar, pleomorphic, mixed ductal / lobular, tubulolobular, signet ring cell and histiocytic type (47). Except the pleomorphic subtype, our information on these variant’s biological and clinical behavior is limited (48–53). Only a few studies showed association between alveolar and solid types with classical type (54).
ILC cells have characteristic cytologic features and a diffuse growth pattern. Mostly, they are histological grade 2 and do not show lymphovascular invasion. Generally, they are ER and PR positive, and they rarely express HER2, p53, EGFR and basal cytokeratin.
Molecular analysis of lobular carcinoma is not studied as much as non-specific ductal carcinoma. They are mostly luminal subtype, but basal and HER2 subtypes were also identified.
Weigelt et al. (39) suggested a close relationship between tubular carcinoma and lobular carcinoma based on their observation of very close hierarchical clustering in lobular and tubular carcinomas. In genomic analysis, lobular carcinomas show a similar genomic profile to low-grade ductal carcinomas, and this genomic profile is very different from high-grade ductal carcinomas. This finding shows a close developmental relationship between “low-grade (low-grade ductal and lobular)” and ER positive tumor types. Results of initial genomic studies detected that boundaries between ductal and lobular carcinomas were unclear, therefore they supported the idea that all low-grade cancers (ductal, lobular and tubular) represent a low-grade tumor family beginning from a general precursor, such as columnar cell lesions (55–58). Expression profile studies of lobular, ductal and low-grade ductal carcinomas were studied with the idea that a classification such as ductal and lobular might not be proper and it was found that they had similar profiles (59–61). However, despite similar profiles were detected in tubular, lobular and low-grade tumors, differences in specific gene expressions suggest that these types are different entities. Especially in lobular tumors, decreased functions of genes associated with cell adhesion and extracellular matrix were described. Thus, these differences determined in gene expression in lobular tumors reflect characteristic differences in the development pattern of invasive lobular carcinomas and the loss of E-cadherin that is a cell adhesion molecule (62, 63).
Including lobular breast cancer in a classification like molecular luminal A, luminal B, HER2 or basal form may cause a lack of understanding in the complex structure and heterogeneity of breast cancer, because lobular breast cancer is a very special subtype in all breast cancers
Studies investigating changes in epigenetic mechanisms such as DNA methylation, and in genetic information carriers that do not code but can lead to functional changes such as microRNA (miRNA) will enable more accurate and complementary description of molecular structure of breast cancer. A comprehensive study that used various technological platforms with different breast cancer types has identified subtype-specific mutations and copy number changes that enabled understanding tumor biology and achieving the targeted treatment by evaluation of many cancer development pathways in different subtypes (64). However, the implementation of treatment programs targeting these changes will depend on an increase in the number of such studies, and results from larger study groups.
Conclusion
Molecular subtyping developed in breast cancer emphasized biological heterogeneity, which has been histopathologically defined by pathologists for a long time. In the last twenty years, identification of HER2 pathway and the relevant use of Herceptin, the use of DNA repair mechanisms and PARP inhibitors were possible by clarification of breast cancer biology with molecular methods and the emergence of new horizons for the development of new therapeutic interventions. The perception on what we know and what we have just defined on genomic architecture underneath different subtypes of breast cancer will probably change with the new generation of molecular methods including next-generation sequencing. We must not forget that molecular classification of breast cancer is still in the development stage and has limitations of today. Old fashioned, traditional histopathological subgrouping has many features for us to prefer this classification especially in special types (such as lobular or ductal, secretory, micropapillary, adenoid cystic carcinoma). In this regard, accepting and applying the conventional histopathological methods and new molecular studies, as a “partnership” seems to be the best way for follow-up in breast cancer. It is clear that we are not in an era where either pathologists can leave their microscope aside and make a classification of breast cancer based on computer, or oncologists can implement follow-up programs just based on molecular classification.
In conclusion, the traditional old-fashioned way should be together with a molecular classification based new-fashion way.
Footnotes
Peer-review: Externally peer-reviewed.
Author contributions: Concept - N.E., S.A.; Design- N.E., S.A., B.Z.; Supervision - N.E., S.A., E.V.; Funding - N.E., E.Y., E.V.; Data Collection &/or Processing - N.E., EY.; Analysis &/or Interpretation - N.E.; Literature Review - N.E., E.Y., B.Z.; Writer - N.E.; Critical Review - E.Y., S.A., E.V.
Conflict of Interest: No conflict of interest was declared by the authors.
Financial Disclosure: The authors declared that this study has received no financial support.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. http://dx.doi.org/10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Singletary SE, Connolly JL. Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J Clin. 2006;56:37–47. doi: 10.3322/canjclin.56.1.37. [DOI] [PubMed] [Google Scholar]
- 3.Ellis IO, Cornelisse CJ, Schnitt SJ, Sasco AJ, Sastre-Garau X, Kaaks R. Invasive breast carcinomas. In: Tavassoli FA, Devilee P, editors. WHO Classification of Tumours Pathology and Genetics of Tumours of the Breast and Female Genital Organs. Lyon: IARC Press; 2003. pp. 13–19. [Google Scholar]
- 4.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. http://dx.doi.org/10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 5.Buerger H, Otterbach F, Simon R, Schäfer KL, Poremba C, Diallo R, Brinkschmidt C, Dockhorn-Dworniczak B, Boecker W. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol. 1999;189:521–526. doi: 10.1002/(SICI)1096-9896(199912)189:4<521::AID-PATH472>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 6.Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Richardson AL, Schnitt SJ, Schmitt FC, Tan PH, Tse GM, Badve S, Ellis IO. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 2010;12:207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakha EA, Reis-Filho JS, Ellis IO. Combinatorial biomarker expression in breast cancer. Breast Cancer Res Treat. 2010;120:293–308. doi: 10.1007/s10549-010-0746-x. http://dx.doi.org/10.1007/s10549-010-0746-x. [DOI] [PubMed] [Google Scholar]
- 8.Denoix PF. Nomenclature classification des cancers [in French] Bull Inst Nat Hyg (Paris) 1952;7:743–748. [Google Scholar]
- 9.International Union Against Cancer. TNM Classification of Malignant Tumours. Geneva, Switzerland: International Union Against Cancer; 1968. [Google Scholar]
- 10.Veronesia U, Vialeb G, Rotmensza N, Goldhirscha A Rethinking TNM: Breast cancer TNM classification for treatment decision-making and research. The Breast. 2006;15:3–8. doi: 10.1016/j.breast.2005.11.011. http://dx.doi.org/10.1016/j.breast.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat. 1992;22:207–19. doi: 10.1007/BF01840834. http://dx.doi.org/10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- 12.Ravdin PM, Siminoff LA, Davis GJ, Mercer MB, Hewlett J, Gerson N, Parker HL. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J J Clin Oncol. 2001;19:980–991. doi: 10.1200/JCO.2001.19.4.980. [DOI] [PubMed] [Google Scholar]
- 13.Goldhirsch A, Glick JH, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Panel members: Meeting highlights: international expert consensus on the primary therapy of early breast cancer 2005. Ann Oncol. 2005;16:1569–1583. doi: 10.1093/annonc/mdi326. http://dx.doi.org/10.1093/annonc/mdi326. [DOI] [PubMed] [Google Scholar]
- 14.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. http://dx.doi.org/10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 15.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. http://dx.doi.org/10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;1:8418–8423. doi: 10.1073/pnas.0932692100. http://dx.doi.org/10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. http://dx.doi.org/10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS, Perou CM. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96–107. doi: 10.1186/1471-2164-7-96. http://dx.doi.org/10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pusztai L, Mazouni C, Anderson K, Wu Y, Symmans WF. Molecular classification of breast cancer: limitations and potential. Oncologist. 2006;11:868–877. doi: 10.1634/theoncologist.11-8-868. http://dx.doi.org/10.1634/theoncologist.11-8-868. [DOI] [PubMed] [Google Scholar]
- 20.Andre F, Pusztai L. Molecular classification of breast cancer: implications for selection of adjuvant chemotherapy. Nat Clin Prac Oncol. 2006;3:621–632. doi: 10.1038/ncponc0636. http://dx.doi.org/10.1038/ncponc0636. [DOI] [PubMed] [Google Scholar]
- 21.Kapp AV, Jeffrey SS, Langerod A, Borresen-Dale AL, Han W, Noh DY, Bukholm IR, Nicolau M, Brown PO, Tibshirani R. Discovery and validation of breast cancer subtypes. BMC Genomics. 2006;7:231. doi: 10.1186/1471-2164-7-231. http://dx.doi.org/10.1186/1471-2164-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackay A, Weigelt B, Grigoriadis A, Kreike B, Natrajan R, A’Hern R, Tan DS, Dowsett M, Ashworth A, Reis-Filho JS. Microarray-based class discovery for molecular classification of breast cancer: analysis of interob-server agreement. J Natl Cancer Inst. 2011;103:662–673. doi: 10.1093/jnci/djr071. http://dx.doi.org/10.1093/jnci/djr071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haibe-Kains B, Desmedt C, Loi S, Culhane AC, Bontempi G, Quackenbush J, Sotiriou C. A three-gene model to robustly identify breast cancer molecular subtypes. J Natl Cancer Inst. 2012;104:311–325. doi: 10.1093/jnci/djr545. http://dx.doi.org/10.1093/jnci/djr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prat A, Parker JS, Fan C, Perou CM. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;135:301–306. doi: 10.1007/s10549-012-2143-0. http://dx.doi.org/10.1007/s10549-012-2143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. http://dx.doi.org/10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 26.Korde LA, Lusa L, McShane L, Lebowitz PF, Lukes L, Camphausen K, Parker JS, Swain SM, Hunter K, Zujewski JA. Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer. Breast Cancer Res Treat. 2010;119:685–699. doi: 10.1007/s10549-009-0651-3. http://dx.doi.org/10.1007/s10549-009-0651-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staaf J, Ringnér M, Vallon-Christersson J, Jönsson G, Bendahl PO, Holm K, Arason A, Gunnarsson H, Hegardt C, Agnarsson BA, Luts L, Grabau D, Fernö M, Malmström PO, Johannsson OT, Loman N, Barkardottir RB, Borg A. Identification of subtypes in human epidermal growth factor receptor 2-positive breast cancer reveals a gene signature prognostic of outcome. J Clin Oncol. 2010;28:1813–1820. doi: 10.1200/JCO.2009.22.8775. http://dx.doi.org/10.1200/JCO.2009.22.8775. [DOI] [PubMed] [Google Scholar]
- 28.Mukohara T. Mechanisms of resistance to anti-human epidermal growth factor receptor 2 agents in breast cancer. Cancer Sci. 2010;102(10):1–8. doi: 10.1111/j.1349-7006.2010.01711.x. http://dx.doi.org/10.1111/j.1349-7006.2010.01711.x. [DOI] [PubMed] [Google Scholar]
- 29.Parra-Palau JL, Pedersen K, Peg V, Scaltriti M, Angelini PD, Escorihuela M, Mancilla S, Sánchez Pla A, Ramón Y, Cajal S, Baselga J, Arribas J. A major role of p95/611-CTF, a carboxy-terminal fragment of HER2, in the down-modulation of the estrogen receptor in HER-2 positive breast cancers. Cancer Res. 2010;70:8537–8546. doi: 10.1158/0008-5472.CAN-10-1701. http://dx.doi.org/10.1158/0008-5472.CAN-10-1701. [DOI] [PubMed] [Google Scholar]
- 30.Sperinde J, Jin X, Banerjee J, Penuel E, Saha A, Diedrich G, Huang W, Leitzel K, Weidler J, Ali SM, Fuchs EM, Singer CF, Köstler WJ, Bates M, Parry G, Winslow J, Lipton A. Quantitation of p95HER2 in parafin sections by sections using a p95-spesific antibody and correlation with outcome in a cohort of trastuzumab-treated breast cancer patients. Clin Cancer Res. 2010;16:4226–4235. doi: 10.1158/1078-0432.CCR-10-0410. http://dx.doi.org/10.1158/1078-0432.CCR-10-0410. [DOI] [PubMed] [Google Scholar]
- 31.Sasso Marianna, Bianchi Francesca, Ciravolo Valentina, Tagliabue Elda. Manuela Campiglio HER2 splice variants and their relevance in breast cancer. Journal of Nucleic Acids Investigation. 2011;2:e9. http://dx.doi.org/10.4081/jnai.2011.e9. [Google Scholar]
- 32.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. http://dx.doi.org/10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 33.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. http://dx.doi.org/10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modified from Schnitt SJ. Will molecular classification replace traditional breast pathology? Int J Surg Pathol. 2010;18:162S–166S. doi: 10.1177/1066896910370771. http://dx.doi.org/10.1177/1066896910370771.
- 35.Correa Geyer F, Reis-Filho JS. Microarray-based gene expression profiling as a clinical tool for breast cancer management: are we there yet? Int J Surg Pathol. 2009;17:285–302. doi: 10.1177/1066896908328577. http://dx.doi.org/10.1177/1066896908328577. [DOI] [PubMed] [Google Scholar]
- 36.Cummings MC, Chambers R, Simpson PT, Lakhani SR. Molecular classification of breast cancer: is it time to pack up our microscopes? Pathology. 2011 Jan;43:1–8. doi: 10.1097/PAT.0b013e328341e0b5. http://dx.doi.org/10.1097/PAT.0b013e328341e0b5. [DOI] [PubMed] [Google Scholar]
- 37.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, Poremba C, Sorensen PH. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–76. doi: 10.1016/s1535-6108(02)00180-0. http://dx.doi.org/10.1016/S1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 38.Persson M, Andrén Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. http://dx.doi.org/10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weigelt B, Horlings HM, Kreike B, Hayes MM, Hauptmann M, Wessels LF, de Jong D, Van de Vijver MJ, Van’t Veer LJ, Peterse JL. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141–150. doi: 10.1002/path.2407. http://dx.doi.org/10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
- 40.Rakha EA, El-Sayed ME, Powe DG, Green AR, Habashy H, Grainge MJ, Robertson JF, Blamey R, Gee J, Nicholson RI, Lee AH, Ellis IO. Invasive lobular carcinoma of the breast: response to hormonal therapy and outcomes. Eur J Cancer. 2008;44:73–83. doi: 10.1016/j.ejca.2007.10.009. http://dx.doi.org/10.1016/j.ejca.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Pestalozzi BC, Zahrieh D, Mallon E, Gusterson BA, Price KN, Gelber RD, Holmberg SB, Lindtner J, Snyder R, Thürlimann B, Murray E, Viale G, Castiglione-Gertsch M, Coates AS, Goldhirsch A International Breast Cancer Study Group. Distinct clinical and prognostic features of infiltrating lobular carcinoma of the breast: combined results of 15 International Breast Cancer Study Group clinical trials. J Clin Oncol. 2008;26:3006–3014. doi: 10.1200/JCO.2007.14.9336. http://dx.doi.org/10.1200/JCO.2007.14.9336. [DOI] [PubMed] [Google Scholar]
- 42.Viale G, Rotmensz N, Maisonneuve P, Orvieto E, Maiorano E, Galimberti V, Luini A, Colleoni M, Goldhirsch A, Coates AS. Lack of prognostic significance of ‘classic’ lobular breast carcinoma: a matched, single institution series. Breast Cancer Res Treat. 2009;117:211–214. doi: 10.1007/s10549-008-0112-4. http://dx.doi.org/10.1007/s10549-008-0112-4. [DOI] [PubMed] [Google Scholar]
- 43.Gruel N, Lucchesi C, Raynal V, Rodrigues MJ, Pierron G, Goudefroye R, Cottu P, Reyal F, Sastre-Garau X, Fourquet A, Delattre O, Vincent-Salomon A. Lobular invasive carcinoma of the breast is a molecular entity distinct from luminal invasive ductal carcinoma. Eur J Cancer. 2010;46:2399–2407. doi: 10.1016/j.ejca.2010.05.013. http://dx.doi.org/10.1016/j.ejca.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Borst MJ, Ingold JA. Metastatic patterns of invasive lobular versus invasive ductal carcinoma of the breast. Surgery. 1993;114:637–641. [PubMed] [Google Scholar]
- 45.Lamovec J, Bracko M. Metastatic pattern of infiltrating lobular carcinoma of the breast: an autopsy study. J Surg Oncol. 1991;48:28–33. doi: 10.1002/jso.2930480106. http://dx.doi.org/10.1002/jso.2930480106. [DOI] [PubMed] [Google Scholar]
- 46.Harris M, Howell A, Chrissohou M, Swindell RI, Hudson M, Sellwood RA. A comparison of the metastatic pattern of infiltrating lobular carcinoma and infiltrating duct carcinoma of the breast. Br J Cancer. 1984;50:23–30. doi: 10.1038/bjc.1984.135. http://dx.doi.org/10.1038/bjc.1984.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakha EA, Ellis IO. Lobular breast carcinoma and its variants. Semin Diagn Pathol. 2010;27:49–61. doi: 10.1053/j.semdp.2009.12.009. http://dx.doi.org/10.1053/j.semdp.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Eusebi V, Magalhaes F, Azzopardi JG. Pleomorphic lobular carcinoma of the breast: an aggressive tumor showing apocrine differentiation. Hum Pathol. 1992;23:655–662. doi: 10.1016/0046-8177(92)90321-s. http://dx.doi.org/10.1016/0046-8177(92)90321-S. [DOI] [PubMed] [Google Scholar]
- 49.Weidner N, Semple JP. Pleomorphic variant of invasive lobular carcinoma of the breast. Hum Pathol. 1992;23:1167–1171. doi: 10.1016/0046-8177(92)90035-2. http://dx.doi.org/10.1016/0046-8177(92)90035-2. [DOI] [PubMed] [Google Scholar]
- 50.Buchanan CL, Flynn LW, Murray MP, Darvishian F, Cranor ML, Fey JV, King TA, Tan LK, Sclafani LM. Is pleomorphic lobular carcinoma really a distinct clinical entity? J Surg Oncol. 2008;98:314–317. doi: 10.1002/jso.21121. http://dx.doi.org/10.1002/jso.21121. [DOI] [PubMed] [Google Scholar]
- 51.Vargas AC, Lakhani SR, Simpson PT. Pleomorphic lobular carcinoma of the breast: molecular pathology and clinical impact. Future Oncol. 2009;5:233–243. doi: 10.2217/14796694.5.2.233. http://dx.doi.org/10.2217/14796694.5.2.233. [DOI] [PubMed] [Google Scholar]
- 52.Simpson PT, Reis-Filho JS, Lambros MB, Jones C, Steele D, Mackay A, Iravani M, Fenwick K, Dexter T, Jones A, Reid L, Da Silva L, Shin SJ, Hardisson D, Ashworth A, Schmitt FC, Palacios J, Lakhani SR. Molecular profiling pleomorphic lobular carcinomas of the breast: evidence for a common molecular genetic pathway with classic lobular carcinomas. J Pathol. 2008;215:231–244. doi: 10.1002/path.2358. http://dx.doi.org/10.1002/path.2358. [DOI] [PubMed] [Google Scholar]
- 53.Sneige N, Wang J, Baker BA, Krishnamurthy S, Middleton LP. Clinical, histopathologic, and biologic features of pleomorphic lobular (ductal-lobular) carcinoma in situ of the breast: a report of 24 cases. Mod Pathol. 2002;15:1044–1050. doi: 10.1097/01.MP.0000027624.08159.19. http://dx.doi.org/10.1097/01.MP.0000030450.20581.E3. [DOI] [PubMed] [Google Scholar]
- 54.Da Silva L, Parry S, Reid L, Keith P, Waddell N, Kossai M, Clarke C, Lakhani SR, Simpson PT. Aberrant expression of E-cadherin in lobular carcinomas of the breast. Am J Surg Pathol. 2008;32:773–783. doi: 10.1097/PAS.0b013e318158d6c5. http://dx.doi.org/10.1097/PAS.0b013e318158d6c5. [DOI] [PubMed] [Google Scholar]
- 55.Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchió C, Reis-Filho JS. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 2010;57:171–192. doi: 10.1111/j.1365-2559.2010.03568.x. http://dx.doi.org/10.1111/j.1365-2559.2010.03568.x. [DOI] [PubMed] [Google Scholar]
- 56.Abdel-Fatah TM, Powe DG, Hodi Z, Lee AH, Reis-Filho JS, Ellis IO. High frequency of coexistence of columnar cell lesions, lobular neoplasia, and low grade ductal carcinoma in situ with invasive tubular carcinoma and invasive lobular carcinoma. Am J Surg Pathol. 2007;31:417–426. doi: 10.1097/01.pas.0000213368.41251.b9. http://dx.doi.org/10.1097/01.pas.0000213368.41251.b9. [DOI] [PubMed] [Google Scholar]
- 57.Simpson PT, Gale T, Reis-Filho JS, Jones C, Parry S, Sloane JP, Hanby A, Pinder SE, Lee AH, Humphreys S, Ellis IO, Lakhani SR. Columnar cell lesions of the breast: the missing link in breast cancer progression? A morphological and molecular analysis. Am J Surg Pathol. 2005;29:734–746. doi: 10.1097/01.pas.0000157295.93914.3b. http://dx.doi.org/10.1097/01.pas.0000157295.93914.3b. [DOI] [PubMed] [Google Scholar]
- 58.Schnitt SJ, Vincent-Salomon A. Columnar cell lesions of the breast. Adv Anat Pathol. 2003;10:113–24. doi: 10.1097/00125480-200305000-00001. http://dx.doi.org/10.1097/00125480-200305000-00001. [DOI] [PubMed] [Google Scholar]
- 59.Gruel N, Lucchesi C, Raynal V, Rodrigues MJ, Pierron G, Goudefroye R, Cottu P, Reyal F, Sastre-Garau X, Fourquet A, Delattre O, Vincent-Salomon A. Lobular invasive carcinoma of the breast is a molecular entity distinct from luminal invasive ductal carcinoma. Eur J Cancer. 2010;46:2399–407. doi: 10.1016/j.ejca.2010.05.013. http://dx.doi.org/10.1016/j.ejca.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 60.Lopez-Garcia MA, Geyer FC, Natrajan R, Kreike B, Mackay A, Grigoriadis A, Reis-Filho JS, Weigelt B. Transcriptomic analysis of tubular carcinomas of the breast reveals similarities and differences with molecular subtype-matched ductal and lobular carcinomas. J Pathol. 2010;222:64–75. doi: 10.1002/path.2743. http://dx.doi.org/10.1002/path.2743. [DOI] [PubMed] [Google Scholar]
- 61.Weigelt B, Geyer FC, Natrajan R, Lopez-Garcia MA, Ahmad AS, Savage K, Kreike B, Reis-Filho JS. The molecular underpinning of lobular histological growth pattern: a genome-wide transcriptomic analysis of invasive lobular carcinomas and grade- and molecular subtype-matched invasive ductal carcinomas of no special type. J Pathol. 2010;220:45–57. doi: 10.1002/path.2629. http://dx.doi.org/10.1002/path.2629. [DOI] [PubMed] [Google Scholar]
- 62.Droufakou S, Deshmane V, Roylance R, Hanby A, Tomlinson I, Hart IR. Multiple ways of silencing Ecadherin gene expression in lobular carcinoma of the breast. Int J Cancer. 2001;92:404–408. doi: 10.1002/ijc.1208. http://dx.doi.org/10.1002/ijc.1208. [DOI] [PubMed] [Google Scholar]
- 63.Rakha EA, Patel A, Powe DG, Benhasouna A, Green AR, Lambros MB, Reis-Filho JS, Ellis IO. Clinical and biological significance of E-cadherin protein expression in invasive lobular carcinoma of the breast. Am J Surg Pathol. 2010;34:1472–1479. doi: 10.1097/PAS.0b013e3181f01916. http://dx.doi.org/10.1097/PAS.0b013e3181f01916. [DOI] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. http://dx.doi.org/10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]