Abstract
Aflatoxins are potent carcinogenic and toxic substances that are produced primarily by Aspergillus flavus and Aspergillus parasiticus. We found that a bacterium remarkably inhibited production of norsolorinic acid, a precursor of aflatoxin, by A. parasiticus. This bacterium was identified as Achromobacter xylosoxidans based on its 16S ribosomal DNA sequence and was designated A. xylosoxidans NFRI-A1. A. xylosoxidans strains commonly showed similar inhibition. The inhibitory substance(s) was excreted into the medium and was stable after heat, acid, or alkaline treatment. Although the bacterium appeared to produce several inhibitory substances, we finally succeeded in purifying a major inhibitory substance from the culture medium using Diaion HP20 column chromatography, thin-layer chromatography, and high-performance liquid chromatography. The purified inhibitory substance was identified as cyclo(l-leucyl-l-prolyl) based on physicochemical methods. The 50% inhibitory concentration for aflatoxin production by A. parasiticus SYS-4 (= NRRL2999) was 0.20 mg ml−1, as determined by the tip culture method. High concentrations (more than 6.0 mg ml−1) of cyclo(l-leucyl-l-prolyl) further inhibited fungal growth. Similar inhibitory activities were observed with cyclo(d-leucyl-d-prolyl) and cyclo(l-valyl-l-prolyl), whereas cyclo(d-prolyl-l-leucyl) and cyclo(l-prolyl-d-leucyl) showed weaker activities. Reverse transcription-PCR analyses showed that cyclo(l-leucyl-l-prolyl) repressed transcription of the aflatoxin-related genes aflR, hexB, pksL1, and dmtA. This is the first report of a cyclodipeptide that affects aflatoxin production.
Aflatoxins are highly toxic, carcinogenic, and teratogenic secondary metabolites that are produced by certain strains of Aspergillus flavus and Aspergillus parasiticus (reviewed in reference 20). Recently, several strains of Aspergillus nomius, Aspergillus pseudotamarii, Aspergillus bombycis, and Aspergillus ochraceoroseus have also been reported to produce aflatoxin. These fungi are ubiquitous and grow on a variety of agricultural products under appropriate temperature and moisture conditions. Aflatoxins have been detected in numerous agricultural commodities, such as cereal grains, whole wheat, rye breads, oil seeds, cottonseed, etc. (25). The toxicity and carcinogenicity of aflatoxins have made contaminated commodities a significant health hazard all over the world. In fact, the incidence of liver cancer is high in regions with high endemic aflatoxin concentrations. Furthermore, the annual costs resulting from crop losses due to aflatoxin contamination and the costs involved in monitoring and disposal of contaminated commodities affect the agricultural economy (34).
Many studies have focused on developing aflatoxin control strategies, including genetic engineering for crop resistance, biological control with competitive, nonaflatoxigenic strains of the fungus A. flavus (6, 9), and regulation of aflatoxin biosynthesis by fungicides, pesticides, inhibitory substances originating from plants, and microbial substances (15, 24, 27). However, most of these strategies have been shown to be limited in effectiveness. Many microorganisms have been studied to control aflatoxin production. Sweedy and Dobson (32) have reported that bacteria, yeast, molds, actinomycetes, and algae can be used to lower aflatoxin levels in foods and feeds. Ono et al. have reported that aflastatin A, which has been isolated from mycelial extracts of Streptomyces sp., effectively inhibits aflatoxin production (17, 23). Saprophytic yeasts, such as Pichia anomala, Candida krusei, and others, have also been shown to inhibit aflatoxin production (14). However, despite the breadth of this research, effective methods to prevent aflatoxin contamination have not been developed.
During continuing research on aflatoxin biosynthesis (36, 37), we noticed that a hydroxyversicolorone (HVN)-accumulating mutant of A. parasiticus lost the ability to form HVN. HVN is an orange pigment that is a precursor of aflatoxins. The color of HVN in the mycelia was lost; i.e., the mycelia became white. Contamination with a certain bacterium was suspected to cause this change in color, and a bacterium was isolated from the culture. This bacterium, which was tentatively designated A1, also remarkably inhibited production of another precursor, norsolorinic acid (NA), in A. parasiticus strain NFRI-95, which is an NA-accumulating strain, as determined by the visual agar plate assay (14). We confirmed that A1 also inhibits aflatoxin production by this fungus.
The objectives of the present study were to identify the bacterial strain, to purify the inhibitory substance from the culture medium of the bacterium, to determine the structure of the inhibitory substance, and to characterize the effects of the inhibitory substance and its isomers and analogues on aflatoxin production. As a result of our research, we obtained a new inhibitor of aflatoxin production, cyclo(l-leucyl-l-prolyl).
MATERIALS AND METHODS
Microorganisms.
The A1 bacterium, which was identified in the present work as Achromobacter xylosoxidans, was used throughout this study. Aflatoxigenic A. parasiticus SYS-4 (= NRRL2999), NFRI-95, and NIAH-26 (38) were also used. The latter two strains were mutants obtained by UV irradiation of A. parasiticus SYS-4. The NFRI-95 strain accumulated NA, a bright red-orange pigment precursor of aflatoxin. A. xylosoxidans subsp. denitrificans JCM5490 and JCM9658 and A. xylosoxidans subsp. xylosoxidans JCM9656, JCM9659, and JCM9660 were obtained from the Japan Collection of Microorganisms, Institute of Physical and Chemical Research (RIKEN), Wako, Japan.
Media.
GY agar (2% glucose, 0.5% yeast extract, 1.5% agar) was used for the visual agar plate assay and the microtiter agar plate assay. GY liquid medium (2% glucose, 0.5% yeast extract) was used for culture of the A1 bacterium. YES liquid medium (2% yeast extract, 20% sucrose) was used for the tip cultures.
Chemicals.
Cyclo(l-Leu-l-Pro), cyclo(d-Leu-d-Pro), cyclo(l-Pro-l-Val), cyclo(l-Gly-l-Leu), cyclo(l-Gly-l-Pro), and cyclo(d-Ala-l-Pro) were obtained from Bachem AG (Bubendorf, Switzerland). Cyclo(l-Leu-d-Pro) was synthesized from Boc-d-proline (Peptide Institute, Inc., Osaka, Japan) and methyl-l-leucine, and cyclo(d-Leu-l-Pro) was synthesized from Boc-l-proline (Peptide Institute, Inc.) and methyl-d-leucine by using the procedure reported by Tani et al. (33). l-Leucine, d-leucine, l-proline, and d-proline were purchased from Sigma Chemical Co. St. Louis, Mo. All other chemicals were reagent grade.
Assays for the inhibitory substance. (i) Visual agar plate assay.
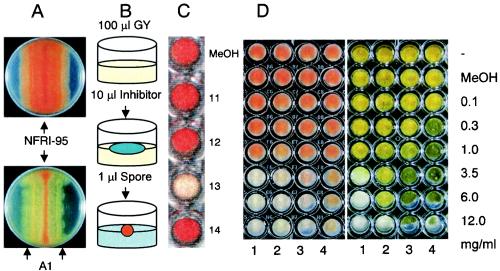
For selection of a bacterium that inhibits aflatoxin production, the visual agar plate assay (14) was used with the NA-accumulating mutant NFRI-95, with minor modifications (Fig. 1A). The spores of the fungus were inoculated in a line at the center of a plate containing GY agar, and an aliquot of a liquid culture of the bacterium was then inoculated in a line 1.5 cm from the centerline. After 3 to 7 days of incubation at 28°C, the effects of the bacterium on either NA accumulation in the mycelium or the growth of the fungus were observed from the underside of the plate. A decrease in the red pigment (NA) in the mycelium of the fungus indicated inhibition of aflatoxin production by the bacterium.
FIG. 1.
Assays for screening bacteria or a substance(s) that inhibits aflatoxin production. (A) Results of a visual agar plate assay with A. parasiticus NFRI-95 without (upper plate) or with (lower plate) bacterial strain A1. (B) Diagram of microtiter agar plate assay. (C) Results of a microtiter agar plate assay performed with fractions 11 to 14 from the HPLC chromatogram (Fig. 2). (D) Microtiter agar plate assay performed with various concentrations of cyclo(l-Leu-l-Pro) (lane 1), cyclo(d-Leu-d-Pro) (lane 2), cyclo(l-Leu-d-Pro) (lane 3), and cyclo(d-Leu-l-Pro) (lane 4). The left panel shows the bottom of the plate, and red indicates NA accumulation. The right panel shows the surface of the plate and morphological changes of the fungus.
(ii) Microtiter agar plate assay.
Based on the visual agar plate assay, we devised a small-scale assay system, in which a 96-well, flat-bottom tissue culture plate (Microtest 96; Becton Dickinson and Company, Franklin Lakes, N.J.) was used as a culture vessel (Fig. 1B and C). Autoclaved GY agar (100 μl) was poured into each well and then solidified. An aliquot (10 to 20 μl) of each fraction from each purification step or the methanol solution containing the inhibitory substance(s) was applied to the agar medium and then incubated for more than 30 min without a lid to let the solution diffuse into the medium and the methanol to evaporate. A spore suspension (1 μl) of A. parasiticus NFRI-95 was inoculated onto the center of the medium. Because the number of spores did not affect the results, we did not adjust it for this method. A small mass of clay or Parafilm was placed at each corner of the plate to produce a thin gap between the lid and the plate, and the gap was sealed with surgical tape (21N, No.12; 12 mm by 9 m; Nichiban Co., Tokyo, Japan,) so that the lid would not open. After incubation at 28°C for 2 or 3 days, the color of the mycelium was observed from the underside of the plate.
(iii) Tip culture method.
A spore suspension (5 μl) of A. parasiticus SYS-4 was inoculated into 250 μl of YES medium supplemented with various concentrations of the inhibitory substance by using a Pipetman tip as a culture vessel (38). After incubation at 28°C for 4 days, the mycelium and culture medium were separated by centrifugation. For detection of aflatoxin, 5 μl of the medium was spotted onto a thin-layer chromatography (TLC) silica gel plate (Silica Gel 60; Merck KGaA, Darmstadt, Germany), which was then developed with a solution containing chloroform, ethyl acetate, and 90% formic acid (6:3:1, vol/vol/vol). Aflatoxins were inspected under long-wavelength UV light (365 nm) and were photographed under UV light (365 nm) with a Fluor-S MultiImager (Bio-Rad Laboratories). To measure the amounts of the aflatoxins, the culture filtrate was extracted with chloroform, and an aliquot of the resulting chloroform extract was injected into a Shimadzu high-performance liquid chromatography (HPLC) apparatus (model LC-10A; Shimadzu Co., Kyoto, Japan) equipped with a silica gel column (0.46 by 15 cm; Shim-pack CLC-SIL; Shimadzu) and a fluorescence monitor (excitation wavelength, 365 nm; emission wavelength, 425 nm; Shimadzu model RF-535) at a flow rate of 1 ml min−1 at room temperature. The solvent system consisted of toluene, ethyl acetate, formic acid, and methanol (178:15:4:3, vol/vol/vol/vol). The retention times of aflatoxins B1, B2, G1, and G2 were compared with those of standard metabolite samples (aflatoxin B-aflatoxin G mixture; Sigma Chemical Co.). To detect precursors of the aflatoxins, precursors in the mycelial mat were extracted with acetone. After the extract was concentrated to dryness, the debris was dissolved in benzene-acetonitrile (98:2, vol/vol) and then analyzed by TLC.
Identification of the A1 bacterium.
The morphological characteristics of the A1 bacterium, which had been cultured on nutrient agar (Oxoid) at 30°C, were observed by microscopy. Gram staining, catalase reactivity, and oxidase reactivity were also determined (1). Final identification of the bacterium was performed by 16S ribosomal DNA (rDNA) sequence analysis. Genomic DNA of the bacterium was prepared with a DNeasy tissue kit (QIAGEN). A fragment of the 16S rDNA of the bacterium was sequenced. The first PCR was carried out by using universal primers Com1-F (CAGCAGCCGCGGTAATAC) and Com2-Ph-R (CCGTCAATTCCTTTGAGTTT) (29), and the 16S rDNA sequence was then determined by using primers rDNA-F1-#221 (AGTTTGATCCTGGCTCAG) and rDNA-R1-#222 (CGCTTGCACCCTACGTA) or primers rDNA-F2-#223 (CGGTCGCAAGATTAAAAC) and rDNA-R2-#224 (CCTTGTTACGACTTCACC). PCR was performed with Ready-to-Go beads (Pharmacia) under the following conditions: 94°C for 5 min, followed by 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 35 cycles and then 72°C for 10 min. All PCR products were ligated into vector TA, cloned, and sequenced. A DNA sequence analysis was performed for both strands by using the M13 reverse and forward primers and an ABI Prism BigDye terminator cycle sequencing Ready Reaction kit (Perkin-Elmer Corp.).
Production of the inhibitory substance.
The A1 bacterium was cultured by using the following culture conditions: (i) GY medium for a culture shaken at 150 rpm and incubated at 37°C for 1 to 4 days; (ii) GY medium, YES medium, or PD medium (2% Bacto Dextrose, 20% potato infusion) (Difco Laboratories, Detroit, Mich.) for a culture shaken at 150 rpm and incubated at 37°C for 3 days; (iii) GY medium for a culture shaken at 150 rpm and incubated at 10, 20, 30, or 37°C for 3 days; or (iv) GY medium for a continuous culture shaken at 150 rpm or a stationary culture incubated at 37°C for 3 days. After each culture, the whole medium was centrifuged at 8,000 × g for 30 min at room temperature. The supernatant was supplemented with 2% glucose and 0.5% yeast extract to compensate for the consumption of nutrients by bacterial growth, and the pH of the medium was adjusted to that of the original medium (pH 6.2 for YES and GY media; pH 5.1 for PD medium) by adding 1 M HCl. After filter sterilization with a 0.22-μm-pore-size Millipore membrane (Millex-GV; Millipore), the resulting solution was used as a medium for an A. parasiticus SYS-4 tip culture. After 4 days of culture, the aflatoxins present in the medium were analyzed by TLC.
Characterization of the inhibitory substance.
The heat stability of the inhibitory substance was examined after the culture medium of the A1 bacterium was incubated at 80°C for 120 min or autoclaved at 121°C for 15 min. The acid and alkaline stabilities of the inhibitory substance were examined by changing the pH of the culture medium to 1.5 or 11 by adding 1 M HCl or 1 M NaOH and incubating the solution at room temperature for 3 h. The pH of the solution was then readjusted to the original pH of the medium. The resulting medium was examined for its effects on aflatoxin production by the tip culture method.
Purification of the inhibitory substance. (i) Preparation of the culture medium.
The A1 bacterium was cultured in GY medium with shaking at 150 rpm at 37°C for 4 days. The total culture volume was 18 liters. The whole culture was then centrifuged at 8,000 × g at room temperature for 30 min. The supernatant was autoclaved and then centrifuged again. The resulting solution was used for purification of the inhibitory substance.
(ii) Diaion HP20 column chromatography.
The culture solution was loaded onto a column packed with Diaion HP20 resin (60 by 200 mm; Mitsubishi Chemical Co., Tokyo, Japan), which had been equilibrated with distilled water. The resin was then washed with 3 liters of water, and the substances bound to the resin were eluted stepwise by using 2 liters each of aqueous 50, 60, 80, and 100% methanol (MeOH) solutions. Each pooled fraction was concentrated to dryness with a rotary evaporator. Each residue was first solubilized with a small volume of methanol, and the remaining debris was then solubilized with water. Inhibitory activities were found in each methanol-solubilized fraction but not in the water-solubilized fraction. The 80% MeOH fraction, which had been solubilized with methanol, showed the strongest activity.
(iii) First TLC.
The 80% MeOH fraction was applied to a silica gel TLC plate and then developed with a solution containing chloroform, ethyl acetate, and 90% formic acid (6:3:1, vol/vol/vol). All areas developed on the TLC plates were divided into 11 regions under 365-nm UV light, and the silica gel was scraped from each region. The substances present in the silica gel were extracted with sixfold amounts of 100% MeOH. Each extract was concentrated to dryness, dissolved in a small amount of MeOH, and subjected to the microtiter agar plate assay. Region 5, which contained a fluorescent band at an Rf of 0.62, showed the strongest inhibitory activity.
(iv) Second TLC.
The resulting region 5 extract was purified further by the same silica gel TLC procedure, except that n-hexane-ethyl acetate-acetic acid (2:1:0.1, vol/vol/vol) was used. The area was divided into nine regions, and region 6, which contained a fluorescent band at an Rf of 0.22, showed the strongest activity.
(v) HPLC.
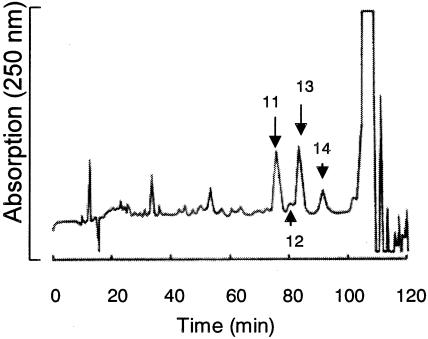
The active fraction was then purified by HPLC (LC-10AD system; Shimadzu) with a Cosmosil 5C18-AR column (10 by 250 mm; Nacalai Tesque, Inc., Tokyo, Japan). The column was developed with a solution containing MeOH, water, and acetic acid (25:74:1, vol/vol/vol) for 80 min, and the concentration of MeOH was then increased to 100% for 40 min. The flow rate was 1.0 ml min−1, and elution was monitored at 250 nm. All eluates were divided into 18 fractions that corresponded to the peaks or areas between two neighboring peaks and were collected. The eluates of the peak at 85.0 min (fraction 13) showed the strongest activity (Fig. 2) and were pooled. Purified inhibitory substance (3.5 mg) was obtained in this way.
FIG. 2.
HPLC chromatogram of the inhibitory substance of A. xylosoxidans A1. Peak 13 corresponds to the purified inhibitory substance.
Physicochemical characterization of the inhibitory substance.
The 1H nuclear magnetic resonance (NMR) spectrum was determined in deuterated chloroform with a JEOL JNM-ECP 500 NMR spectrometer. Chemical shifts were referenced to CDCl3 (δH 7.26, δC 77.0). Mass spectra were obtained with a JEOL AX505HA spectrometer (direct probe). 3-Nitrobenzyl alcohol was used as a matrix for fast atom bombardment mass spectrometry, and for electron impact ionization mass spectrometry the ionization voltage was 70 eV. Specific rotations were measured with a Horiba SEPA-200 polarimeter.
RT-PCR.
A. parasiticus SYS-4 (= NRRL2999) was cultured in YES broth or YES broth supplemented with 3.5 mg of cyclo(l-Leu-l-Pro) ml−1. After the mycelia were cultured for 63 h, they were disrupted with TRI REAGENT (Sigma-Aldrich, Inc.) by using FastPrep FP100A (Q-BIO gene; Bio 101). Total RNA was prepared according to the manufacturer's instructions (Sigma-Aldrich, Inc.) and then treated with RNase-free DNase (Gibco BRL). Reverse transcription (RT)-PCR was carried out by using the resulting total RNA and an RT-PCR kit (ReverTra Dash; Toyobo Co., Ltd., Osaka, Japan). The primers used were aflR-BamHI-F (CGCGGATCCATGGTTGACCATATCTCCCC) and aflR-HindIII-R (CCCCAAGCTTCATTCTCGATGCAGGTAATC) for aflR (accession no. AF441437) (35), HexB-F1 (CTGCGGGTGGAGCTGCA) and HexB-R1 (CAAGCTCCAAGGGCGGC) for the hexanoate synthase gene (accession no. AF391094) (13), pKSL1-F1 (CCAGGACAGCCCTATTCTAG) and pKSL1-R1 (GGAGTCCAGTGGTATTCAGC) for the polyketide synthetase gene, pksL1 (accession no. L42766) (10), MT-1wholeF1 (ACAAATACCCCTGGCTCAGG) and MT-1wholeR1 (ACCTGTTCCATCAAATCGTC) for the O-methyltransferase I gene, dmtA (accession no. AB022906) (22), and CMDF1 (GGTGATGGCCAGATCACCAC) and CMDR1 (CCGATGGAGGTCATGACGTG) for the calmodulin gene (accession no. AY017584) (41).
Feeding experiment.
By using the tip culture method, A. parasiticus NIAH-26 was cultured in YES medium supplemented with 40 μM sterigmatocystin in the presence or absence of 2 mg of cyclo(l-Leu-l-Pro) ml−1 at 28°C for 4 days. Aflatoxin formation was measured by extraction of the medium with chloroform, followed by HPLC analysis, as described above.
Nucleotide sequence accession number.
The genomic nucleotide sequence data for the 16S rRNA gene of A. xylosoxidans A1 have been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB161691.
RESULTS
Isolation and identification of bacteria that inhibit aflatoxin production by A. parasiticus.
The A. parasiticus hvn-1 mutant (36) lost HVN formation activity after several successive cultures. One bacterium (designated A1) was obtained from the culture. This organism also showed inhibitory activity for NA production by A. parasiticus NFRI-95 in the visual agar plate assay (Fig. 1A). This bacterium did not appear to inhibit growth of the fungus, as the fungus grew beyond the bacterial colony. After the visual agar plate assay, we transferred spores, as well as mycelia containing spores in the inhibition zone, to a new plate and then cultured the transplanted fungus and monitored the accumulation of pigments in the mycelia. The resulting colonies accumulated the pigment, indicating that the inhibitor(s) did not affect the next generation of the fungus (data not shown).
The A1 bacterium was a short rod-shaped organism that was gram negative, asporogenous, mobile, catalase positive, and oxidase positive. We determined a 1,485-nucleotide sequence for the 16S rRNA of this bacterium. A FASTA search of the GenBank database demonstrated that this sequence exhibited 99.933% sequence identity with the 16S rRNA gene of either A. xylosoxidans subsp. xylosoxidans (accession no. AF411021) or A. xylosoxidans strain AU0665 (accession no. AF411019). Only one base (thymine at position 444) in the A1 sequence (accession no. AB161691) was different from the cytosine in the two previously described sequences. These results indicate that A1 belongs to A. xylosoxidans. We designated this organism A. xylosoxidans NFRI-A1.
To determine whether the inhibition of aflatoxin production is unique to the A1 bacterium or is common in this species, we used other strains belonging to A. xylosoxidans in the visual agar plate assay. All five strains used, A. xylosoxidans subsp. denitrificans JCM5490 and JCM9658 and A. xylosoxidans subsp xylosoxidans JCM9656, JCM9659, and JCM9660, showed the same inhibition of NA production by A. parasiticus NFRI-95 (data not shown).
Microtiter agar plate assay.
We devised a small-scale assay method for detection of the inhibition of aflatoxin production (Fig. 1B). A well of a microtiter plate was used as a culture vessel for A. parasiticus NFRI-95. Because a culture medium depth greater than 2 mm was required to detect stable accumulation of NA in the mycelia, 100 μl of GY agar (depth, 4 mm) was poured into each well. The extract from a culture of A. xylosoxidans NFRI-A1 was applied onto solidified GY agar, and the A. parasiticus mutant NFRI-95 was then inoculated. Incubation for 2 to 3 days was found to be appropriate for sensitive detection of the inhibitors, whereas culture for even 1 day usually provided the same results. We observed a red-orange color for the mycelia from the reverse side of the plate, indicating NA accumulation. In contrast, from the upper side, we observed the effects of the substance on fungal growth, as well as morphological changes, such as conidiospore formation (Fig. 1C and D).
In this method, the sample was solubilized or suspended with methanol. High concentrations of methanol on the surface of the medium inhibited fungal growth. Also, methanol vapor from the inhibitor solution often inhibited growth of the fungus when many wells were used for the assays. We produced a gap between the plate and the lid by putting clay or Parafilm between the plate and the lid. Because methanol vapor could get out from inside the plate through the surgical tape, we could later detect clear signs of inhibition.
Culture conditions for production of the inhibitory substance.
Inhibitory activity was detected in the culture medium. We cultured A. xylosoxidans NFRI-A1 under various conditions, and then the culture media of the bacterium, which had been supplemented with nutrients, were used for the tip culture method with A. parasiticus SYS-4, followed by TLC analyses. When the A1 bacterium was cultured in GY liquid medium for 1 to 4 days, the medium of the 4-day culture showed the strongest inhibition of aflatoxin production. The A1 bacterium produced significant amounts of the inhibitory substance(s) at temperatures from 20 to 37°C. All media tested supported production of inhibitory substances. Aeration (shaking cultures and stationary cultures) did not significantly affect the production of inhibitory substances.
Treatment of cell-free supernatant fluids at pH 1.5 or 11 for 3 h, heat treatment at 80°C for 120 min, or autoclaving at 121°C for 15 min did not significantly affect the inhibitory activity, indicating that the inhibitory substance may be stable. We used the following conditions to purify the inhibitor(s): a culture shaken at 150 rpm at 37°C for 4 days.
Purification of the inhibitory substance.
Eighteen liters of A. xylosoxidans NFRI-A1 culture medium was applied to a Diaion HP20 column. The inhibitory activity bound to the column under aqueous conditions. The activity was then recovered in the 60 to 80% methanol fraction, and approximately 210 mg of residue was obtained following drying. Although the 80 to 100% methanol fraction also showed significant inhibitory activity, the activity was much weaker than that of the 60 to 80% methanol fraction.
The resulting solubilized 60 to 80% methanol fraction was further purified by TLC. Approximately 34 mg of residues was obtained from region 5 with a fluorescent band at an Rf of 0.62 on the TLC plate. The residues were purified further by the second TLC. Two of the nine regions on the TLC plate, region 6 with a fluorescent band at an Rf of 0.22 (about 13.7 mg) and region 5 with a fluorescent band at an Rf of 0.31 (about 7.6 mg), showed inhibitory activity. After the second TLC of the former region (Rf, 0.22), the inhibitory substance was purified further by HPLC (Fig. 2). Approximately 3.5 mg of inhibitory substance was obtained with a purity of more than 99% (Fig. 2, fraction 13). Although we also obtained other inhibitory compounds from the region with an Rf of 0.31 using HPLC, the amounts of this region were too small (less than 1 mg) to characterize.
Identification of the inhibitory substance.
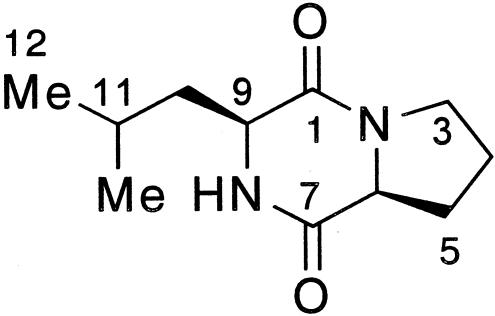
The molecular mass of the purified inhibitory substance was determined to be 210 Da based on the electron impact ionization mass spectrometry and fast atom bombardment mass spectrometry data. The 1H NMR spectrum showed the following resonances: δ 0.96 (3H, d, J = 6.4 Hz, 11-CH3), 1.00 (3H, d, J = 6.4 Hz, H-12), 1.52 (1H, ddd, J = 5.0, 9.6, 14.8 Hz, H-10), 1.74 (1H, m, H-11), 1.90 (1H, m, H-4), 1.98-2.09 (2H, m, H-4, H-10), 2.13 (1H, m, H-5), 2.35 (1H, dddd, J = 3.2, 8.0, 8.0, 12.8 Hz, H-5), 3.56 (2H, m, H-3), 4.01 (1H, dd, J = 3.4, 9.6 Hz, H-9), 4.11 (1H, t, J = 8.0 Hz, H-6), and 5.86 (1H, br.s, NH). The 13C NMR spectrum showed the following 11 resonances: δ 21.2 (11-CH3), 22.7 (C-4), 23.3 (C-12), 24.7 (C-11), 28.1 (C-5), 38.6 (C-10), 45.4 (C-3), 53.4 (C-9), 59.0 (C-6), 166.1 (C-1), and 170.1(C-7). These data and the two-dimensional NMR (H, H-correlation spectroscopy, heteronuclear multiple quantum coherence, heteronuclear multiple bond connectivity) data indicated that the substance was cyclo(Leu-Pro) (Fig. 3). The absolute structure was unambiguously determined to be (6S,9S) by comparing the NMR data and specific rotation with those of commercially available cyclo(l-Leu-l-Pro) and cyclo(d-Leu-d-Pro) and synthetic cyclo(d-Leu-l-Pro) and cyclo(l-Leu-d-Pro). The NMR data for the inhibitory substance were completely in accordance with the data for cyclo(l-Leu-l-Pro) and cyclo(d-Leu-d-Pro) but not with the data for cyclo(d-Leu-l-Pro) and cyclo(l-Leu-d-Pro). The specific rotation of the inhibitory substance, [α]D24 −124° (c 0.4, ethanol), was almost the same as that of cyclo(l-Leu-l-Pro), [α]D24 −109° (c 0.4, ethanol), but it was quite different from that of cyclo(d-Leu-d-Pro), [α]D24 +152° (c 0.4, ethanol). These results indicate that the purified inhibitory substance is cyclo(l-Leu-l-Pro).
FIG. 3.
Structure of cyclo(l-Leu-l-Pro).
Characterization of the inhibition activities.
We investigated the inhibitory activity of commercially obtained cyclo(l-Leu-l-Pro) for NA accumulation by the A. parasiticus NFRI-95 mutant using the microtiter agar plate assay (Table 1). This substance at a concentration greater than 3.5 mg ml−1 was found to completely inhibit accumulation of NA by A. parasiticus NFRI-95. It also partially inhibited the accumulation of NA at a concentration of 1.0 mg ml−1, suggesting that this concentration may be close to the 50% inhibitory concentration. Cyclo(d-Leu-d-Pro), a stereoisomer of cyclo(l-Leu-l-Pro), showed activity similar to that of cyclo(l-Leu-l-Pro) (Fig. 1D). l-Leucine, d-leucine, l-proline, or d-proline or combinations of these amino acids did not show any inhibitory activity, indicating that the inhibition was specific to the cyclodipeptide. Four other cyclodipeptides containing one of the amino acids of cyclo(l-leu-l-Pro) were also examined (Table 1). Cyclo(l-Leu-l-Gly) did not show any activity until the concentration was 3.5 mg ml−1, and it weakly inhibited NA production by the fungus at a concentration of 6.0 mg ml−1. The other two cyclodipeptides, cyclo(l-Gly-l-Pro) and cyclo(d-Ala-l-Pro), showed no inhibition. In contrast, cyclo(l-Pro-l-Val) inhibited the accumulation of NA even at a concentration of 0.3 mg ml−1, which was lower than the concentration at which the cyclo(Leu-Pro) showed inhibition. These results suggest that a cyclodipeptide structure consisting of one hydrophobic amino acid and a proline may contribute to the inhibitory activity.
TABLE 1.
Inhibition of NA accumulation by strain NFRI-95 by cyclo(l-Leu-l-Pro)-related substances as determined by the microtiter agar plate assaya
| Expt | Chemical(s) | Inhibition at the following concn:
|
|||||
|---|---|---|---|---|---|---|---|
| 0.1 mg ml−1 | 0.3 mg ml−1 | 1.0 mg ml−1 | 3.5 mg ml−1 | 6.0 mg ml−1 | 12.0 mg ml−1 | ||
| 1 | Cyclo(l-Leu-l-Pro) | − | − | ± | + | + | ND |
| Cyclo(d-Leu-d-Pro) | − | − | ± | + | + | ND | |
| 2 | Cyclo(l-Leu-l-Pro) | − | − | ± | + | + | ND |
| l-Leucine | − | − | − | − | − | ND | |
| d-Leucine | − | − | − | − | − | ND | |
| l-Proline | − | − | − | − | − | ND | |
| d-Proline | − | − | − | − | − | ND | |
| l-Leu + l-Pro | − | − | − | − | − | ND | |
| d-Leu + d-Pro | − | − | − | − | − | ND | |
| l-Leu + d-Pro | − | − | − | − | − | ND | |
| d-Leu + l-Pro | − | − | − | − | − | ND | |
| 3 | Cyclo(l-Leu-l-Pro) | − | − | ± | + | + | ND |
| Cyclo(l-Pro-l-Val) | − | ± | + | + | + | ND | |
| Cyclo(l-Leu-l-Gly) | − | − | − | − | ± | ND | |
| Cyclo(l-Gly-l-Pro) | − | − | − | − | − | ND | |
| Cyclo(d-Ala-l-Pro) | − | − | − | − | − | ND | |
| 4 | Cyclo(l-Leu-l-Pro) | − | − | ± | + | + | + |
| Cyclo(d-Leu-d-Pro) | − | − | ± | + | + | + | |
| Cyclo(d-Pro-l-Leu) | − | − | − | ± | + | + | |
| Cyclo(l-Pro-d-Leu) | − | − | − | ± | ± | ± | |
All chemicals were dissolved or suspended in methanol. Addition of only methanol did not inhibit NA accumulation. −, no effect; ±, partial inhibition; +, inhibition; ND, not determined.
We compared the effects of four isomers, cyclo(l-Leu-l-Pro), cyclo(d-Leu-d-Pro), cyclo(l-Leu-d-Pro), and cyclo(d-Leu-l-Pro), in more detail (Fig. 1D and Table 1). Cyclo(l-Leu-l-Pro) completely inhibited accumulation of NA by A. parasiticus NFRI-95 at a concentration of 3.5 mg ml−1 and also inhibited spore formation. Cyclo(d-Leu-d-Pro) also showed complete inhibition of NA accumulation at the same concentration, whereas the effect on spore formation appeared to be weaker because the fungus could produce spores even at a concentration of 6 mg ml−1. In contrast, the other two isomers, cyclo(l-Leu-d-Pro) and cyclo(d-Leu-l-Pro), had much lower activities. Cyclo(l-Leu-d-Pro) could completely inhibit NA accumulation at a concentration of 12.0 mg ml−1, whereas cyclo(d-Leu-l-Pro) could not completely inhibit NA accumulation even at the highest concentrations (Table 1 and Fig. 1D). Interestingly, these isomers affected the colors of the spores, and the higher concentrations changed the spore color from light green to dark green.
Effects of cyclo(l-Leu-l-Pro) on aflatoxin production.
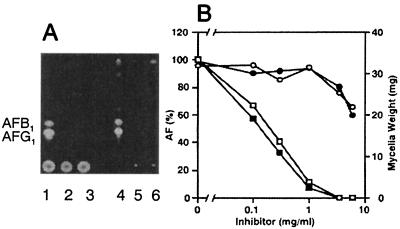
A. parasiticus SYS-4 (= NRRL2999) was incubated with various concentrations of either cyclo(l-Leu-l-Pro) or cyclo(d-Leu-d-Pro) in tip cultures. As determined by this method, either inhibitor at a concentration of 3.5 mg ml−1 completely inhibited aflatoxin production (Fig. 4A, lanes 2 and 3), whereas the fungal growth was only slightly affected at this concentration. In contrast, with both substances at a concentration of 6 mg ml−1, mycelial weight was significantly decreased, indicating that a high concentration of these substances can also inhibit fungal growth. Addition of these substances did not change the pH of the medium. TLC also showed that there was no accumulation of any intermediates related to the aflatoxin production pathway in the mycelia (Fig. 4A, lanes 5 and 6).
FIG. 4.
Inhibition of aflatoxin production and growth of A. parasiticus SYS-4 (= NRRL2999) by cyclo(l-Leu-l-Pro) and cyclo(d-Leu-d-Pro) as determined by the tip culture method. (A) A. parasiticus SYS-4 was cultured in YES medium (lanes 1 and 4) or YES medium supplemented with 3.5 mg of cyclo(l-Leu-l-Pro) ml−1 (lanes 2 and 5) or with 3.5 mg of cyclo(d-Leu-d-Pro) (lanes 3 and 6) ml−1 at 28°C for 4 days. The culture medium (lanes 1 to 3) or the mycelial extract (lanes 4 to 6) was analyzed by TLC. (B) A. parasiticus SYS-4 was cultured with various concentrations of cyclo(l-Leu-l-Pro) (○ and □) or cyclo(d-Leu-d-Pro) (• and ▪) at 28°C for 4 days. Wet weight of the mycelia (in milligrams per 250 μl of culture medium) and aflatoxin (AF) production in the culture filtrates were analyzed by HPLC; the values for 100% total aflatoxins were 81.9 ± 6.5 mg ml−1 (□) and 106.7 ± 13.8 mg ml−1 (▪).
With both substances aflatoxin production decreased at concentrations ranging from 0 to 1.0 mg ml−1 (Fig. 4B) in tip cultures. The 50% inhibitory concentrations of cyclo(l-Leu-l-Pro) and cyclo(d-Leu-d-Pro) were almost same, 0.20 and 0.13 mg ml−1, respectively. In contrast, fungal growth was not affected at these concentrations, indicating that these cyclodipeptides specifically inhibited aflatoxin production. However, higher concentrations (more than 6 mg ml−1) of the substances inhibited fungal growth (Fig. 4B).
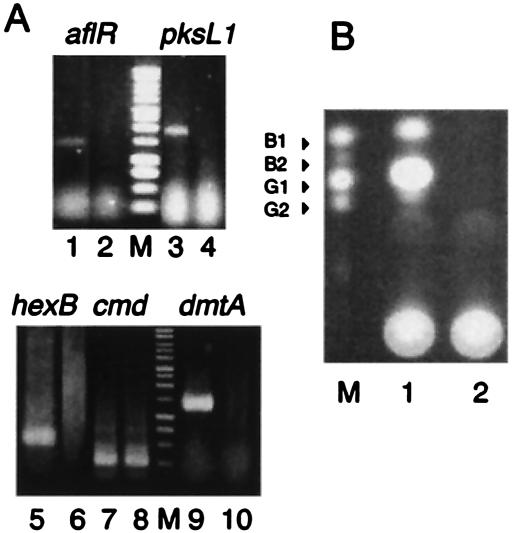
Inhibition of the transcription of aflatoxin-related genes.
The effects of cyclo(l-Leu-l-Pro) on transcription were analyzed by RT-PCR. A. parasiticus SYS-4 was cultured in YES medium with or without cyclo(l-Leu-l-Pro) at a concentration of 3.5 mg ml−1 by the tip culture method. Expression of the aflatoxin-related genes aflR, hexB, dmtA, and pks was detected in the absence of the inhibitor (Fig. 5A). However, expression of these genes was completely inhibited by cyclo(l-Leu-l-Pro) at a concentration of 3.5 mg ml−1, whereas fungal growth was only slightly affected at this concentration. Complete inhibition of aflatoxin production by the inhibitor at a concentration of 3.5 mg ml−1 was also confirmed by TLC of the culture filtrate (Fig. 5B). Expression of the cmd gene, a constitutively expressed calmodulin gene that is not related to aflatoxin production, was not inhibited in the presence of cyclo(l-Leu-l-Pro) (Fig. 5A).
FIG. 5.
RT-PCR of the aflatoxin-related genes. (A) Total RNA was prepared from mycelia of A. parasiticus SYS-4 (= NRRL2999) which had been cultured in YES medium without cyclo(l-Leu-l-Pro) (lanes 1, 3, 5, 7, and 9) or with 3.5 mg of cyclo(l-Leu-l-Pro) ml−1 (lanes 2, 4, 6, 8, and 10) by the tip culture method, and RT-PCR for aflR (lanes 1 and 2), pksL1 (lanes 3 and 4), hexB (lanes 5 and 6), dmtA (lanes 9 and 10), and cmd (lanes 7 and 8) were then performed. The sizes of the RT-PCR products were as follows: aflR, 1,134 bp; pksL1, 1,922 bp; hexB, 585 bp; dmtA, 1,190 bp; and cmd, 328 bp. Lane M contained a 1-kb DNA ladder marker (Promega). (B) Culture medium (5 μl) of A. parasiticus SYS-4 which had been cultured in the absence (lane 1) or presence (lane 2) of cyclo(l-Leu-l-Pro) was analyzed by TLC. Lane M contained standard samples of aflatoxins B1, B2, G1, and G2.
We also investigated effect of cyclo(l-Leu-l-Pro) on enzyme activity. When sterigmatocystin was added to the culture medium in the feeding experiments, 56.4 ± 5.7 μg of total aflatoxins was produced in 200 μl of the medium. In contrast, addition of 2.0 mg of cyclo(l-Leu-l-Pro) ml−1 caused a drastic decrease in aflatoxin production to 7.1% of the amount in the absence of the inhibitor (4.0 ± 6.4 μg of total aflatoxins in the culture medium). Inhibition of the production of aflatoxin from sterigmatocystin may have been due to the inhibition of expression of the enzyme genes by cyclo(l-Leu-l-Pro).
DISCUSSION
A. xylosoxidans was isolated for the first time from human ear discharge, and the genus Achromobacter Yabuuchi and Yano 1981, with the type species A. xylosoxidans, was recently emended by Yabuuchi et al. (39). Many beneficial functions of Achromobacter sp. have been reported, including stimulation of ionic transport to promote plant growth (2) and production of glutaryl-3-deacetoxy-7-aminocephalosporanic acid acylase, which is an important enzyme in the production of the antibiotic cephalosporin (11). In the present study, we showed for the first time that A. xylosoxidans can specifically inhibit aflatoxin production and that the inhibitory activities are common in A. xylosoxidans.
Cyclic dipeptides (2,5-diketopiperazines) are among the most common peptide derivatives found in nature, and they have been shown to exhibit antiviral, antibacterial, antifungal, and antitumor activities (7, 12, 16, 18). Recently, cyclo(l-Phe-l-Pro) and cyclo(l-Phe-trans-4-OH-l-Pro) were isolated from Lactobacillus plantarum MiLAB 393 (30). Among the many cyclodipeptides isolated from nature, cyclo(l-Leu-l-Pro) has been isolated from many living organisms, including Streptomyces (28), Rosellinia necatrix (5), a marine sponge (Rhaphisia pallida) (31), and a marine bacterium (Halobacillus litoralis) (40). It has been reported that cyclo(Pro-Leu) retards the growth of rice seedlings and roots (7) and that it is effective against vancomycin-resistant enterococci and leukemic cells (28). Because of their many biological functions, cyclic peptides have attracted considerable attention.
In this work we demonstrated for the first time that cyclo(l-Leu-l-Pro) specifically inhibits secondary metabolism at a low concentration, although it also inhibits fungal growth at higher concentrations. The inhibitory activity of cyclo(l-Leu-l-Pro) is dependent on its cyclic structure, because either single amino acid or a combination of the two amino acids (Table 1) did not result in any inhibition at the concentrations investigated. Also, proline and a hydrophobic amino acid in the cyclodipeptide seemed to be critical to the inhibitory activity, as other cyclodipeptides without proline did not show the same activity. The requirement for leucine or valine for activity suggests that a hydrophobic interaction may be involved in the activity.
Because the structure of cyclo(l-Leu-l-Pro) does not resemble the structure of any aflatoxin precursor (37), this molecule may not inhibit an enzyme involved in aflatoxin biosynthesis through competition with a precursor for the enzyme. RT-PCR results showed that cyclo(l-Leu-l-Pro) represses the transcription of genes involved in the aflatoxin gene cluster. It remarkably inhibited expression of the aflR gene, which encodes a transcription factor that positively regulates expression of the structural genes (26, 35) (Fig. 5). An absence of aflR expression caused a lack of expression of the structural genes hexB (13), dmtA (22), and pksL1 (10). In fact, in a feeding experiment conversion of sterigmatocystin to aflatoxins was drastically decreased, indicating that most of the enzymes involved in the pathway from ST to aflatoxins were lost. These results suggest that cyclo(l-Leu-l-Pro) may affect a certain unknown step involved in signal transduction, causing expression of aflR (4), or may directly affect the expression of the aflR gene by an unknown mechanism. The detailed mechanism of the inhibition of both aflatoxin biosynthesis and fungal growth by cyclo(l-Leu-l-Pro) remains to be elucidated.
Chirality plays a crucial role in biochemical systems, and enantiomers often have very different physiological behaviors (3). However, cyclo(l-Leu-l-Pro) and cyclo(d-Leu-d-Pro) have similar inhibitory activities (Table 1), although cyclo(l-Leu-l-Pro) appears to be slightly stronger than cyclo(d-Leu-d-Pro) in inhibition of spore formation (Fig. 4). These results might be explained by the relationship of this activity with the lipophilic nature of the molecules. In contrast, another enantiomer, cyclo(l-Leu-d-Pro) or cyclo(d-Leu-l-Pro), exhibited lower inhibition than cyclo(l-Leu-l-Pro) or cyclo(d-Leu-d-Pro) (Fig. 4). The more restricted mobility of the proline ring in the former enantiomers might be related to these differences (8). In the cyclopeptides containing a proline, the cis-trans isomerism of the N-alkylamide bond in the proline has been implicated in the receptor-mediated biological activity of proline-containing compounds (3, 12). The relationship between the configuration of the molecules and these activities may be important in clarifying the mechanism of inhibition by cyclodipeptides.
Cyclo(l-Leu-l-Pro) also inhibited fungal growth at a concentration higher than 6 mg ml−1 (Fig. 4B). Antifungal activities of other cyclic dipeptides have been reported previously (18, 19). Although some mechanisms for antifungal activity have been suggested (3, 21), they have not been clarified. This work also showed that the isomers cyclo(l-Leu-d-Pro) and cyclo(d-Leu-l-Pro) affected the color of the spores, whereas cyclo(l-Leu-l-Pro) and cyclo(d-Leu-d-Pro) did not affect the pigmentation (Fig. 1D). Furthermore, the effect of these substances on sclerotium formation could provide information about the mechanisms of these substances in cells. A study of these secondary metabolic processes is now in progress in our laboratory.
In purifying the inhibitor, we found that the culture medium of the A1 bacterium contained at least three inhibitory substances other than cyclo(l-Leu-l-Pro). The amounts of these substances were not large enough for further characterization. Although the combination of these unknown substances and cyclo(l-Leu-l-Pro) might result in synergetic inhibitory activity, detailed relationships among the substances remain to be studied.
The biosynthetic mechanism and the secretion mechanism of the cyclodipeptide of A. xylosoxidans NFRI-A1 are still unknown. It has been reported that cyclodipeptides might be degradation products of proteins following spontaneous cyclization (21). However, it has been found that many cyclodipeptides play various biological roles. These substances are now regarded as important metabolic substances rather than as protein artifacts.
For purification of the inhibitor from the bacterial culture, a simple, sensitive, and small-scale detection system is of primary importance. Although the visual agar plate assay provided sensitive and clear results, the experimental scale seemed to be too large even if we used a small plate (diameter, 6 cm). Instead, we devised a small-scale assay system using a microtiter plate. The microtiter plate is quite useful. Magnusson et al. have independently reported use of a similar microtiter plate well assay to monitor antifungal activity of the metabolites produced by Lactobacillus coryniformis subsp. coryniformis (18, 19). We have already isolated other inhibitory substances from soil bacteria by using the microtiter agar plate assay. We hope that some of our results will soon be useful in preventing aflatoxin contamination.
Acknowledgments
We thank J. W. Bennett for supplying an A. parasiticus HVN-accumulating mutant (hvn-1) and M. Kito, H. Arai, and H. Hatabayashi for technical assistance. The FASTA search of the GenBank database was performed with the assistance of the Computer Center for Agriculture, Forestry and Fisheries Research, MAFF, Japan.
This work was supported in part by grant-in-aid BDP-04-VI-1-1 (Bio-Design Program) from the Ministry of Agriculture, Forestry and Fisheries.
REFERENCES
- 1.Barrow, G. I., and R. K. A. Feltham. 1993. Cowan and Steel's manual for the identification of medical bacteria, 3rd ed. Cambridge University Press, Cambridge, United Kingdom.
- 2.Bertrand, H., C. Plassard, X. Pinochet, B. Touraine, P. Normand, and J. C. Cleyet-Marel. 2000. Stimulation of the ionic transport system in Brassica napus by a plant growth-promoting rhizobacterium (Achromobacter sp.). Can. J. Microbiol. 46:229-236. [DOI] [PubMed] [Google Scholar]
- 3.Brandl, C. J., and C. M. Deber. 1986. Hypothesis about the function of membrane-buried proline residues in transport proteins. Proc. Natl. Acad. Sci. USA 83:917-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo, A. M., R. A. Wilson, J. W. Bok, and N. P. Keller. 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66:447-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y.-S. 1960. Studies on the metabolic products of Rosellinia necatrix Berlese. I. Isolation and characterization of several physiologically active neutral substances. Bull. Agric. Chem. Soc. Jpn. 24:372-381. [Google Scholar]
- 6.Cotty, P. J. 1994. Influence of field application of an atoxigenic strain of Aspergillus flavus on the population of A. flavus infecting cotton bolls and on the aflatoxin content of cottonseed. Phytopathology 84:1270-1277. [Google Scholar]
- 7.Cronan, J. M., T. R. Davidson, F. L. Singleton, R. R. Colwell, and J. H. Cardellina. 1998. Plant growth promoters isolated from a marine bacterium associated with Palythoa sp. Nat. Prod. Lett. 11:271-278. [Google Scholar]
- 8.Deslauriers, R. 1976. Influence of d and l amino-acid residues on the confirmation of peptides in solution: a carbon-13 nuclear magnetic resonance study of cyclo(prolyl-leucyl). Biopolimers 15:1677-1679. [DOI] [PubMed] [Google Scholar]
- 9.Dorner, J. W., R. J. Cole and, and D. T. Wicklow. 1999. Aflatoxin reduction in corn through field application of competitive fungi. J. Food Prot. 62:650-656. [DOI] [PubMed] [Google Scholar]
- 10.Feng, G. H., and T. J. Leonard. 1995. Characterization of the polyketide synthase gene (pksL) required for aflatoxin biosynthesis in Aspergillus parasiticus. J. Bacteriol. 177:6247-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzosi, G., E. Battistel, I. Gagliardi, and W. Van der Goes. 1995. Screening and characterization of microorganisms with glutaryl-7ADCA acylase activity. Appl. Microbiol. Biotechnol. 43:508-513. [DOI] [PubMed] [Google Scholar]
- 12.Graz, M., A. Hunt, H. Jamie, G. Grant, and P. Milne. 1999. Antimicrobial activity of selected cyclic dipeptides. Pharmazie 54:772-775. [PubMed] [Google Scholar]
- 13.Hitchman, T. S., E. W. Schmidt, F. Trail, M. D. Rarick, J. E. Linz, and C. A. Townsend. 2001. Hexanoate synthase, a specialized type I fatty acid synthase in aflatoxin B1 biosynthesis. Bioorg. Chem. 29:293-307. [DOI] [PubMed] [Google Scholar]
- 14.Hua, S.-S. T., J. L. Baker, and M. Flores-Espiritu. 1999. Interactions of saprophytic yeasts with a nor mutant of Aspergillus flavus. Appl. Environ. Microbiol. 65:2738-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua, S.-S. T., O.-K. Grosjean, and J. L. Baker. 1999. Inhibition of aflatoxin biosynthesis by phenolic compounds. Lett. Appl. Microbiol. 29:289-291. [DOI] [PubMed] [Google Scholar]
- 16.Katz, E., and A. L. Demain. 1977. The peptide antibiotics of Bacillus: chemistry, biogenesis, and possible functions. Bacteriol. Rev. 41:449-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo, T., M. Sakurada, S. Okamoto, M. Ono, H. Tsukuji, A. Suzuki, H. Nagasawa, and S. Sakuda. 2001. Effects of aflastatin A, an inhibitor of aflatoxin production, on aflatoxin biosynthetic pathway and glucose metabolism in Aspergillus parasiticus. J. Antibiot. 54:650-657. [DOI] [PubMed] [Google Scholar]
- 18.Magnusson, J., and J. Schnurer. 2001. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 67:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnusson, J., K. Strom, S. Roos, J. Sjogren, and J. Schnurer. 2003. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 219:129-135. [DOI] [PubMed] [Google Scholar]
- 20.Massey, T. E., R. K. Steward, J. M. Daniels, and L. Liu. 1995. Biochemical and molecular aspects of mammalian susceptibility to aflatoxin B1 carcinogenicity. Proc. Soc. Exp. Biol. Med. 208:2213-2227. [DOI] [PubMed] [Google Scholar]
- 21.Milne, P. J., A. L. Hunt, K. Rostoll, J. J. Van Der Walt, and C. J. M. Graz. 1998. The biological activity of selected cyclic dipeptides. J. Pharm. Pharmacol. 50:1331-1337. [DOI] [PubMed] [Google Scholar]
- 22.Motomura, M., N. Chihaya, T. Shinozawa, T. Hamasaki, and K. Yabe. 1999. Cloning and characterization of the O-methyltransferase I gene (dmtA) from Aspergillus parasiticus associated with the conversion of demethylsterigmatocystin to sterigmatocystin and dihydrodemethylsterigmatocystin to dihydrosterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 65:4987-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono, M., S. Sakuda, A. Suzuki, and A. Isogai. 1997. Aflastatin A, a novel inhibitor of aflatoxin production by aflatoxigenic fungi. J. Antibiot. 50:111-117. [DOI] [PubMed] [Google Scholar]
- 24.Park, D. L. 1993. Controlling aflatoxin in food and feed. Food Technol. 47:92-96. [Google Scholar]
- 25.Payne, G. A. 1998. Aflatoxin in maize. Crit. Rev. Plant Sci. 10:423-440. [Google Scholar]
- 26.Payne, G. A., G. J. Nystrom, D. Bhatnagar, T. E. Cleveland, and C. P. Woloshuk. 1993. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl. Environ. Microbiol. 59:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao, G. P., and H. N. Sigh. 1992. Fungitoxic evaluation of essential oils extracted from higher plants against some sugarcane pathogens in vitro. Trop. Sci. 32:377-382. [Google Scholar]
- 28.Rhee, K.-H. 2002. Isolation and characterization of Streptomyces sp. KH-614 producing anti-VRE (vancomycin-resistant enterococci) antibiotics. J. Gen. Appl. Microbiol. 48:321-327. [DOI] [PubMed] [Google Scholar]
- 29.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strom, K., J. Sjogren, A. Broberg, and J. Schnurer. 2002. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(l-Phe-l-Pro) and cyclo(l-Phe-trans-4-OH-l-Pro) and 3-phenyllactic acid. Appl. Environ. Microbiol. 68:4322-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su, J., Y. Zhong, L. Zeng, H. Wu, X. Shen, and K. Ma. 1996. A new N-carboxyindole alkaloid from the marine sponge Rhaphisia pallida. J. Nat. Prod. 59:504-506. [Google Scholar]
- 32.Sweeny, M. J., and A. D. W. Dobson. 1998. Mycotoxin production by Aspergillus, Fusarium and Penicillium species. Int. J. Food Microbiol. 43:141-158. [DOI] [PubMed] [Google Scholar]
- 33.Tani, H., T. Honma, Y. Fujii, K. Yoneyama, and H. Nakajima. 2003. A plant growth retardant related to chlamydocin and its proposed mechanism of action. Phytochemistry 62:1133-1140. [DOI] [PubMed] [Google Scholar]
- 34.Van Egmond, H. P. 1995. Mycotoxins: regulations, quality assurance and reference materials. Food Addit. Contam. 12:321-330. [DOI] [PubMed] [Google Scholar]
- 35.Woloshuk, C. P., K. R. Fouz, J. F. Brewer, D. Bahtnagar, T. E. Cleveland, and G. A. Payne. 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yabe, K., N. Chihaya, S. Hamamatsu, E. Sakuno, T. Hamasaki, H. Nakajima, and J. W. Bennett. 2003. Enzymatic conversion of averufin to hydroxyversicolorone and elucidation of a novel metabolic grid involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 69:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yabe, K., and H. Nakajima. 2004. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 64:745-755. [DOI] [PubMed] [Google Scholar]
- 38.Yabe, K., H. Nakamura, Y. Ando, N. Terakado, H. Nakajima, and T. Hamasaki. 1988. Isolation and characterization of Aspergillus parasiticus mutants with impaired aflatoxin production by a novel tip culture method. Appl. Environ. Microbial. 54:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yabuuchi, E., Y. Kawamura, Y. Kosako, and T. Ezaki. 1998. Emendation of genus Achromobacter and Achromobacter xylosoxidans (Yabuuchi and Yano) and proposal of Achromobacter ruhlandii (Packer and Vishniac) comb. nov., Achromobacter piechaudii (Kiredjian et al.) comb. nov., and Achromobacter xylosoxidans subsp. denitrificans (Ruger and Tan) comb. nov. Microbiol. Immunol. 42:429-438. [DOI] [PubMed] [Google Scholar]
- 40.Yang, L., R. Tan, Q. Wang, W. Huang, and Y. Yin. 2002. Antifungal cyclopeptides from Halobacillus litoralis YS3106 of marine origin. Tetrahedron Lett. 43:6545-6548. [Google Scholar]
- 41.Yasui, K., K. Kitamoto, K. Gomi, C. Kumagai, Y. Ohya, and G. Tamura. 1995. Cloning and nucleotide sequence of the calmodulin-encoding gene (cmdA) from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 59:1444-1449. [DOI] [PubMed] [Google Scholar]