Abstract
McMurdo Station, Antarctica, has discharged untreated sewage into McMurdo Sound for decades. Previous studies delineated the impacted area, which included the drinking water intake, by using total coliform and Clostridium perfringens concentrations. The estimation of risk to humans in contact with the impacted and potable waters may be greater than presumed, as these microbial indicators may not be the most appropriate for this environment. To address these concerns, concentrations of these and additional indicators (fecal coliforms, Escherichia coli, enterococci, coliphage, and enteroviruses) in the untreated wastewater, water column, and sediments of the impacted area and drinking water treatment facility and distribution system at McMurdo Station were determined. Fecal samples from Weddell seals in this area were also collected and analyzed for indicators. All drinking water samples were negative for indicators except for a single total coliform-positive sample. Total coliforms were present in water column samples at higher concentrations than other indicators. Fecal coliform and enterococcus concentrations were similar to each other and greater than those of other indicators in sediment samples closer to the discharge site. C. perfringens concentrations were higher in sediments at greater distances from the discharge site. Seal fecal samples contained concentrations of fecal coliforms, E. coli, enterococci, and C. perfringens similar to those found in untreated sewage. All samples were negative for enteroviruses. A wastewater treatment facility at McMurdo Station has started operation, and these data provide a baseline data set for monitoring the recovery of the impacted area. The contribution of seal feces to indicator concentrations in this area should be considered.
Research on the continent of Antarctica requires the establishment and maintenance of permanent research and support facilities. The human activities associated with these facilities generate solid and liquid wastes, one of which is human fecal material. The Antarctic continent and its surrounding waters are considered unique and pristine, and the Antarctic Treaty System (46) requires complete removal of human wastes from field facilities. However, it does not prohibit the discharge of these same types of waste from permanent facilities into coastal waters. McMurdo Station, a U.S. facility that supports the largest community on the continent, has released untreated sewage into the waters of McMurdo Sound since its establishment in 1955. Though the extreme conditions of this region may be presumed to be inhibitory to the survival of fecal and nonfecal bacteria, studies have shown that low water temperatures (ca. −1.8°C) enhance survival of allochthonous bacteria (27, 42, 43) and reduce rates of bacterivory (36) and degradation of anthropogenic organic material (33).
Previous studies used only total coliforms and Clostridium perfringens to delineate the area of McMurdo Sound impacted by the sewage discharge, which included the drinking water intake (21, 32, 37). Concerns about the associated risks of contamination of the drinking water system and divers have been raised, as no rigorous survey of the microbiological quality of the distribution system had been attempted and total coliforms and C. perfringens may not be the most appropriate microbial indicators for the Antarctic environment. To address these issues, we used a suite of microbial indicators (total and fecal coliforms, enterococci, Escherichia coli, coliphage, C. perfringens, human enteric viruses) to assess the extent of fecal contamination in the water column and sediments impacted by the discharged sewage. We also evaluated the drinking water treatment facility and distribution system. To determine the potential contribution of Weddell seals (Leptomychotes weddellii) to the microbial indicator populations of McMurdo Sound, we analyzed their feces for fecal coliforms, E. coli, enterococci, and C. perfringens.
Recently, a wastewater treatment facility was put into operation at McMurdo Station. This facility is the first of its design on the continent and is designed to totally eliminate the contaminating concentrations of organic matter and nutrients and fecal bacteria into McMurdo Sound. In addition to this being the first comprehensive study of microbial indicators in this region of Antarctica and of Weddell seal feces, the data provide a baseline microbial indicator data set for the water column and sediments that can be used for assessing the recovery of the impacted area.
MATERIALS AND METHODS
Sample sites and collection.
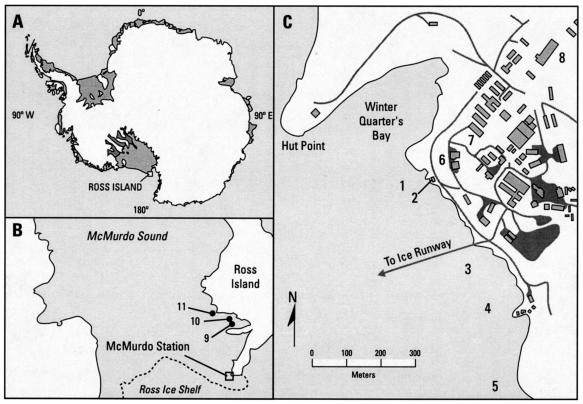
Water column, sediment, drinking source water, drinking water, and untreated wastewater samples were collected at various sites at McMurdo Station and the surrounding area during the months of October and November 1999 and 2000 (Table 1 and Fig. 1). Seal scat samples, which are fecal stools left on the ice surface by unidentified seals for undetermined times, were collected from the sea ice in the areas of Little and Big Razorback Islands (Table 1 and Fig. 1). Unless otherwise noted, all analyses were conducted in laboratories within McMurdo Station's Albert P. Crary Science and Engineering Center. The untreated wastewater samples were collected through a covered opening that had been cut into the discharge pipe. This collection site was approximately 3 m downstream of a masticator and approximately 65 m upstream of the discharge point in McMurdo Sound. The raw sewage collected at this site was diluted approximately sixfold with marine water that had been discharged from the flowthrough aquaria located in the Albert P. Crary Science and Engineering Center. Though the discharge from the aquaria was consistent during the sample periods in this study, the volume of wastewater was variable but cyclic and contained not only human waste but also gray water from all buildings at McMurdo Station.
TABLE 1.
Location of sample sites within or near McMurdo Station, Antarctica
| Sample site | Site description | Latitude | Longitude | Distance from outfall (m) |
|---|---|---|---|---|
| 1 | Sewage outfall | 77°50′52"S | 166°39′12"E | 0 |
| 2 | Masticator | NAa | NA | NA |
| 3 | Transition South | 77°50′59"S | 166°39′39"E | 332 |
| 4 | Drinking water intake | 77°51′04"S | 166°39′53"E | 444 |
| 5 | Cape Armitage | 77°51′18"S | 166°39′54"E | 822 |
| 6 | Drinking water plant | NA | NA | NA |
| 7 | Dormitory | NA | NA | NA |
| 8 | Vehicle shop | NA | NA | NA |
| 9 | Big Razorback Island | 77°68′S | 166°78′E | 1,829 |
| 10 | Little Razorback Island | 77°40′S | 166°31′E | 1,905 |
| 11 | Cape Evans | 77°38′S | 166°24′E | 2,332 |
NA, not applicable.
FIG. 1.
Samples were collected on the continent of Antarctica (A), along the coast of Ross Island (B), and along the coast of and within McMurdo Station (C). See Table 1 for a description of numbered sample locations.
Drinking water treatment facility and distribution system samples.
The drinking water treatment facility utilizes reverse osmosis (RO) to produce approximately 1.5 × 106 gallons day−1. To reduce the corrosivity of the product water, carbon dioxide, limestone contactors, and soda ash are used to collectively increase the alkalinity and raise the pH to approximately 9.0 prior to distribution of the potable water (1). Samples to assess the integrity of the drinking water treatment process and microbiological quality of the potable water were collected from the drinking water intake within the treatment facility immediately after RO filtration but just prior to liquid chlorine disinfection and from two sites within the distribution system that represented dead ends or low-use areas (Fig. 1 and Table 1). Grab samples (1 liter) were collected at each site, serially diluted, if appropriate, and prepared for the recovery of total and fecal coliforms, E. coli, enterococci, C. perfringens, and total culturable bacteria as described below.
Large volumes of water were collected from the post-RO filtration and distribution sites by using standard methods for the recovery of enteric viruses (11) using an electropositive cartridge filter (1-MDS filter; Cuno, Inc.) (44). Dechlorination was not necessary, as the post-RO sample was collected prior to application of the disinfectant. When appropriate, sodium thiosulfate was injected into the sample collection line upstream of the cartridge filter by using a venturi-type proportioning injector to inject sodium thiosulfate to a final concentration of 10 mg liter−1, based on a flow rate of 25 liters min−1.
Wastewater samples.
Grab samples (1 liter) were collected, serially diluted with filter-sterilized source water, and then used for the recovery of the suite of bacterial indicators described for the drinking water samples. Divers also collected grab samples from the accumulated sewage solids at the end of the discharge pipe and sediment cores from the edge of this same area. Water column grab samples were also collected approximately 0.3 m above the sediment surface where the sediment cores were collected. Each water column sample was serially diluted with filter-sterilized source water and then used for the recovery of the suite of bacterial indicators described for the drinking water samples plus coliphage.
Sediment core samples were collected with sterile modified syringes (2.5-cm diameter) (30, 31). Each sediment core was first processed by extruding the upper 5.0 cm of each core into a sterile bottle containing 10 ml of filter-sterilized source water containing, as final concentrations, 0.1% Tween 80 (vol/vol) and 15% glycerol (vol/vol) (34). The samples were then vortexed for 30-s intervals for 5 min, followed by centrifugation for 5 min at 500 × g. The supernatants were transferred to sterile tubes and serially diluted with filter-sterilized source water. The resulting suspensions were used to recover the same suite of indicators described for the wastewater samples. The residual sediment material was dried at 105°C for 48 h and subsequently used to obtain the dry weight for each sample.
A second sediment core was collected for the recovery of enteric viruses. The upper 5.0 cm of this core was extruded into a sterile bottle containing 60 ml of 3% (wt/vol) beef extract (pH 9.0) and gently mixed for 15 min. The suspensions were centrifuged for 5 min at 500 × g, and the supernatant was transferred to a sterile bottle. Gentamicin and kanamycin were both added at a final concentration of 50.0 μg ml−1. These samples were immediately frozen and stored at −70°C until shipment to a stateside laboratory.
In addition to these samples, large volumes of wastewater were also collected from the wastewater stream immediately after the masticator using the enteric virus method described for the drinking water samples, except that an electropositive cartridge filter (Filterite Corp.) was used (44).
Water column and sediment samples.
Water and sediment core samples were collected by divers using sterile 1-liter bottles and modified syringes as described above. All samples were kept from freezing by placing them in a closed cooler during transit to the laboratory. All water and core samples were immediately processed as described above for the wastewater samples and used for the recovery of the suite of bacterial indicators as described for the drinking water samples. Selected sites were also sampled for the recovery of enteric viruses using the methods described above for wastewater samples.
Seal scat samples.
Random seal fecal scats were collected from the surface of the sea ice with sterile spatulas and transferred to sterile tubes. Since these samples had to be collected in a manner that did not disturb or disrupt seal activity or behavior, all samples were collected from areas where seals routinely hauled out (e.g., cracks in the sea ice) but were not present at the time of sampling. All tubes were stored in a closed cooler and kept frozen until receipt at the laboratory, where the samples were immediately placed at room temperature for slow thawing. Each thawed scat sample was split and weighed. One subsample was directly sampled by using a sterile swab and streaking the appropriate medium for the recovery of C. perfringens. The same sample was then processed further by removing and resuspending approximately 1.0 g of the fecal material in 10.0 ml of phosphate-buffered saline supplemented with 0.1% Tween 80 and 15.0% glycerol as final concentrations. The samples were vortexed and centrifuged for 5 min at 5,000 × g, and the supernatants were transferred to sterile tubes. The supernatants were serially diluted in supplemented phosphate-buffered saline and used to inoculate the appropriate media for the recovery and enumeration of fecal coliforms, E. coli, enterococci, C. perfringens organisms, and total culturable bacteria as described below. The residual fecal material was dried to obtain the dry weight for each sample as described above.
The second subsample (approximately 5.0 g) was suspended in 3.0% beef extract (pH 9.0), vortexed, and immediately frozen and stored at −70°C. These samples were collectively shipped stateside for processing for the recovery of enteric viruses as described below.
The following assays were performed on the respective samples described above. Unless otherwise stated, the membrane filtration technique was used.
Total coliforms.
m-Endo agar (Difco) was used for the recovery and enumeration of total coliforms (16). Inoculated filters were incubated at 35°C for 18 to 24 h. Colonies exhibiting a green-gold metallic sheen were counted as total coliforms.
Fecal coliforms.
m-FC agar (Difco) was used for the recovery and enumeration of fecal coliforms (12). Inoculated filters were incubated in a 44.5°C water bath for 18 to 24 h. Colonies exhibiting a blue coloration were counted as fecal coliforms.
E. coli.
EC medium with methylumbelliferyl-β-glucuronide (Difco), to which agar was added (15 g liter−1), was used for the recovery and enumeration of E. coli (12). All plates were incubated for 18 to 24 h at 35°C. Those colonies that presented a blue fluorescence under long-wave UV light were counted as E. coli.
Enterococci.
mE (Difco) and EIA (Difco) agars were used in a two-step procedure for the recovery and enumeration of enterococci (13). Inoculated filters were incubated on mE agar at 41°C for 48 h. Each filter on mE agar that was positive for growth was then transferred to an EIA agar plate and incubated at 41°C for 20 to 30 min. Those colonies that developed a black or reddish-brown precipitate on the underside of the filter were counted as enterococci.
C. perfringens.
m-CP medium (Accumedia Manufacturers, Inc.) was used for the recovery and enumeration of C. perfringens organisms following the protocol of Bisson and Cabelli (6) as modified by Armon and Payment (3). All uninoculated m-CP medium plates were reduced prior to use by storing them in an anaerobic chamber for at least 24 h. Water, sediment, and sewage solids from sewage effluent samples were processed using the membrane filtration technique. Selected seal scat samples were subsampled with sterile swabs and used to directly inoculate plates of m-CP medium. All plates were incubated for 24 to 36 h at 45°C in an anaerobic atmosphere. Isolated colonies that exhibited a characteristic salmon-pink color after exposure to sodium hydroxide fumes were counted as C. perfringens.
Total culturable bacteria.
Marine agar 2216 (Difco) and R2A agar (Difco) were used for the recovery of total culturable bacteria (TCB) from marine and nonmarine samples, respectively (14). Plates were inoculated using the spread plate technique and incubated in the dark at 21 to 23°C for 3 to 5 days. All colonies were counted.
Coliphage.
Coliphage samples were collected from the masticator, sewage outfall, Transition South, and Cape Evans (Table 1) and processed for the recovery and enumeration of F+ RNA coliphage using the oligonucleotide probes and protocol described by Hsu et al. (34) and modified by Griffin et al. (26). The oligonucleotide probes were labeled with digoxigenin and detected colorimetrically as described by Hsu et al. (34).
Enteric viruses.
Large-volume samples for the detection and enumeration of enteric viruses were collected from the drinking water treatment plant at a location immediately following RO treatment, the distribution system, untreated sewage, the drinking water intake, sewage outfall, Transition South, and Little Razorback Island. All samples were processed using standard adsorption and elution methods (11). Eluted viruses from these samples were concentrated using an aluminum hydroxide adsorption-precipitation method (11).
Sediment samples for enterovirus analysis were collected from sites at the drinking water intake, sewage outfall, Transition South, and Little Razorback Island. Seal scat samples from Little Razorback Island were also collected. All samples were immediately stored at −80°C and maintained at this temperature during shipment to a stateside laboratory, where the samples were processed for recovery of enteric viruses using a modified standard protocol (11). Briefly, AlCl3 was added to a final concentration of 1.5 mM to each sediment or scat sample, and the samples were vortexed and centrifuged for 20 min at 5,000 × g. The supernatants were transferred to fresh tubes, and after their pHs were adjusted to 3.5 with 1.0 N HCl they were gently mixed for 30 min and then centrifuged for 10 min at 10,000 × g. The supernatants were discarded, and the pellets were resuspended in 1/20 of the original volumes with 0.45 N Na2HPO4. Gentamicin and kanamycin were both added at a final concentration of 50.0 μg ml−1 in both types of sample concentrates. Buffalo Green Monkey kidney cell lines were inoculated with the concentrates and incubated at 37°C for at least 14 days. During this period, the cells lines were microscopically examined periodically for the presence of cytopathic effects (CPE). Those samples that demonstrated CPE were processed further with an integrated cell culture-PCR method as described by Reynolds et al. (39). Those samples that did not produce CPE on the first passage were used to inoculate a second Buffalo Green Monkey kidney cell line. If after the second passage the sample did not produce CPE, that sample was considered negative for the presence of infectious enteric viruses. The equivalent weights, as wet weight, processed for both types of samples were 2 g.
Potable water and distribution system chemical data.
Data for potable water temperature, pH, calcium, total alkalinity, and total dissolved solids were collected during both seasons by the drinking water treatment facility staff. Routine sample collection included finished drinking water collected within the treatment plant, a dormitory, and a vehicle shop, located approximately 100 and 400 m from the treatment facility, respectively. All analyses were conducted as required for potable water stateside and according to the methods described in Standard Methods for the Examination of Water and Wastewater (15).
Statistical analyses.
When appropriate, data were log10 transformed using the format log10 (x + 1). Geometric mean and data comparisons using analysis of variance (α = 0.05) were performed using Minitab, release 13 (Minitab, Inc.).
RESULTS
Source water, drinking water facility, and distribution system.
The mean concentrations of each indicator (≤6 CFU 100 ml−1) and TCB (53 CFU ml−1) in the water column samples collected at the drinking water intake in McMurdo Sound were similar to concentrations recovered from the pre-RO sample site within the treatment facility (≤12 CFU 100 ml−1 and 86 CFU ml−1, respectively) (Table 2). The sediment samples collected from this area had low indicator concentrations (<65 CFU g [dry weight]−1), except for C. perfringens (5.00 × 102 g [dry weight]−1) (Table 2). The drinking water treatment process and distribution system samples of McMurdo Station were negative for all indicators (Table 2). The TCB concentrations within the distribution system averaged 26 CFU ml−1, with an individual sample containing 1.60 × 103 CFU ml−1.
TABLE 2.
Microbial indicator occurrence in samples collected at McMurdo Station and local areasa
| Sample site | Sample type | No. of samples | Total coliforms | Fecal coliforms | E. coli | Enterococci | C. perfringens | TCB | Coliphage |
|---|---|---|---|---|---|---|---|---|---|
| Drinking water source | Water column | 3 | 6 (2-11) | 4 (1-6) | 2 (1-2) | 6 (1-64) | 2 (0-3) | 53 (50-72) | NDb |
| Sediment | 3 | 2 (0-2) | 2 (0-2) | 2 (0-2) | 6 (1-64) | 34 (0-5.00 × 102) | 3.68 × 102 (3.47 × 102-4.00 × 102) | ND | |
| Intake (pre-RO) | Nonpotable | 2 | 12 (7-16) | 8 (3-16) | 2 (0-5) | 4 (2-5) | 0 | 86 (43-1.73 × 102) | ND |
| Treatment plant | Potable | 4 | 0 | 0 | 0 | 0 | 0 | 0 | ND |
| Distribution system | Potable | 7 | 1 (0-1) | 0 | 0 | 0 | 0 | 26 (0-1.60 × 103) | ND |
| Masticator | Untreated sewage | 3 | 1.28 × 106 (6.30 × 105-2.60 × 106) | 2.15 × 105 (1.40 × 105-5.90 × 105) | 5.00 × 104 (n = 1)c | 1.09 × 105 (2.90 × 104-6.40 × 105) | 3.03 × 104 (3.00 × 102-1.00 × 105) | 5.75 × 107 (3.90 × 107-2.20 × 108) | 2.63 × 103 (1.02 × 103-1.30 × 104) |
| Sewage outfall | Water column | 4 | 1.63 × 102 (77-3.40 × 102) | 21 (2-1.40 × 102) | 15 (1-43) | 22 (30-89) | 4 (0-39) | 4.59 × 102 (90-3.31 × 103) | 0 |
| Settled solids | 1 | 8.70 × 105 | 3.50 × 105 | 3.60 × 105 | 7.30 × 106 | 9.90 × 105 (n = 4)c (1.00 × 105-1.50 × 107) | 2.00 × 105 | ND | |
| Sediment | 3 | 5.52 × 102 (60-5.00 × 103) | 1.04 × 104 (0-1.07 × 105) | 65 (0-1.16 × 104) | 2.22 × 103 (60-8.99 × 104) | 7.78 × 102 (0-9.13 × 104) | 1.75 × 105 (90-2.45 × 107) | 0 | |
| Transition South | Water column | 2 | 1.37 × 102 (n = 1)c | 83 (55-1.10 × 102) | 49 (10-87) | 48 (23-72) | 0 | 2.07 × 102 (2.00 × 102-2.14 × 102) | 0 |
| Sediment | 2 | 0 (n = 1) | 4 (0-8) | 4 (1-8) | 10 (0-20) | 3.55 × 102 (20-1.02 × 103) | 1.16 × 103 (8.12 × 102-1.50 × 103) | 0 | |
| Cape Armitage | Sediment | 1 | 0 | 0 | 0 | 0 | 3.52 × 102 (n = 2)c (4-7.00 × 102) | 4.30 × 103 | ND |
| Big Razorback Island | Water column | 1 | 0 | 0 | 1 | 3 | 0 | 5.76 × 102 | ND |
| Sediment | 1 | 4.10 × 102 | 1.00 × 104 | 6.40 × 103 | 90 | 0 | 1.00 × 104 | ND | |
| Little Razorback Island | Water column | 1 | ND | 0 | 0 | 0 | 0 | 4.08 × 102 | 1 |
| Sediment | 1 | ND | 5 | 0 | 0 | 3.15 × 102 | 8.61 × 104 | 3 | |
| Cape Evans | Water column | 1 | 0 | 0 | 0 | 0 | 0 | 1.30 × 102 | 0 |
| Sediment | 1 | 0 | 0 | 0 | 0 | 0 | 3.40 × 102 | ND |
Values are expressed as CFU 100 ml−1 for water column, nonpotable, potable, settled solids, and untreated sewage samples; CFU g (dry weight)−1 for sediment samples; CFU ml−1 for TCB; and PFU 100 ml−1 for coliphage. Values in parentheses list the range of values for the geometric means.
ND, not determined.
Values represent the numbers of samples for the respective analyses.
The chemical data from the two sample sites in the distribution system indicate that a chemically stable system has been established, as the chemical parameters listed in Table 3, except for water temperature, were statistically equivalent (P > 0.05) to those recorded at the treatment plant. The Langlier index values for these samples indicate that the potable water is balanced to slightly scale forming. There was, however, a statistically significant (P < 0.001) loss of chlorine residual over the relatively short distribution distances. The percent hypochlorous acid concentration in the potable water samples was calculated using the average pH values from Table 3 and hypochlorous acid ionization constants for the respective temperatures by using the equation [1 + (Ki/H+)]−1 × 100 = percent hypochlorous acid, where Ki is the hypochlorous acid ionization constant (moles liter−1) and H+ is the negative antilog of the average pH value. The calculated hypochlorous acid concentrations in samples from the treatment plant, dormitory, and vehicle shop were 0.06 mg liter−1, 0.03 mg liter−1, and 0.02 mg liter−1, respectively. These concentrations are only 9.43, 8.15, and 7.90% of the respective measured chlorine residuals.
TABLE 3.
Mean values for drinking water chemical parameters at McMurdo Station
| Sample site | No. of samples | Temp (°C) | pH | Ca2+ (mg liter−1) | Alkalinitya (mg liter−1) | TDSb (mg liter−1) | Langlier index | Chlorine residualc (mg liter−1) |
|---|---|---|---|---|---|---|---|---|
| Treatment plant | 7 | 7.00 ± 0.82 | 8.71 ± 0.42 | 33.20 ± 6.88 | 83.20 ± 18.64 | 361.4 ± 49.81 | 0.50 ± 0.48 | 0.60 ± 0.03 |
| Dormitory | 7 | 12.70 ± 0.76 | 8.71 ± 0.27 | 34.30 ± 6.88 | 78.60 ± 16.00 | 380.0 ± 58.02 | 0.57 ± 0.31 | 0.41 ± 0.05 |
| Vehicle shop | 7 | 20.00 ± 3.46 | 8.65 ± 0.21 | 35.00 ± 4.79 | 81.10 ± 16.57 | 385.7 ± 55.33 | 0.65 ± 0.30 | 0.30 ± 0.09 |
Total alkalinity was measured by CaCO3 concentration.
TDS, total dissolved solids.
Chlorine residual was measured as free chlorine.
Untreated sewage and sewage-impacted sites.
The untreated sewage contained relatively high concentrations of total (1.28 × 106 CFU 100 ml−1) and fecal (2.15 × 105 CFU 100 ml−1) coliforms, E. coli (5.00 × 104 CFU 100 ml−1), enterococci (1.09 × 105 CFU 100 ml−1), and C. perfringens (3.03 × 104 CFU 100 ml−1). Coliphage were also detected (2.63 × 103 PFU 100 ml−1), and all of the isolates hybridized with probe IIa, IIb, or III, indicating they were from a human source (26). The indicator and TCB concentrations were reduced by 3.5 to 5.1 log in the water column at the sewage discharge site relative to the untreated sewage (Table 2). No F+ coliphage were recovered from the discharge site samples (Table 2).
In general, sediment samples collected at the sewage outfall retained greater concentrations of all of the indicators than the overlying water column (Table 2). The total coliform and E. coli concentrations were moderately elevated (3.4- and 4.3-fold, respectively), while the fecal coliform (2.7 log), enterococcus (2.0 log), C. perfringens (2.3 log), and TCB (2.6 log) concentrations were significantly greater. No F+ coliphage were recovered from these samples.
The sewage solids that accumulate at the discharge site were significantly enriched for all of the indicators except for total and fecal coliforms (Table 2). The mean concentrations of total and fecal coliforms were similar to those found in the untreated sewage, while E. coli, enterococci, and C. perfringens concentrations were 7.2-, 67.0-, and 32.7-fold greater, respectively. Compared to water column and sediment data from this area, the concentrations of indicators recovered from sewage solids were, on average, approximately 5 and 3 logs greater, respectively (Table 2). There were no significant differences in the TCB values from the sewage solids and sediments collected in the outfall area (Table 2).
As the distance from the discharge site within the impacted area increased, the concentrations of indicators significantly declined (Table 2). The concentrations of all indicators recovered from the water column at Transition South were very similar to those recovered at the sewage outfall. However, in the sediments at this site the concentrations of indicators were significantly reduced (2.6-log reduction), with the exceptions of E. coli (16.3-fold reduction) and C. perfringens (2.2-fold reduction), compared to the outfall sediments. The sediment sample collected from the site furthest from the impacted area, Cape Armitage, was negative for all of the indicators except C. perfringens (Table 2).
Nonimpacted sites.
The concentrations of the indicators in water column samples collected from Big and Little Razorback Islands and Cape Evans were all ≤3 CFU 100 ml−1 or not detected (Table 2). Interestingly, the Little Razorback Island water column samples were positive for the presence of coliphage, and the isolate hybridized with probes IIa and IIb.
Sediment samples collected from sites that were >1,800 m from the sewage outfall were negative for the presence or had very low concentrations (≤5 CFU g [dry weight]−1) of total and fecal coliforms, E. coli, and enterococci, with the exception of Big Razorback Island (Table 2). This site was positive for all of the indicators, with the exception of C. perfringens, at concentrations similar to those found in sediments at the sewage outfall site (Table 2). In contrast to the Big Razorback Island site, sediments from Little Razorback Island were positive for C. perfringens at concentrations that were similar to those found in the sewage outfall sediments (Table 2). In agreement with the water column results from Little Razorback Island, the sediments were positive for the presence of coliphage (3 PFU 100 ml−1), and all coliphage hybridized with probe IIa or IIb.
Seal scat.
Weddell seal scat was collected from five sites along tidal cracks that developed just off the coast of Ross Island in the general vicinities of McMurdo Station and Little Razorback Island. Of the seven scat samples that were sampled and streaked using sterile swabs, only one was positive for C. perfringens. However, the extraction method increased the recovery of indicator bacteria from scat materials (n = 4), as C. perfringens (range, 3 to 2.02 × 106 CFU g [dry weight]−1), fecal coliforms (3 to 1.4 × 104 CFU g [dry weight]−1), E. coli (0 to 1.21 × 104 CFU g [dry weight]−1), enterococci (1.21 × 102 to 3.10 × 105 CFU g [dry weight]−1), and TCB (5.02 × 105 to 2.66 × 108 CFU g [dry weight]−1) were present in concentrations similar to those in the untreated sewage (Table 2).
Enteric viruses.
Samples collected from the RO system (range of equivalent volume processed, 7 to 28,503 liters; n = 4), distribution system (1,142 to 11,210 liters; n = 2), untreated sewage (24 to 33 liters; n = 3), water columns at the drinking water intake (1.8 liters; n = 1), sewage outfall (1 to 40 liters; n = 2), Transition South (2 liters; n = 1), and Little Razorback Island (2 liters; n = 1) were negative for infectious enteric viruses. Sediment samples (2 g [wet weight]) from the drinking water intake (n = 1), sewage outfall (n = 1), and Transition South (n = 1) and sediment (n = 1) and seal scat (2 g [wet weight]; n = 3) samples collected at Little Razorback Island were also negative for enteric viruses.
DISCUSSION
McMurdo Station currently supports between 200 and 1,500 residents, with the greatest populations occurring during the months of August through February. Until recently, the facility discharged between 58,000 and 300,000 liters of untreated sewage day−1 into McMurdo Sound, with the months of September through February having the greatest discharge volumes (2). During this period, McMurdo Sound is covered with a 3- to 5-m-thick ice sheet that prevents wind-driven mixing of these waters and extensive redistribution of the sewage from the discharge site.
Previous studies, using only total coliforms or C. perfringens, delineated the area and severity of contamination around the submerged sewage effluent pipe off the coast of McMurdo Station (21, 32, 37). These studies determined that an area that extended for approximately 1 km along the shoreline of McMurdo Station and seaward for at least 300 m had been severely impacted by the sewage discharge. The impacted area included the location of the drinking water source water intake and multiple areas of routine diving activities. One of these studies also monitored current patterns under the sea ice in the impacted area and found that prevailing currents moved away from the drinking water intake (37). However, they also documented that these currents experienced episodic and strong reversals that impacted the drinking water intake area. These current patterns were implicated as a mechanism for the distribution of human fecal waste and the associated microbial indicators during periods when the overlying water is covered by sea ice.
The concentrations of C. perfringens and total coliforms recovered from the water column and sediments at the drinking water intake during this study were relatively low but still presented a persistent challenge to the drinking water treatment process, as demonstrated by the pre-RO data (Table 2). The RO process was operating efficiently during our study, as no indicators were recovered in the post-RO water (Table 2). However, within the distribution system there was one total coliform-positive sample and one sample with a TCB concentration of 1.60 × 103 CFU ml−1 (Table 2). TCB concentrations of >500 CFU ml−1 are inhibitory to the recovery of total coliforms from drinking water samples (23), suggesting that sections of McMurdo Station's distribution system may be susceptible to bacterial regrowth events that are not detected by routine total coliform analyses. Additionally, greater than 90% of the measured total chlorine residual is in a nonbactericidal form that is inefficient at inactivating bacteria within the distribution system. Though the RO treatment may be operating efficiently, indicator and other bacteria can be introduced into a distribution system through pipe breaks and repairs and transient pressure differentials (35). A more comprehensive monitoring program of the distribution system should be considered.
There were no infectious enteroviruses recovered from any of the samples, including the untreated raw sewage. Concentrations of infectious enteroviruses have been shown to vary significantly in untreated and biologically treated wastewater due to the occurrence of the virus being directly related to a limited number of active enterovirus carriers in the general population (29). The relatively small and healthy population of McMurdo Station may significantly reduce the probability of having a source of infectious enteroviruses large enough to detect with the method used in this study.
Coliphage genotypes that are most often associated with human feces were recovered from the untreated sewage. Though the coliphage concentrations in the untreated sewage were relatively low, they are within a range commonly recovered from the same types of samples from temperate climates (8, 28). Interestingly, coliphage were not recovered from the discharge site, which is only 65 m downstream from the untreated sewage sample site. The inability to recover coliphage at the sewage outfall may be due to a dilution effect reducing the concentration to below the detection limit (35 PFU 100 ml−1) of our assay.
The sewage solids at the discharge site act as a reservoir and source for all of the indicator bacteria and TCB, especially C. perfringens (Table 2). The microorganisms and dissolved and particulate organic components that are continuously transported from the discharge site by the currents in this area provide a source of bacteria and nutrients to the sediment systems along the coastline of McMurdo Station. Indicator and pathogenic bacteria associated with raw or partially treated sewage have been shown to settle into marine sediments, where they accumulate and reach higher densities than in the overlying water column (10, 18, 24, 25). Additionally, lower water temperatures and elevated nutrient concentrations, like those in the immediate area of sewage outfalls, have also been shown to enhance the survival of bacteria in the water column and sediments of southern polar regions (19, 20, 33, 42).
Unexpectedly, the water column and sediment samples from Big and Little Razorback Islands were positive for one or more of the indicators used in this study, while the Cape Evans site was negative for all of the indicators (Table 2). All three sites were considered to be negative control sites prior to our sampling events. Based on coliphage probe hybridization, the coliphage recovered from Little Razorback Island were most likely from a human source (17, 26, 34). The only human activities in this immediate area are in seasonal camps that are built on the sea ice to provide living and research facilities for between 4 and 20 residents. These camps have been positioned in the same general area for years between the months of August and February. There are no waste treatment or containment facilities at these sites. Outhouses, positioned over holes drilled into but not through the sea ice, serve as the only toilet facilities. The obvious implication of the fecal indicator occurrence in the water column and sediments from these sites that are so distant from the sewage outfall is that large quantities of human fecal material collected in these sea ice holes are released during the melting of the sea ice. The fact that group II probes did hybridize with the coliphage does not unequivocally prove that the source of the coliphage is human, as the host specificity of F+ RNA coliphage among mammals (i.e., humans, pigs, cattle, poultry) has been shown to be less absolute than previously thought (41).
Though the yearly thaw of the sea ice and the eventual release of the human fecal wastes may contribute to indicator presence in this area, the possible contribution of indicators from seal feces cannot be dismissed. The number of Weddell seals in the area of Little and Big Razorback Islands is estimated to be 223 (M. Cameron, personal communication). The scat samples contained concentrations of the indicators that were similar to those recovered from the untreated sewage (Table 2). The inconsistencies within the indicator concentrations in the scat samples are attributed to their being dry, frozen, and exposed to sunlight for unknown periods of time prior to collection. Accordingly, the indicator concentrations in the scat samples are considered to be conservative estimates, as these conditions have been shown to reduce the culturability of indicators (5, 40, 45).
This is the first report on the occurrence of C. perfringens and the other indicators in this Antarctic marine mammal. C. perfringens has been isolated from healthy arctic hooded seals (Cystophora cristata) (4). Assuming that the concentrations of the indicators in the scat samples are conservative estimates of concentrations in fresh Weddell seal feces, this group of mammals represents a significant source of microbial indicators traditionally used to monitor for the presence of human fecal wastes.
During February 2003, a wastewater treatment facility was brought online at McMurdo Station, and it is monitored according to the Clean Water Act (22) (K. Nickle, personal communication). The facility is currently running at only 55.0% of its designed capacity and removing biological oxygen demand (99% reduction), total suspended solids (98% reduction), phosphates (50 to 65% reduction), nitrates (90 to 95% reduction), and ammonia (90 to 95% reduction) from the treatment stream prior to discharge into McMurdo Sound. All solids recovered from the treatment are transported stateside for disposal.
The data generated from this study provide a baseline for the microbiological quality of the water column and sediments in the area impacted by the release of untreated human sewage from McMurdo Station. Monitoring studies of the recovery of the impacted area should employ a suite of microbial indicators as in this study, as different indicators have different decay rates (as determined by culturability) (7, 9, 18, 30, 38). In general, the bacterial indicators can be categorized in regard to their survival in marine systems as follows: total coliforms < fecal coliforms and E. coli < enterococci < C. perfringens. Interestingly, the distribution of indicators based on occurrence and concentration in the sediments with increasing distance from the discharge site follows this order. The reductions in concentrations of the respective indicators over time and distance from the discharge site could therefore be used as a metric of recovery within the impacted area to pre-sewage discharge conditions. For example, due to their rapid decay rates in marine waters, the occurrence of total coliforms in the water column was indicative of very recent fecal contamination in that area, most likely carried from the discharge site. Sediments at the Cape Armitage site, which receives the least amount of fecal material within the impacted area, were negative for all of the indicators except for C. perfringens, indicating that this area has received less fecal material and/or that the length of time between exposures was greater than the decay rates for the other indicators.
Future studies using microbial indicators of human fecal pollution to assess the extent and impact of human fecal discharges at McMurdo Station or other coastal research facilities on Antarctica should include an assessment of the possible impact of indigenous mammals, such as Weddell seals. Our data indicate that these mammals are a source of microbial indicators that have traditionally been used to assess the contribution of human fecal waste into coastal waters.
Acknowledgments
This study was supported by National Science Foundation Office of Polar Programs grant OPP 02-31870 to J.T.L.
We acknowledge C. Gerba and K. Reynolds (University of Arizona) for enterovirus analyses, M. Cameron (National Marine Mammal Laboratory) and his field crew for the collection of seal fecal samples, R. Robbins (Raytheon Polar Services Co. [RPS]) and his group for dive support, K. Nickle (RPS) for discussions of the wastewater treatment facility, and the employees of the National Science Foundation and RPS who provided logistic and technical assistance. We also acknowledge B. Boynton (U.S. Geological Survey) for assistance with the graphics.
REFERENCES
- 1.Antarctic Support Associates. 1999. United States Antarctic program drinking water monitoring summary. File 070-2696299-L. Antarctic Support Associates, Denver, Colo.
- 2.Antarctic Support Associates. 1999. United States Antarctic program wastewater monitoring summary. File 070A-2696399-L. Antarctic Support Associates, Denver, Colo.
- 3.Armon, R., and P. Payment. 1988. A modified m-CP medium for enumerating C. perfringens from water samples. Can. J. Microbiol. 34:78-79. [DOI] [PubMed] [Google Scholar]
- 4.Aschfalk, A., and W. Müller. 2001. Clostridium perfringens toxin types in hooded seals in the Greenland Sea, determined by PCR and ELISA. J. Vet. Med. 48:765-769. [DOI] [PubMed] [Google Scholar]
- 5.Barcina, I., J. Gonzalez, J. Iriberri, and L. Egea. 1990. Survival strategy of Escherichia coli and Enterococcus faecalis in illuminated fresh and marine systems. J. Appl. Bacteriol. 68:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Bisson, J., and V. Cabelli. 1979. Membrane filtration enumeration method for Clostridium perfringens. Appl. Environ. Microbiol. 37:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabelli, V. 1983. Health effects criteria for marine recreational waters. EPA-600/1-80-03. U.S. Environmental Protection Agency, Cincinnati, Ohio.
- 8.Calci, K., W. Burkhardt III, W. Watkins, and S. Rippey. 1998. Occurrence of male-specific bacteriophage in feral and domestic animal wastes, human feces, and human-associated wastewaters. Appl. Environ. Microbiol. 64:5027-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberlain, C., and R. Mitchell. 1978. A decay model for enteric bacteria in natural waters, p. 325-348. In R. Mitchell (ed.), Water pollution microbiology. John Wiley & Sons, Inc., New York, N.Y.
- 10.Chung, H., and M. Sobsey. 1993. Survival of F-specific coliphages, Bacteroides fragilis phages, hepatitis A virus (HAV) and poliovirus 1 in seawater and sediment. Water Sci. Technol. 27:425-428. [Google Scholar]
- 11.Clesceri, L., A. Greenberg, and A. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, p. 115-131. American Public Health Association, Washington, D.C.
- 12.Clesceri, L., A. Greenberg, and A. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, p. 63-65. American Public Health Association, Washington, D.C.
- 13.Clesceri, L., A. Greenberg, and A. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, p. 74-78. American Public Health Association, Washington, D.C.
- 14.Clesceri, L., A. Greenberg, and A. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, p. 34-41. American Public Health Association, Washington, D.C.
- 15.Clesceri, L., A. Greenberg, and A. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, p. 1-90. American Public Health Association, Washington, D.C.
- 16.Clesceri, L., A. Greenberg, and A. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, p. 56-62. American Public Health Association, Washington, D.C.
- 17.Cole, D., S. C. Long, and M. D. Sobsey. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 69:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies, C., J. Long, M. Donald, and N. Ashbolt. 1995. Survival of fecal microorganisms in marine and freshwater sediments. Appl. Environ. Microbiol. 61:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delille, D. 1995. Seasonal changes of subantarctic benthic bacterial communities. Hydrobiologia 310:47-57. [Google Scholar]
- 20.Delille, D., and E. Perret. 1989. Influence of temperature on the growth potential of southern polar marine bacteria. Microb. Ecol. 18:117-123. [DOI] [PubMed] [Google Scholar]
- 21.Edwards, D., G. McFeters, and M. Venkatesan. 1998. Distribution of Clostridium perfringens and fecal sterols in a benthic coastal marine environment influenced by the sewage outfall from McMurdo Station, Antarctica. Appl. Environ. Microbiol. 64:2596-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Federal Register. 1972. Federal Water Pollution Control Act. Fed. Regist. 101-607:1-230. [Google Scholar]
- 23.Federal Register. 1989. National primary drinking water rules and regulations. Fed. Regist. 54:27486-27541. [Google Scholar]
- 24.Gerba, C., and J. McLeod. 1976. Effect of sediments on the survival of Escherichia coli in marine waters. Appl. Environ. Microbiol. 32:114-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goyal, S., C. Gerba, and J. Melnick. 1977. Occurrence and distribution of bacterial indicators and pathogens in canal communities along the Texas coast. Appl. Environ. Microbiol. 34:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin, D., P. Stokes, J. Rose, and J. Paul. 2000. Bacterial indicator occurrence and the use of an F+ specific RNA coliphage assay to identify fecal sources in Homosassa Springs, Florida. Microb. Ecol. 39:56-64. [DOI] [PubMed] [Google Scholar]
- 27.Halton, J., and W. Nehlsen. 1968. Survival of E. coli in zero degree centigrade sea water. J. Water Pollut. Control Fed. 40:865-868. [PubMed] [Google Scholar]
- 28.Havelaar, A., K. Furuse, and W. Hogeboom. 1986. Bacteriophages and indicator bacteria in human and animal feces. J. Appl. Microbiol. 60:255-262. [DOI] [PubMed] [Google Scholar]
- 29.Havelaar, A., M. van Olphen, and Y. Drost. 1993. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl. Environ. Microbiol. 59:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill, R., I. Knight, M. Anikis, and R. Colwell. 1993. Benthic distribution of sewage sludge indicated by Clostridium perfringens at a deep-ocean dump site. Appl. Environ. Microbiol. 59:47-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill, R., W. Straube, A. Palmisano, S. Gibson, and R. Colwell. 1996. Distribution of sewage indicated by Clostridium perfringens at a deep-water disposal site after cessation of sewage disposal. Appl. Environ. Microbiol. 62:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howington, J., G. McFeters, J. Barry, and J. Smith. 1992. Distribution of the McMurdo Station sewage plume. Mar. Pollut. Bull. 25:324-327. [Google Scholar]
- 33.Howington, J., G. McFeters, W. Jones, and J. Smith. 1994. The effect of low temperature on BOD in Antarctic seawater. Water Res. 28:2585-2587. [Google Scholar]
- 34.Hsu, F., Y. Shieh, M. Beekwilder, and M. Sobsey. 1995. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl. Environ. Microbiol. 61:3960-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeChevalier, M., R. Gullick, M. Karim, M. Friedman, and J. Funk. 2003. The potential for health risks from intrusion of contaminants into the distribution system from pressure transients. J. Water Health 1:3-14. [PubMed] [Google Scholar]
- 36.McClintock, J. 1994. Trophic biology of Antarctic shallow-water echinoderms. Mar. Ecol. Prog. Ser. 111:191-202. [Google Scholar]
- 37.McFeters, G., J. Barry, and J. Howington. 1993. Distribution of enteric bacteria in Antarctic seawater surrounding a sewage outfall. Water Res. 27:645-650. [DOI] [PubMed] [Google Scholar]
- 38.Omura, T., M. Onuma, and Y. Hashimoto. 1982. Viability and adaptability of E. coli and enterococcus group to salt water with high concentrations of sodium chloride. Water Sci. Technol. 14:115-126. [Google Scholar]
- 39.Reynolds, K., C. Gerba, and I. Pepper. 1996. Detection of infectious enteroviruses by an integrated cell culture-PCR procedure. Appl. Environ. Microbiol. 62:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rozen, Y., and S. Belkin. 2001. Survival of enteric bacteria in seawater. FEMS Microbiol. Rev. 25:513-529. [DOI] [PubMed] [Google Scholar]
- 41.Schaper, M., J. Jofre, M. Uys, and W. Grabow. 2002. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J. Appl. Microbiol. 92:657-667. [DOI] [PubMed] [Google Scholar]
- 42.Smith, J., J. Howington, and G. McFeters. 1994. Survival, physiological response, and recovery of enteric bacteria exposed to polar marine environment. Appl. Environ. Microbiol. 60:2977-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, J., and G. McFeters. 1993. Survival and recoverability of enteric bacteria exposed to the Antarctic marine environment. Antarct. J. U.S. 28:120-123. [Google Scholar]
- 44.Sobsey, M., and J. Glass. 1980. Poliovirus concentration from tap water with electropositive adsorbent filters. Appl. Environ. Microbiol. 40:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solic, M., and N. Krstulovic. 1992. Separate and combined effects of solar radiation, temperature, salinity, and pH on the survival of faecal coliforms in seawater. Mar. Pollut. Bull. 24:411-416. [Google Scholar]
- 46.U.S. Department of State. 2002. Handbook of the Antarctic Treaty System, p. 1-1005. U.S. Department of State, Washington, D.C.