Abstract
A combination of experimental and theoretical approaches was used to investigate the role of nutrient starvation as a potential trigger for biofilm detachment. Experimental observations of detachment in a variety of biofilm systems were made with pure cultures of Pseudomonas aeruginosa. These observations indicated that biofilms grown under continuous-flow conditions detached after flow was stopped, that hollow cell clusters were sometimes observed in biofilms grown in flow cells, and that lysed cells were apparent in the internal strata of colony biofilms. When biofilms were nutrient starved under continuous-flow conditions, detachment still occurred, suggesting that starvation and not the accumulation of a metabolic product was responsible for triggering detachment in this particular system. A cellular automata computer model of biofilm dynamics was used to explore the starvation-dependent detachment mechanism. The model predicted biofilm structures and dynamics that were qualitatively similar to those observed experimentally. The predicted features included centrally located voids appearing in sufficiently large cell clusters, gradients in growth rate within these clusters, and the release of most of the biofilm with simulated stopped-flow conditions. The model was also able to predict biofilm sloughing resulting solely from this detachment mechanism. These results support the conjecture that nutrient starvation is an environmental cue for the release of microbes from a biofilm.
The accumulation of microorganisms on surfaces to form biofilms is increasingly being recognized as an important strategy of microbial survival in natural and engineered environments (6, 7). Biofilm accumulation is determined by the balance of attachment, growth, and detachment processes. Of these phenomena, the least understood is detachment. Detachment refers to the release of microbial cells and their associated matrix polymers from the biofilm to the bulk fluid bathing the film. Some of the factors that have been suggested to be important in biofilm detachment include matrix-degrading enzymes (1, 3, 14, 30), microbially generated gas bubbles (16), nutrient levels and microbial growth status (2, 12, 18, 22, 23, 25), availability of multivalent cross-linking cations (2, 4, 29), fluid shear stress (18-20, 27), contact attrition (5), quorum-sensing signals (1, 9), and the activation of a lytic bacteriophage (32).
Some of these detachment mechanisms are purely physical, but others may be predominantly biological. With regard to the biological pathways to biofilm detachment, it is interesting to consider the possible triggers of the detachment process. What are the environmental cues that initiate the progression that eventually leads to the release of cells? Two possible triggers for detachment are included in the factors listed above. These are the accumulation of a metabolic product and the depletion of a metabolic substrate. We focused on these triggers in particular because they have the potential to explain the observation of biofilm cell cluster hollowing (13, 15, 17, 21, 24, 28).
One intriguing mechanism of biofilm detachment invokes control by a quorum-sensing signal. Dissolution of the biofilm matrix and release of bacteria are hypothesized to be triggered when the signal molecule, an excreted bacterial metabolite, accumulates to a threshold concentration. Such a mechanism leads directly to the prediction that natural signal molecules or their analogues could be used to disperse biofilms and clean surfaces of biofouling (9). This mechanism of biofilm detachment was originally suggested by the observation that arresting the flow of a medium to a biofilm system caused the biofilm to spontaneously detach within a few days (8). The cessation of flow presumably allowed the detachment signal molecule to accumulate, eventually reaching a concentration sufficient to trigger dispersion of the biofilm. The theoretical implications of this quorum-sensing mechanism were investigated with a computer model (11). The model results indicated that high concentrations of the signal molecule excreted by bacteria occur in the center of cell clusters where diffusive egress is most restricted. This location is the same region in the biofilm that is likely to be nutrient deficient. This information leads to an alternative explanation of biofilm detachment based on a starvation response.
The purpose of the work reported here was to investigate, through a combination of experimental and theoretical approaches, the role of nutrient starvation in biofilm detachment. We hypothesized that localized nutrient depletion in a biofilm induces starvation in some cells and that these cells detach when the starvation persists for a sufficient period of time. Our investigation of this hypothesis involved two approaches. First, a cellular automata computer model of biofilm development was used to explore the behavior predicted by this detachment mechanism. Simulations from this model were compared to experimental observations of biofilm structure and detachment in Pseudomonas aeruginosa biofilms grown in a variety of biofilm reactors. The second approach was to perform preliminary laboratory experiments designed to discriminate the quorum-sensing hypothesis of biofilm detachment from the nutrient starvation hypothesis.
MATERIALS AND METHODS
Experimental methods.
Three different systems were used to produce biofilms of P. aeruginosa strain PA01: drip-flow reactors, glass capillary reactors, and colony biofilms. Biofilms grown in drip-flow reactors were used to investigate the roles of stopped flow and nutrient starvation on biofilm detachment. Biofilms grown in glass capillary reactors were examined directly by confocal laser scanning microscopy to observe hollow cell clusters. Colony biofilms were examined by transmission electron microscopy for signs of cell lysis.
Biofilms were grown in continuous drip-flow reactors by allowing minimal medium to flow dropwise over inclined stainless steel coupons. The reactor system has been described elsewhere (26). The medium flow rate was 50 ml h−1, the surface area of the coupons was 9.4 cm2, and the temperature was 22°C. The medium (33) contained glucose at 0.1 g/liter as the sole carbon and energy source. Biofilms were also grown in glass capillary tubes under continuous-flow conditions as described previously (34). Each tube had a square cross section facilitating direct microscopic observation through the tube wall. The nominal inside dimension of the tube was 0.9 mm. The inoculum was strain PA01 carrying an inducible green fluorescent protein (31). Biofilms were grown in 0.1× tryptic soy broth at a flow rate of 20 ml h−1 for 24 h at 37°C. Biofilms were counterstained by injection of a solution of rhodamine B at 50 mg liter−1 into the capillary tube to show the extent of the biomass and then were examined by confocal laser scanning microscopy. Colony biofilms were grown as described by Walters et al. (31). Colony biofilms were grown on tryptic soy agar for 48 h and then were processed for transmission electron microscopy.
After 4 days of growth in a drip-flow reactor, biofilm detachment was stimulated by stopping the flow of medium, laying the reactor flat, and covering the biofilm with 15 ml of medium to keep it hydrated. The system was allowed to stand with no mixing for 3 days. Slides were removed, allowed to drain to remove any detached cells from the biofilm, and scraped to enumerate attached bacteria. Detachment was also stimulated by subjecting a 4-day-old mature biofilm to glucose starvation by switching the influent to the same medium lacking glucose and to pure water. Slides were removed and scraped to enumerate attached bacteria.
Analytical methods applied to biofilms grown as described above included enumeration of viable and total cells, determination of biofilm thickness, confocal laser scanning microscopy, and transmission electron microscopy. Viable cell numbers were determined by scraping biofilms from the coupons used in the drip-flow reactors, homogenization to disperse cell aggregates, serial dilution, and plating on R2A agar (Becton Dickinson, Sparks, Md.). Total cell numbers were determined by direct microscope enumeration of bacteria in homogenates that had been deposited onto membranes by filtration. Biofilm thickness was determined by image analysis of 4′,6′-diamidino-2-phenylindole-stained frozen sections (10). Biofilms in glass capillary tubes were examined with a Leica TCS NT confocal laser scanning microscope. Excitation lines were at 488 and 568 nm, and emission was collected at 500 to 530 nm (green channel) and at 585 to 615 nm (red channel). Transmission electron microscopy of colony biofilms was performed as described by Walters et al. (31).
Computational methods.
A full mathematical description of the model used in this study has been presented elsewhere (11). Therefore, only a qualitative description of the model is presented here; modifications other than the detachment mechanism were as follows: the limiting substrate was switched from glucose to oxygen, and the node spacing (volume element side length) was changed to 1.71 μm to correspond to an intrinsic cell density of 350 mg liter−1, an individual cell volume of 0.5 μm3, and a cell volume fraction of 0.1.
The bacterium-level automata model, BacLAB, simulates the stochastic behavior of a bacterial biofilm on a submerged flat surface (substratum). BacLAB uses a hybrid modeling approach that linearly separates biofilm processes according to the natural time scales on which the processes occur. A conventional deterministic differential equation approach is used in modeling chemical diffusion and reaction, while a stochastic cellular automation approach is used to model bacterial cell division, detachment, and movement.
To simulate detachment resulting from starvation, a mechanism consisting of two parameters was implemented in the BacLAB model: minimum local nutrient concentration, CS,min, and duration of time below that concentration, tdetach. When the local nutrient concentration for a bacterium falls below CS,min, a counter for that bacterium records the cumulative number of time steps during which the bacterium is exposed to a low-nutrient enviroment. The counter begins at zero and continues until it reaches tdetach, at which point the cell detaches. If at any time step the local nutrient concentration for a bacterium rises above CS,min, the counter is decremented either until it returns to zero or the local nutrient concentration again falls below CS,min.
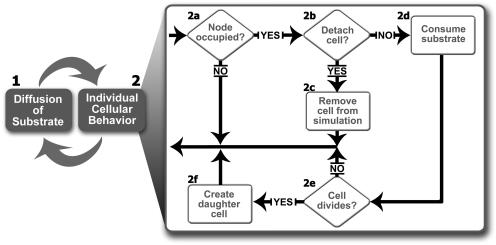
Each simulation begins by inoculating the substratum with randomly placed spherical colonies. The algorithm that defines how BacLAB then proceeds is illustrated in Fig. 1; the numbered processes are as follows. Step 1, the differential equation step, involves diffusion of the substrate from the bulk fluid into the biofilm. In step 2, the cellular automata step, the following operations are performed on each volume element in the simulation. Step 2a determines whether the volume element is occupied by a bacterium. If the volume element is unoccupied, nothing further is done with it for the current time step. If the volume element is occupied, more calculations are performed. Step 2b determines whether the bacterium meets the requirements for detachment as described above. In step 2c, if the requirements for detachment are satisfied, the bacterium is removed from the simulation. Additionally, any freely floating bacteria in other volume elements that can no longer be traced back to the substratum as a result of the removal of the initial bacterium are identified and removed. In step 2d, the bacterium is allowed to consume the substrate based on the local concentration of the substrate, the duration of the time step, and the kinetics associated with the bacterium. Step 2e determines whether the bacterium has consumed enough substrate to divide. In step 2f, a new bacterium neighboring the parent is created, and the excess substrate (not required for the creation of the daughter cell) is split between the parent and daughter cells.
FIG. 1.
General procedure followed in a typical BacLAB simulation. See the text for an explanation of the sequence of operations.
Computer experiment.
To evaluate the effect of detachment resulting from starvation on biofilm development, we conducted 10 replicate computer experiments simulating the starvation-induced detachment mechanism. Each experiment simulated 500 h of biofilm development on a rectangular surface measuring 512 by 512 μm according to the procedure outlined in Fig. 1. The values for node spacing in the x, y, and z dimensions were the same; each model cell defined a cube with a side of 1.71 μm. Table 1 shows the values used to model the kinetics and solute transport for a typical biofilm. These representative values fit the ranges available from the literature, with the exception of those pertaining to detachment, for which no literature values are available.
TABLE 1.
Parameter values
| Parameter | Value |
|---|---|
| Maximum specific growth rate | 0.3 h−1 |
| Time step | 1 h |
| Bulk substrate concn | 8 g m−3 |
| Diffusivity of substrate in aqueous phase (including liquid, channels, and voids) | 7.20 × 10−6 m2 h−1 |
| Relative effective diffusivity of substrate in biofilm | 0.55 |
| CS,min | 1 g m−3 |
| Monod half-saturation coefficient | 0.1 g m−3 |
| Volume element side length | 1.71 μm |
| Avg cell mass | 1.75 × 10−13 g |
| No. of initial colonies | 28 |
| No. of nodes in x direction | 300 |
| No. of nodes in y direction | 300 |
| No. of nodes in z direction | 300 |
| Radius of initial colonies | 8.55 μm |
| tdetach | 24 h |
| Yield coefficient | 0.24 g gs−1 |
The computer model was also used to simulate a stopped-flow experiment in which the nutrient concentration in the bulk water above the boundary layer was set to zero at a specified time step.
Computational resources.
BacLAB was written in C++ and compiled by using the GCC complier collection (release 3.2.2) from the Free Software Foundation. All experiments were run on a dual-AMD XP 2800+ workstation with 2 gigabytes of RAM. A single experiment typically required 24 computer hours to simulate 500 biofilm hours and utilized approximately 700 megabytes of RAM. These simulations took approximately three times longer than quorum-sensing mechanism simulations (11). The longer computation time was the result of solving for solute concentrations iteratively on a finer time grid so as to avoid numerical artifacts in the critical regions, where solute concentrations were nearly zero.
RESULTS AND DISCUSSION
A combination of theoretical and experimental results bearing on the detachment of microorganisms from biofilms is presented below. The model simulations are not intended to provide quantitative matches to the experimental data. Rather, the simulations are offered as illustrations of general biofilm structures or behaviors that arise from starvation-dependent detachment. The agreement or lack of agreement between theory and experiment should be evaluated at a qualitative level.
Detachment from stopping the flow of medium or omitting nutrients from the medium.
After 4 days of growth in the drip-flow reactor, P. aeruginosa formed milky biofilms on the stainless steel slides used as the attachment surface. The areal cell density at this stage of development was approximately 109 cells cm−2. The log10 total areal cell density was 9.56 ± 0.13 (mean and standard deviation). The log10 viable areal cell density was 9.25 ± 0.21. The biofilms were approximately 300 μm thick. Biofilm detachment could be stimulated by stopping the flow of medium, laying the reactor flat, and covering the biofilm with 15 ml of medium to keep it hydrated. The system was allowed to stand, with no mixing at all, for 3 days. During this interval, the biofilm became increasingly translucent and the metal substratum became progressively more visible. Slides were removed and scraped to enumerate attached bacteria. By both total cell counting and viable cell counting, it was determined that stopped-flow conditions produced at least a 1-log-unit reduction in biofilm cell numbers (Table 2). This means that more than 90% of the cells detached. To confirm this phenomenon, the number of cells in the fluid bathing the biofilm was also measured (Table 3). These data show that the decrease in the number of attached cells was accompanied by a concomitant and balancing increase in the number of freely floating cells.
TABLE 2.
Detachment from P. aeruginosa biofilmsa
| Condition | Flow | Mean ± SD log10 reduction in cell numbers
|
|
|---|---|---|---|
| Total | Viable | ||
| Flow stopped | Static | 1.18 ± 0.30 | 1.21 ± 0.28 |
| Glucose omitted | Shear | 1.31 ± 0.16 | 1.02 ± 0.14 |
| All nutrients omitted | Shear | 1.37 ± 0.18 | 1.36 ± 0.10 |
Data are from triplicate measurements.
TABLE 3.
Cell numbers in biofilm and planktonic phases before and after P. aeruginosa biofilm detachment
| Cells | Mean ± SD log10 cell numbers in the indicated samples
|
|||||
|---|---|---|---|---|---|---|
| Before detachment
|
After detachment
|
|||||
| Biofilm | Planktonic | Total | Biofilm | Planktonic | Total | |
| Total | 10.6 ± 0.1 | 9.4 ± 0.1 | 10.7 | 9.7 ± 0.2 | 11.0 ± 0.4 | 11.0 |
| Viable | 10.4 ± 0.2 | 9.4 ± 0.1 | 10.5 | 9.6 ± 0.2 | 10.5 ± 0.2 | 10.5 |
The time course of biofilm detachment after stopping the flow of medium in a drip-flow reactor is charted in Fig. 2 along with the detachment predicted by BacLAB under simulated stopped-flow conditions. In both the model and the experiment, nearly 90% of the cells within a P. aeruginosa biofilm were lost during the first day. One difference between the model and the experiment is that the model predicted continued detachment, whereas a tenacious layer of cells remained attached according to experimental data. Approximately 2 to 7% of the biofilm that was originally present remained for at least another 96 h in the experiment. While the simulations correctly predicted the rapid release of most of the cells from the surface, they did not capture the long-term retention of a residual fraction of the cells.
FIG. 2.
Biofilm detachment after stopping of the flow of medium in a drip-flow reactor (circles) and as predicted by BacLAB (average of six simulations) (solid line). Approximately 90% of the biomass detached after 1 day under static conditions.
Detachment of the biofilm in the drip-flow reactor system was visually confirmed by microscopic examination of frozen cross sections. The observations were also consistent with a loss of approximately 90% of the biofilm after stopping the flow for 3 days. The biofilm thickness decreased from 334 ± 34 μm (mean and standard deviation) before detachment to 27 ± 10 μm after detachment.
While these experimental data confirmed the observation of biofilm detachment in response to stopping the flow of medium, they do not indicate whether the detachment was a response to the accumulation of a signal molecule or whether it was just a reaction to nutrient starvation. To answer this question, we subjected 4-day-old mature biofilms to glucose starvation by switching the influent to the same medium lacking glucose. The continuous flow in these experiments should have prevented the accumulation of a detachment signal molecule. Furthermore, reduced synthesis of a detachment signal molecule would be expected in the absence of the sole carbon source. After 3 days of starvation under these continuous-flow conditions, biofilms detached to a similar degree as in the static assay (Table 2). The same results were obtained when pure water was used during the starvation period in place of glucose-free medium. These data support the hypothesis that biofilm detachment is a response to nutrient depletion. While these data do not disprove triggering of the mechanism of detachment by a metabolic product, they also do not provide support for this mechanism.
Observation and prediction of hollowing of biofilm cell clusters.
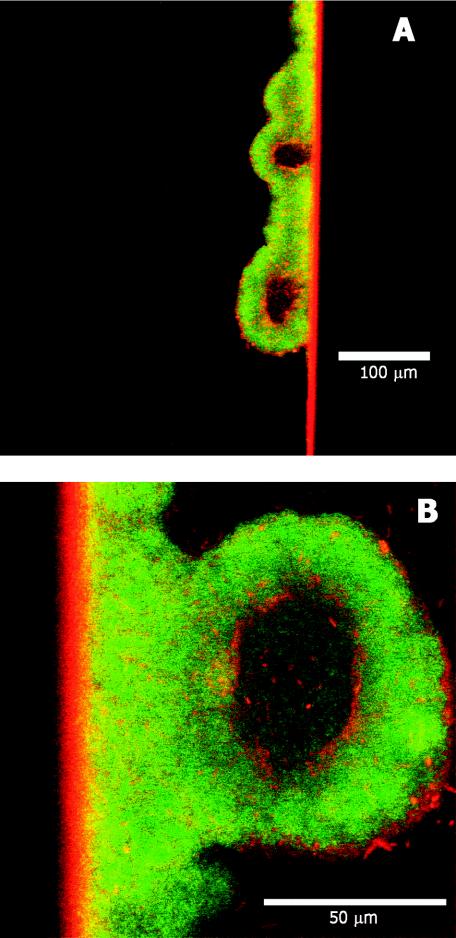
When P. aeruginosa biofilms were grown in continuous-flow capillary reactors, hollow cell clusters were sometimes observed by confocal laser scanning microscopy (Fig. 3). The hollowing appeared to be spatially organized with two key features: (i) hollowing only occurred in cell clusters that had reached a sufficient size, and (ii) voids were always centrally located at the substratum. These features are qualitatively consistent with the conjecture that detachment depends on nutrient availability.
FIG. 3.
Confocal laser scanning microscopy of hollow P. aeruginosa biofilm cell clusters. Biofilms were grown in glass capillary tubes under continuous flow. The bacteria contained green fluorescent protein and appeared green. The specimen was counterstained with rhodamine B (red), the primary utility of which was in locating the glass wall. Shown are two different locations of the same specimen.
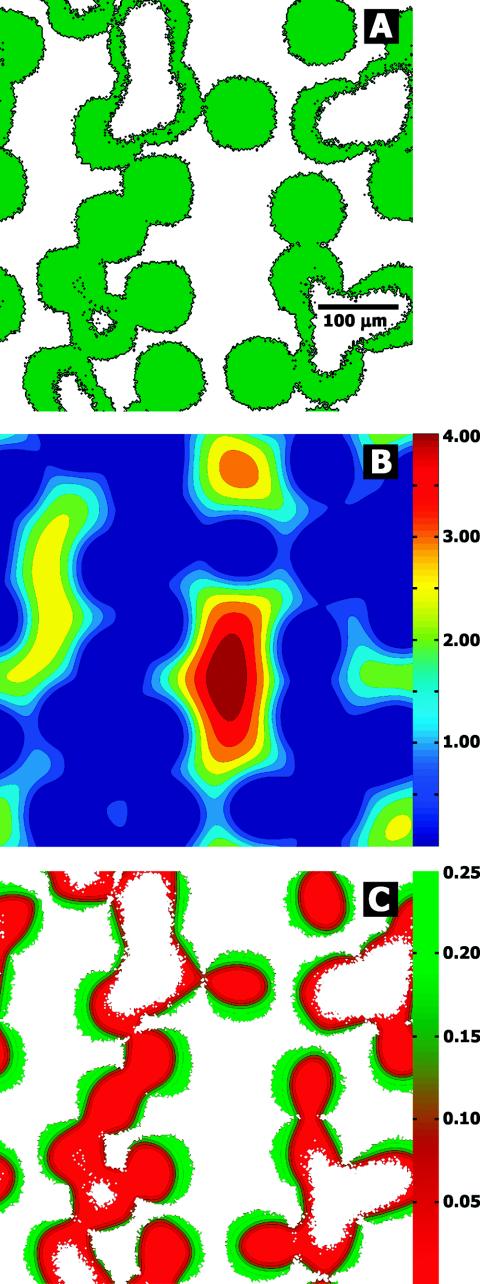
The results of the BacLAB simulations predicted biofilm cell cluster hollowing with the same features. Hollowing occurred at the substratum in the center of large clusters (Fig. 4A). The hollow regions in the biofilm corresponded to areas of low nutrient concentrations (Fig. 4B). The model predicted that cells near the bulk fluid interface were growing much more rapidly than cells deeper in the biofilm (Fig. 4C). The simulations indicated that the transition from high to low growth rates was spatially sharp, occurring over a distance of a few tens of micrometers at most (Fig. 4C).
FIG. 4.
Representative BacLAB simulation predicting hollow biofilm cell clusters. All three panels show patterns at the base of the biofilm across the square substratum area. The distribution of biomass at the substratum (green in panel A) suggests that larger clusters develop hollow interiors. Also shown are the predicted distributions of oxygen concentrations, in milligrams liter−1, at the substratum (B) and the specific growth rates, in hours−1, at the substratum (C). These simulations predicted sharp gradients in the concentration of the metabolic substrate (B) and the growth status of cells in the biofilm (C).
The presence of voids within biofilm cell clusters was reinforced by microscopic observation of the rapid motility of bacterial cells inside some cell clusters. (A movie of seething within a P. aeruginosa biofilm cell cluster can be viewed at http://www.erc.montana.edu/Res-Lib99-SW/Movies/Database/MD_DisplayScript.asp.) The rapid motility of these cells suggests that the local viscosity in the cluster interior has been reduced. To achieve such a reduction in viscosity, most of the cell mass and extracellular matrix material in the cluster void would have to have been degraded or released into the bulk fluid.
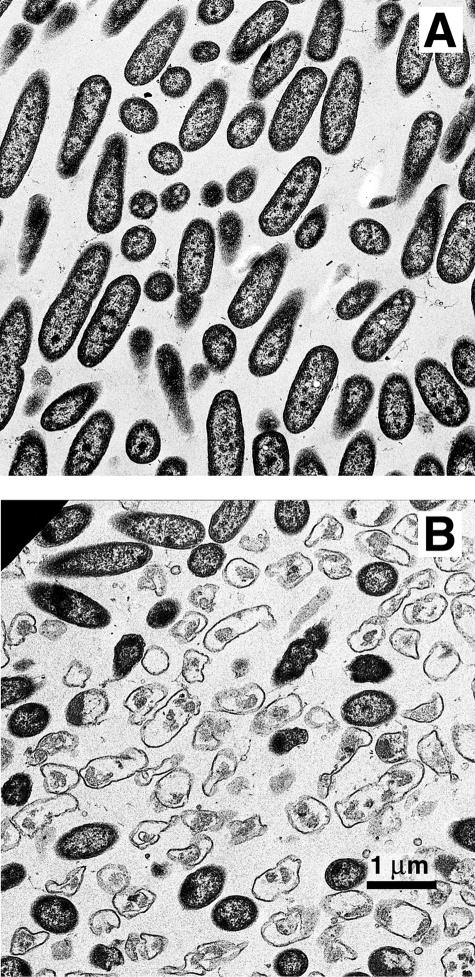
Others (17, 21, 24, 28, 32; L. Purevdorj-Gage and P. Stoodley, unpublished data) have observed hollowing of P. aeruginosa biofilm cell clusters similar to that described here. Webb et al. (32) have also presented strong evidence that this behavior is associated with the activation of a bacteriophage. This evidence suggests the following mechanism of hollowing in these biofilms. At a certain stage of biofilm development, the phage is activated in the center of some cell clusters. The phage causes some, but not all, cells to lyse. Enzymes, possibly polysaccharide lyases, proteases, or nucleases, are released and locally thin the extracellular polymeric substance matrix. Some of the cells survive the phage and either are released through a pore to the bulk fluid or are trapped inside the void, where they become motile and seethe inside the hollow cell cluster. According to this hypothesis, the observed hollowing would result from a combination of cell lysis, extracellular matrix degradation, and cell dispersal. In support of a role for cell lysis, we observed lysis in the internal strata of P. aeruginosa colony biofilms (Fig. 5). Because there is no flow in the colony biofilm system, lysed cells are retained and can be identified by transmission electron microscopy. Webb et al. (32) have proposed that the activation of the phage in P. aeruginosa biofilms is a response to localized oxidative stress.
FIG. 5.
Transmission electron microscopic analysis of biofilms. (A) Little lysis is evident in cells near the air interface of the biofilm. (B) Cell lysis is observed in the interior of P. aeruginosa colony biofilms.
We suggest that the activation of a phage or other lytic processes within biofilms could be a consequence of prolonged nutrient starvation. In other words, nutrient starvation might be the trigger for the detachment process. Computer model simulations of biofilm development based on a conjecture of starvation-dependent detachment generate biofilm structures that resemble those observed in experiments. This mechanism can explain the detachment that is observed when the flow of medium is stopped. The simulations are also consistent with the widespread, if anecdotal, observation of biofilm sloughing, a phenomenon that other models of biofilm detachment have failed to capture. Recent work with Shewanella oneidensis biofilms suggests that oxygen limitation triggers the detachment of biofilms formed by this microorganism (K. M. Thormann, S. M. Saville, S. Shukula, and A. M. Spormann, submitted for publication).
Some recent experiments with mutants of P. aeruginosa that are incapable of synthesizing homoserine lactone signal molecules supported the interpretation that quorum sensing does not trigger detachment in this microorganism. Wilson et al. (35) compared detachment rates for wild-type (PAO1) and signaling-deficient mutant (JP1) strains of P. aeruginosa. The mutant is unable to make N-3-oxo-dodecanoyl homoserine lactone, which has been shown to influence the structure of the biofilm (8). Wilson et al. (35) determined that detachment patterns were similar for the two strains. Other experiments specifically tested whether the same signaling mutant was capable of the cell cluster-hollowing mode of detachment (Purevdorj-Gage and Stoodley, unpublished). These investigators found no effect of quorum sensing on what they termed “seeding dispersal.”
Prediction of biofilm sloughing.
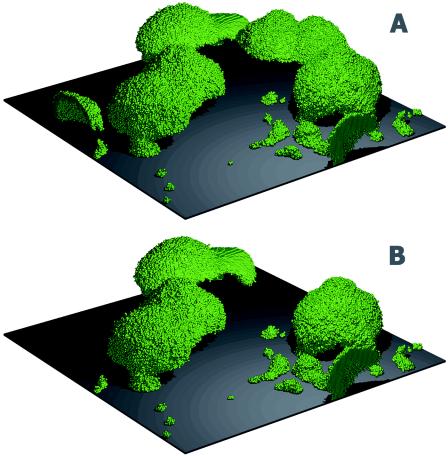
An interesting observation from the BacLAB simulations was the prediction of sloughing. In the early stages of biofilm development, cell clusters developed hollow interiors through lysis and detachment, as described above. The remaining biomass then appeared as “mushrooms” or large chunks of biofilm loosely tethered to the substratum. At some point, the few bacteria anchoring the biofilm became nutrient starved and detached, releasing large chunks of biomass back into the bulk fluid (Fig. 6).
FIG. 6.
BacLAB simulation showing biofilm sloughing. Biofilm structures at 235 h (A), before the sloughing event, and at 240 h (B), after the sloughing event, are shown. An entire cell cluster near the rear corner of the simulated area disappeared in the interval between the two time points. (The entire simulation can be viewed at http://www.erc.montana.edu/Res-Lib99-SW/Movies/Database/MD_DisplayScript.asp.)
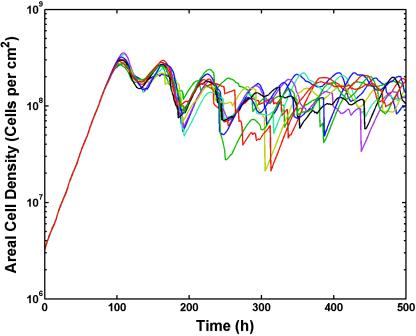
Biofilm sloughing occurred regularly in BacLAB simulations. Of the 10 replicate simulations conducted, 6 had sloughing events that resulted in a loss of 50% or more of the biomass (>0.301-log-unit reduction) in one time step (Fig. 7). Smaller detachment events releasing multicellular aggregates occurred continually, offsetting the ongoing growth of biofilm cells and allowing the biofilm to reach an oscillatory steady state. While it was beyond the scope of this work to investigate the detached particle size distribution, it is clear that detachment resulting from nutrient starvation would be expected to lead to the release of everything from single cells to nearly 70% of the biofilm biomass in a single event (27).
FIG. 7.
BacLAB replicate simulations showing sloughing as revealed by sharp decreases in areal cell density. The parameter settings were identical in these 10 simulations; only the random initial distributions of cells on the substrata differed in the 10 runs. Sloughing events, defined as a loss of 50% of biofilm biomass in a single time step (1 h), occurred in 6 of the 10 simulations.
Surely there are multiple pathways to biofilm detachment—simple physical shearing by fluid flow, cell lysis, and enzymatic dissolution of matrix material. Detachment is a critical process governing biofilm structure and the rate and extent of biofilm accumulation. It regulates the dissemination of microorganisms from a contaminated surface. New experimental and computational studies are needed to understand and isolate the factors that control biofilm detachment.
REFERENCES
- 1.Allison, D. G., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 2.Applegate, D. H., and J. D. Bryers. 1991. Effects of carbon and oxygen limitations and calcium concentrations on biofilm removal processes. Biotechnol. Bioeng. 37:17-25. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caccavo, F., B. Frolund, F. V. Kloeke, and P. H. Nielsen. 1996. Deflocculation of activated sludge by the dissimilatory Fe(III)-reducing bacterium Shewanella alga BrY. Appl. Environ. Microbiol. 62:1487-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, H. T., B. E. Rittmann, D. Amar, R. Heim, O. Ehlinger, and Y. Lesty. 1991. Biofilm detachment mechanisms in a liquid-fluidized bed. Biotechnol. Bioeng. 38:499-506. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., and P. S. Stewart. 2001. Battling biofilms. Sci. Am. 285:74-81. [DOI] [PubMed] [Google Scholar]
- 8.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-299. [DOI] [PubMed] [Google Scholar]
- 9.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 48:87-102. [DOI] [PubMed] [Google Scholar]
- 10.Huang, C.-T., P. S. Stewart, and G. A. McFeters. 1998. The study of microbial biofilms by classical fluorescence microscopy, p. 411-429. In M. H. F. Wilkinson and F. Schut (ed.), Digital image analysis of microbes: imaging, morphometry, fluorometry and motility techniques and applications. John Wiley & Sons, Chichester, England.
- 11.Hunt, S. M., M. A. Hamilton, J. T. Sears, G. Harkin, and J. Reno. 2003. A computer investigation of chemically mediated detachment in bacterial biofilms. Microbiology 149:1155-1163. [DOI] [PubMed] [Google Scholar]
- 12.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan, J. B., M. F. Meyenhofer, and D. H. Fine. 2003. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohashi, A., and H. Harada. 1994. Characterization of detachment mode of biofilm developed in an attached-growth reactor. Water Sci. Technol. 30:35-45. [Google Scholar]
- 17.Parsek, M. R., and C. Fuqua. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peyton, B. M., and W. G. Characklis. 1993. Statistical analysis of the effect of substrate utilization and shear stress on the kinetics of biofilm detachment. Biotechnol. Bioeng. 41:728-735. [DOI] [PubMed] [Google Scholar]
- 19.Picioreanu, C., M. C. M. van Loosdrecht, and J. J. Heijnen. 2001. Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol. Bioeng. 72:205-218. [PubMed] [Google Scholar]
- 20.Rittmann, B. E. 1982. The effect of shear stress on biofilm loss rate. Biotechnol. Bioeng. 24:501-506. [DOI] [PubMed] [Google Scholar]
- 21.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawyer, L. K., and S. W. Hermanowicz. 1998. Detachment of biofilm bacteria due to variations in nutrient supply. Water Sci. Technol. 37:211-214. [Google Scholar]
- 23.Speitel, G. E., and F. A. Digiano. 1987. Biofilm shearing under dynamic conditions. J. Environ. Eng. ASCE 113:464-475. [Google Scholar]
- 24.Stapper, A. P., G. Narasimhan, D. E. Ohman, J. Barakat, M. Hentzer, S. Molin, A. Kharazmi, N. Hoiby, and K. Mathee. 2004. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J. Med. Microbiol. 53:679-690. [DOI] [PubMed] [Google Scholar]
- 25.Stewart, P. S. 1993. Model of biofilm detachment. Biotechnol. Bioeng. 41:111-117. [DOI] [PubMed] [Google Scholar]
- 26.Stewart, P. S., J. Rayner, F. Roe, and W. M. Rees. 2001. Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J. Appl. Microbiol. 91:525-532. [DOI] [PubMed] [Google Scholar]
- 27.Stoodley, P., S. Wilson, L. Hall-Stoodley, J. D. Boyle, H. M. Lappin-Scott, and J. W. Costerton. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turakhia, M. H., K. E. Cooksey, and W. G. Characklis. 1983. Influence of a calcium-specific chelant on biofilm removal. Appl. Environ. Microbiol. 46:1236-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vats, N., and S. F. Lee. 2000. Active detachment of Streptococcus mutans cells adhered to Epon-hydroxylapatite surfaces coated with salivary proteins in vitro. Arch. Oral Biol. 45:305-314. [DOI] [PubMed] [Google Scholar]
- 31.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wentland, E. J., P. S. Stewart, C. T. Huang, and G. A. McFeters. 1996. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol. Prog. 12:316-321. [DOI] [PubMed] [Google Scholar]
- 34.Werner, E. M., F. Roe, A. Bugnicourt, M. Franklin, A. Heydorn, S. Molin, B. Pitts, and P. S. Stewart. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, S., M. A. Hamilton, G. C. Hamilton, M. R. Schumann, and P. Stoodley. 2004. Statistical quantification of detachment rates and size distributions of cell clumps from wild-type (PAO1) and cell signaling mutant (JP1) Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:5847-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]