Abstract
The organisms of a bluish-green layer beneath the shards of a gypsum rock were characterized by molecular techniques. The cyanobacterial consortium consisted almost exclusively of Chroococcidiopsis spp. The organisms of the shards expressed nitrogenase activity (C2H2 reduction) aerobically and in light. After a prolonged period of drought at the rock, the cells were inactive, but they resumed nitrogenase activity 2 to 3 days after the addition of water. In a suspension culture of Chroococcidiopsis sp. strain PCC7203, C2H2 reduction required microaerobic conditions and was strictly dependent on low light intensities. Sequencing of a segment of the nitrogenase reductase gene (nifH) indicated that Chroococcidiopsis possesses the alternative molybdenum nitrogenase 2, expressed in Anabaena variabilis only under reduced O2 tensions, rather than the widespread, common molybdenum nitrogenase. The shards apparently provide microsites with reduced light intensities and reduced O2 tension that allow N2 fixation to proceed in the unicellular Chroococcidiopsis at the gypsum rock, unless the activity is due to minute amounts of other, very active cyanobacteria. Phylogenetic analysis of nifH sequences tends to suggest that molybdenum nitrogenase 2 is characteristic of those unicellular or filamentous, nonheterocystous cyanobacteria fixing N2 under microaerobic conditions only.
Several kinds of nitrogenases occur in prokaryotes (6, 20). In addition to the long-known conventional molybdenum-containing nitrogenase, the N2-fixing bacterium Azotobacter vinelandii can express an enzyme with vanadium in the prosthetic group when grown in a medium lacking Mo. When both Mo and V are lacking in the culture medium, A. vinelandii synthesizes a nitrogenase with Fe as the only metal in its active center. The distribution of alternative nitrogenases in microorganisms is limited. In general, alternative nitrogenases have been observed only in laboratory cultures, and their ecological role in natural habitats is unknown.
With regard to cyanobacteria, the occurrence of a vanadium nitrogenase has been inferred from both physiological and gene characterization studies of the heterocystous cyanobacterium Anabaena variabilis (15, 36). Circumstantial evidence suggests that A. variabilis also possesses an iron nitrogenase (16), but this has not yet been corroborated by molecular data. A vanadium nitrogenase has also been detected only in Anabaena isolates from the water fern Azolla and in Nostoc sp. strain strain PCC6720 (Anabaenopsis circularis) (36). In addition, cyanobacteria may possess two sets of genetically different Mo-containing nitrogenases, a unique feature which has been clearly shown for A. variabilis (32, 37). The well-characterized conventional molybdenum nitrogenase is present in heterocysts of filamentous cyanobacteria. The other protein complex is expressed in vegetative cells and heterocysts of A. variabilis under reduced O2 tension and resembles the nitrogenase from the filamentous, nonheterocystous cyanobacterium Plectonema boryanum (34). Again, this second molybdenum nitrogenase has been observed only in laboratory-grown cultures.
This laboratory screened several unusual habitats in Central Europe for the occurrence of nitrogenase types. The rock Sachsenstein at Bad Sachsa in the southern part of the German Harz mountains was one of the sites examined. It is composed of more than 90% CaSO4 (gypsum). This 40-m-high cliff surface is friable and covered with flakes or shards of different sizes and depths. Organisms occurring there have to cope with large fluctuations in water supply, huge variations in light intensity, and a minimal humus layer. Because gypsum is highly insoluble, incoming dust brought by rainfall might provide the elements for growth of the organisms. It is suspected, though not proven, that the bioavailability of nitrogen or molybdenum limits life at this site.
The Sachsenstein and other gypsum rocks in the vicinity have long been known for the occurrence of rare plants and have recently been shown to contain unusual arbuscular mycorrhizal fungi as well (19). Beneath the shards of the vegetation-free rim of the cliff, roughly 2 to 5 cm from the surface, we noticed a distinct bluish-green layer, apparently composed mainly of cyanobacteria. These and other prokaryotes could also be unusual, and it therefore seemed worthwhile to analyze the microbial composition. The N demand of the organisms under the shards could be met by N2 fixation. Consequently, the shards were analyzed for nitrogenase activity, nitrogenase genes, and 16S rRNA composition. In addition, organisms isolated from the shards were screened for possession of the different nitrogenase types.
MATERIALS AND METHODS
Location and sampling.
The gypsum rock called Sachsenstein is a cliff of 90% pure CaSO4 near Bad Sachsa at the southwest ridge of the Harz Mountains (52°04′N, 10°55′E). Soil parameters have been described previously (19). The surface of the largely vegetation-free rim of the rock is friable, and its shards show beneath an endolithic blue-green layer consisting of cyanobacteria and other organisms. Fragments of the rock (shards of approximately 2 by 2 by 1 cm), obtained by splitting off the blue-green layer largely free from uncolored rock parts, were collected by hand, eventually by breaking the shards into smaller pieces.
Isolation of strains and cultivation.
Samples from the gypsum rock were collected in October 2001, March 2002 (for isolation of cyanobacteria only), March 2003, and June 2003. Rock fragments were incubated on 1.5% agar plates supplemented with BG11 medium (28) for 10 to 14 days at 20 to 25°C and 10 μE · m−2 · s−1. Cyanobacterial strains were isolated after several passages first on agar plates and then in liquid culture with reduced light (plates and vessels wrapped in soft paper) to favor the growth of cyanobacteria and to retard that of green algae. When no growth of other bacteria was observed after incubation for 1 week on 1.5% agar plates containing 5% Luria-Bertani medium and BG11 medium, the cultures were regarded as axenic. The DNA of the new isolates was characterized for 16S ribosomal DNA and nitrogenase genes. The reference strain Chroococcidiopsis sp. strain PCC7203 (Chroococcidiopsis thermalis, PCC reference strain) was grown in batch cultures in BG11 medium at 25°C under continuous illumination with 30 μE · m−2 · s−1 and aeration with a mixture of air and CO2 (98:2, vol/vol). For C2H2 reduction, the cultures were first washed and then grown for at least 48 h in BG11 medium without nitrate. Anaerobic culture conditions were achieved by gassing with N2 for 5 h prior to the assay.
DNA isolation.
Genomic DNA of cyanobacterial liquid cultures was isolated as described in reference 35. Total DNA of the gypsum shards was isolated from 200 mg of the bluish-green material removed from the shards by using the UltraClean soil DNA kit (Mo Bio) according to the manufacturer's recommendations.
PCR and sequencing.
To identify the cyanobacteria at the Bad Sachsa stand, a sample of the bluish-green layer under the shards was transferred to agar plates containing BG11 medium and grown for 2 weeks under continuous light at 25°C. The cell material from these enrichment cultures or from isolated axenic strains was boiled for 5 min, and the lysate was used as the template for PCR. A segment of the 16S rRNA gene spanning nucleotides 359 to 1387 (Escherichia coli K-12 numbering) was amplified by using the cyanobacterium-specific primer CYA359F (24) and the universal primer 1387R (23). PCR products were cloned with the pGEM-T Easy vector system (Promega). The plasmid DNA from the clones was isolated by using standard minipreparation protocols and characterized by restriction analysis. DNAs of those clones with different restriction patterns were sequenced.
For microorganisms other than cyanobacteria, the general primer pair NifH-F-NifH-R (29), spanning nucleotides 34 to 491 of Sinorhizobium meliloti nifH (GenBank accession no. V01215), served to amplify a segment of the nitrogenase reductase gene nifH by using total DNA either from the shards or from liquid enrichment cultures of heterotrophic bacteria kept in the dark. Amplification products of Chroococcidiopsis-related strains were obtained by use of the cyanobacterial enrichment cultures described above and the primer pair HC1f (5′ TCCACTCGCTTGATGTTGCAC 3′) and HC1r (5′ CATTTCACCAGAGGTGACG 3′). These primers were designed from an internal part of the partial nifH sequence from Chroococcidiopsis sp. strain PCC7203 obtained with NifH-F and NifH-R in this work. Hot-start PCR was performed with the following protocol for the 16S rRNA gene: cycles 1 and 2, 20 s at 94°C, 30 s at 66°C, and 30 s at 72°C; cycles 3 to 5, 20 s at 94°C, 30 s at 64°C, and 35 s at 72°C; cycles 6 to 9, 20 s at 94°C, 30 s at 62°C, and 40 s at 72°C; cycles 10 to 16, 20 s at 94°C, 30 s at 60°C, and 45 s at 72°C; cycles 17 to 40, 20 s at 94°C, 40 s at 56°C, and 60 s at 72°C; final elongation, 10 min at 72°C. The hot-start PCR protocol for nifH has been described previously (29).
Computing programs.
Sequence data were compared with the National Center for Biotechnology Information data bank by using the BLAST program (2). Sequences of the PCR products were always analyzed by editing out the primers. Alignments were done with the ClustalX program (38). Phylograms were constructed by the neighbor-joining method with 1,000 replicate trees (30), correcting for multiple substitutions by Kimura's two-parameter method (16a). These analyses were performed with the TREECON software package (39).
C2H2 reduction by the shards from the gypsum rock.
Approximately 1.2 kg of gypsum shards, distinctly colored with a bluish-green layer of cyanobacteria, were transferred immediately upon collection at the cliff site to 1-liter flasks, which were then covered with Suba-Seal septa and supplemented with 10 ml of C2H2 (corresponding to approximately 1.7% of the gas phase of the flasks). The flasks were transported to the laboratory within 4 h; they were kept either in the light or in the dark (wrapped with aluminum foil) during transportation. Upon arrival at the laboratory, they were immediately analyzed for their gas contents. They were then exposed to room temperature (20 to 25°C) and daylight, conditions as comparable to natural conditions as possible. Controls were kept in the dark. For the wetting experiment, 20 ml of distilled water was injected through the Suba-Seal septa. Reduced O2 tension in the flasks was achieved by evacuating the flasks with a pump, flushing with argon, and reinjecting 10 ml of C2H2. The C2H2 and C2H4 contents in the flasks were determined by gas chromatography using a Carbosieve SII column (100/120 mesh, 10 ft by 1/8 in.) at 210°C for the column and 275°C for both the flame ionization detector and the injector. For determination of H2 and O2 contents, a gas chromatograph equipped with a molecular sieve 5A column (45/60 mesh, 2 m by 1/8 in.) and a thermal conductivity detector were used, as described previously (15). Temperatures were 120°C for the column and 150°C for both detector and injector. Quantitative results were obtained by relating the peak heights to a standard curve. Addition of standards for H2, O2, C2H2, or C2H4 made certain that the right gases had been detected.
C2H2 reduction of isolated cultures.
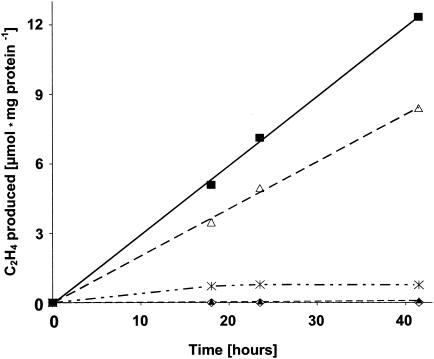
Microaerobically grown Chroococcidiopsis sp. strain PCC7203 was concentrated by centrifugation at low speed (3,000 × g) at room temperature. Cells with a content of about 0.2 mg of protein, determined by the Lowry method (21a), were anaerobically assayed in 7-ml Fernbach flasks covered with Suba-Seal septa and supplemented with 1 ml of C2H2. Gases in the flasks were removed or injected through the diaphragms of the Suba-Seal septa with gas-tight Hamilton syringes. In order to produce different light intensities, the flasks were incubated in a growth chamber at different distances from the light source (see the legend to Fig. 3).
FIG. 3.
C2H2 reduction by Chroococcidiopsis sp. strain PCC7203. Cells had been grown in BG11 medium but without combined nitrogen for at least 2 days and then under continuous gassing with N2 for 5 h prior to the experiment. Assays were performed in 7.0-ml Fernbach flasks containing 3 ml of a Chroococcidiopsis suspension with 0.6 mg of protein in total. The gas phase was argon-C2H2 (85:15, vol/vol). Values are averages from three different determinations. The flasks were placed at different positions from the light source. Light intensity at the different positions was 300 (⋄), 70 (✻), or 30 (▪) μE · m−2 · s−1; for Fernbach flasks partly wrapped with a layer of aluminium foil, light intensity was estimated to be between 2 and 5 μE · m−2 · s−1 (▵). Fernbach flasks kept in the dark (▴) were also assayed.
Nucleotide sequence accession numbers.
Nucleotide sequence accession numbers in GenBank were as follows: for the 16S rRNA gene segments, AY422693 (Chroococcidiopsis sp. strain Bad Sachsa), AY422694 (Nostoc sp. strain Bad Sachsa), AY422695 (Gloeothece sp. strain Bad Sachsa), AY422696 (BSC-16-1), and AY422697 (BSC-16-2); for the nifH gene segments, AY422706 (Chroococcidiopsis sp. strain PCC7203), AY422698 (BS-5), AY422699 (BS-HD1), AY422700 (BS-H4), AY422701 (BS-H1), AY422702 (BS-H2), AY422703 (BS-HD5), AY422704 (BS-A4), AY422705 (BSC-2), and AJ620858 (Chroococcidiopsis sp. strain Bad Sachsa).
RESULTS
Composition of the bluish-green layer beneath the shards.
The bluish-green layer under the shards microscopically showed coccoidal and rod-like eubacteria and a few spirillum-type microorganisms. Among the few eukaryotic algae present, Chlamydomonas spp. and other undetermined species were detected. The bluish-green color of the layer was undoubtedly due to cyanobacteria, among which Chroococcidiopsis was by far the dominant genus (Table 1). Chroococcidiopsis was seen as aggregates of 16 cells (or multiples thereof), sometimes of 4 cells, and also, more abundantly, as single cells in the suspensions examined under the microscope. Baeocytes (18) were also present. Whether the larger single cells occurred naturally or were artificially created by suspending the material could not be determined. As indicated by the results of the five representative microscopic determinations given in Table 1, some samples contained Nostoc sp. (similar to Nostoc punctiforme) filaments of variable cell length and size. In suspension cultures or on plates, this cyanobacterium formed fairly large colonies (as huge globules of hundreds of filaments) which were only very occasionally seen in the samples taken directly from the gypsum rock. Additionally, at least two species of the Oscillatoriales were detected, but again, these were found only occasionally in the samples from the gypsum rock.
TABLE 1.
Occurrence of different cyanobacteria under the shards of the gypsum rock of Bad Sachsaa
| Determination no. or parameter | No. of:
|
|||
|---|---|---|---|---|
| Chroococcidiopsis colonies | Chroococcidiopsis single cells | Nostoc filaments | Oscillatorialesb filaments | |
| 1 | 3 | 0 | 0 | 0 |
| 2 | 16 | 5 | 0 | 2 |
| 3 | 2 | 0 | 0 | 3 |
| 4 | 21 | 139 | 6 | 137 |
| 5 | 32 | 25 | 1 | 6 |
| Total | 74 | 169 | 7 | 148 |
| Diam (μm) of cells | 3.5-5 | ∼3.5 | 0.5-2.5 | 0.5-5 |
| No. of cells per colony or filament | 16 or multiples of 16 | 10 (avg) | >20; smaller at the tip | |
In the samples examined, Chroococcidiopsis occurred both as aggregates consisting regularly of 16 cells and as single cells. Diameters of cells were determined microscopically. About 100 mg of the blue-green layer of organisms under the gypsum shards was suspended in 0.5 ml of H2O, and the cyanobacteria were counted in a 50-μl aliquot. The five determinations are representative of five different spots of the gypsum rock collected in October 2001. The counts in March and June 2003 (not documented) confirmed the observation that Chroococcidiopsis is absolutely the dominating cyanobacterium occurring beneath the shards. Our morphological identification of the cyanobacteria was controlled by D. Mollenhauer, Biebergemünd, Germany.
Two different types.
The composition of organisms, the cell sizes, and the lengths of filaments differed too much among shard samples (Table 1) to allow a statistical evaluation of the data. However, there was no doubt that Chroococcidiopsis prevailed almost exclusively among the cyanobacteria on the gypsum rock. Table 1 includes only those organisms which could be removed from the shards by scratching.
N2 fixation (C2H2 reduction) activities of gypsum rock shards.
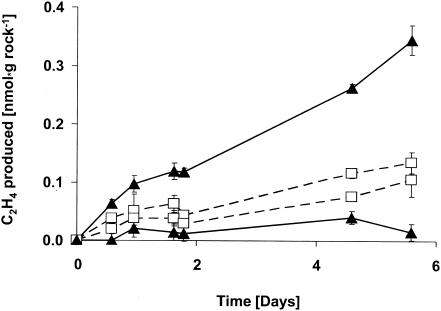
Due to their endolithic mode of life, the cyanobacteria could not be scratched off the shards without risking loss of viability. Therefore, shards from the gypsum rock were transferred to the assay flasks as a whole and exposed to C2H2 directly at the stand at Bad Sachsa in order to determine activity under conditions that were as natural as possible. The shards consisted mainly of gypsum and a very small amount (far less than 1%, by weight) of cyanobacteria. The cyanobacterial layer under the shards was heterogeneous from shard to shard and therefore had to be assayed in large flasks (1.2 kg of small shards was collected and assayed in 1-liter bottles) in order to detect C2H4 production with accuracy and to obtain an average value with rates above the detection limit. The shards with the bluish-green layer beneath reduced C2H2 to C2H4 almost linearly up to 5 days (Fig. 1). Light stimulation was often, but not consistently, seen in the experiments that were performed under laboratory conditions (daylight, natural light-dark cycles, room temperature). For example, as shown in Fig. 1, of two flasks collected a few meters apart in March 2003, C2H2 reduction was distinctly light stimulated in one but not in the other. Experiments with shards collected in October 2001 gave similar activities, and light was slightly stimulatory in two samples assayed (data not shown).
FIG. 1.
C2H2 reduction by organisms beneath shards from the gypsum rock, sampled in March 2003. Shards from the gypsum rock with a distinct bluish-green layer beneath were transferred to 1-liter flasks, covered with Suba-Seal septa, and supplemented with 10 ml of C2H2 at the field site (zero point on the abscissa of the graph). Two flasks were kept in continuous daylight (▴), whereas the other two (□) were assayed in the dark (wrapped with aluminum foil). Standard deviations (error bars) refer to different C2H4 determinations in the gas chromatograph (n = 2). Each point in this graph (and in Fig. 2 and 3) reflects the sum of the gas produced from the beginning till the determination.
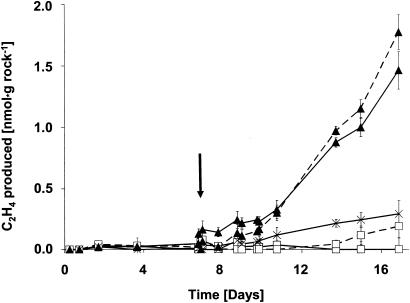
The material collected in October 2001 and March 2003 was wet, and the cyanobacterial layer was densely bluish-green, likely due to high rainfall before collection. In contrast, an unusual drought period of about 6 weeks preceded our visit to the gypsum rock in June 2003. The organisms were hardly visible then as a faint greenish layer, apparently due to their poikilohydric nature. The cyanobacteria in the greenish layer consisted almost entirely of Chroococcidiopsis (>98%). However, much smaller and less densely packed colonies were seen, and the color of the cells was not as intense as in October 2001 or March 2003. The rest consisted of Gloeothece (far less than 1%) and a few Oscillatoria filaments (also far less than 1%). The amount of other photosynthetic organisms (diatoms, green algae) was negligible. The shards collected in June 2003 and transferred to the flasks showed no C2H2 reduction in the first 7 days (Fig. 2). Distilled water was then injected through the Suba-Seal septa to moisture the shards. In flasks kept in the light, C2H2 reduction commenced after 2 to 3 days and then proceeded linearly during the following days both in air and under reduced O2 tensions (Fig. 2). Sample flasks kept in the dark did not reduce significant amounts of C2H2 but formed H2 under microaerobic conditions. This rate was approximately 60 times higher than the light-dependent C2H2 reduction activity, determined 4 days after wetting (data not shown). The flasks kept in the dark consumed O2, whether wetted or not, resulting in almost complete anaerobiosis after 2 weeks.
FIG. 2.
C2H2 reduction by organisms beneath shards from the gypsum rock after a prolonged period of drought. Shards were sampled in June 2003, after a period of drought lasting at least 6 weeks. The experimental conditions were the same as those for Fig. 1. Since no activity was obtained during the first 7 days, moisture was added to the flasks (arrow). After the 7th day, conditions in the flasks were as follows: 2% O2, natural light-dark cycle, moisture added (solid triangles with dashed lines); air, natural light-dark cycle, moisture added (solid triangles with solid lines); 2% O2, dark, moisture added (open squares with dashed lines); air, dark, moisture added (open squares with solid lines); air, natural light-dark cycle, but no moisture added (starbursts with solid lines).
N2 fixation activities determined in a laboratory culture.
The Chroococcidiopsis isolate from the gypsum rock grew poorly in liquid culture and therefore was not suitable for analysis. Consequently, the experiments were performed with the Chroococcidiopsis reference strain PCC7203. C2H2 reduction in 7.0-ml Fernbach flasks was found to be strikingly affected by the light intensity (Fig. 3). Flasks with densely packed cell suspensions showed no C2H4 formation at 300 μE · m−2 · s−1. The activity increased with decreasing light intensity. The rate amounted to about 300 nmol h−1 · mg of protein−1 at an intensity of 30 μE · m−2 · s−1 at the surface of the vessel. The light intensity within cell suspensions inside the flasks could not be determined but might have been considerably lower. C2H4 formation was lower in a flask partially wrapped with aluminium foil to achieve an estimated fivefold reduction of the light intensity. Samples kept in the dark showed almost no activity (Fig. 3). When Chroococcidiopsis sp. strain PCC7203, reducing C2H2 in dim light overnight, was exposed to high light intensities (1,800 μE · m−2 · s−1), activity immediately ceased (data not shown). The cells continuously reduced C2H2 in dim light over days. Thus, for photosynthesis and N2 fixation to proceed in Chroococcidiopsis, separation of both processes from each other by light-dark rhythms is not required.
Characterization of organisms from beneath the shards by sequencing of a 16S rRNA gene segment.
Organisms growing under shards were enriched on agar plates in BG11 medium. From this enrichment, three axenic cultures and two other different cyanobacterial clones were characterized by PCR amplification, cloning, and sequencing of a segment of the 16S rRNA gene. Because these sequences were deposited in GenBank, their position within a phylogram is not shown here. The first sequence, derived from Chroococcidiopsis sp. strain Bad Sachsa, was 95% identical, on the DNA level, to Chroococcidiopsis sp. strain BB96.1 SAG 2026 (accession no. AJ344555) and 94% identical to the well-characterized Chroococcidiopsis sp. strain 029 (4). The second sequence had the highest identities (98%) to Gloeothece membranacea PCC6501 (accession no. X78680) and the next highest (92%) to Synechocystis sp. strain PCC6702 (accession no. AB041936); thus, the corresponding axenic isolate was termed Gloeothece sp. strain Bad Sachsa. The third cyanobacterial sequence obtained belonged to the Nostoc group, with highest identities (98%) to symbiotic cyanobacteria of lichens (Nephroma and Peltigera spp.; accession no. AF506258 and AF506247, respectively) or the cycad symbiont N. punctiforme PCC73102 (AF027655). The corresponding axenic isolate was thus termed Nostoc sp. strain Bad Sachsa. The fourth and fifth sequences obtained from enrichment cultures showed the highest identities with Oscillatoriales: one (BSC-16-1) showed 92% identity to Phormidium murrayi (accession no. AF218374), and the other (BSC-16-2) showed 93% identity to Leptolyngbya sp. strain CNNP1-B3-C9 (accession no. AY239600). The results of sequencing corroborated the morphological determinations of the cyanobacteria isolated.
Determination of the nifH sequences in bacteria from shards.
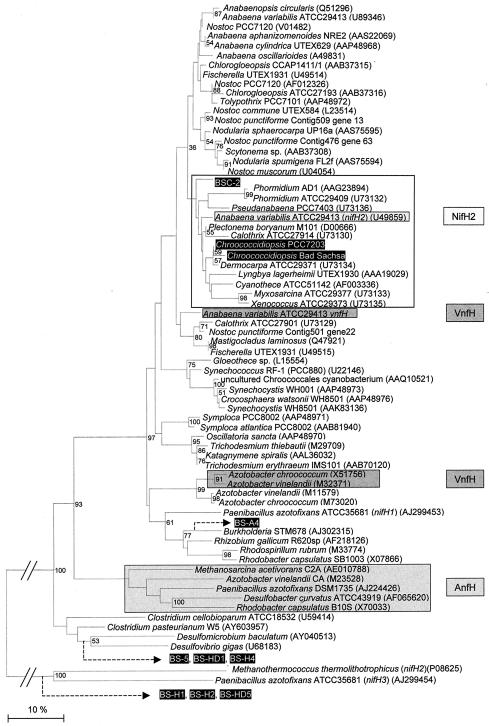
As indicated in the dendrogram of all deduced partial NifH sequences obtained from uncultured bacteria from Bad Sachsa and selected sequences from GenBank (Fig. 4), none of the sequences from Bad Sachsa is found in the literature to date. The Bad Sachsa sequences are related to nifH copies of Paenibacillus azotofixans (thus to aerobic, spore-forming bacteria) and of sulfate-reducing bacteria (Desulfomicrobium baculatum and Desulfovibrio gigas). Most cyanobacterial nitrogenase reductase sequences (Fig. 4) of the selected, well-conserved gene region cluster together and are not completely in parallel with published 16S rRNA gene dendrograms (as exemplified by the different nifH and vnfH copies of A. variabilis). The unicellular, aerobically N2 fixing organisms Synechococcus sp. strain RF-1, Synechocystis sp. strain WH8501, and Gloeothece sp. may form their own, independent cluster with respect to nifH; similarly, the sequences of the filamentous cyanobacteria Trichodesmium spp. and Symploca atlantica stand far apart from other filamentous cyanobacteria (Fig. 4). Molybdenum nitrogenase 1 is restricted to A. variabilis and other heterocystous cyanobacteria in the dendrogram. Both new cyanobacterial nifH sequences from the gypsum rock and the newly obtained sequence from Chroococcidiopsis sp. strain PCC7203 are most closely related to nifH from a Dermocarpa sp. and, together with P. boryanum nifH and A. variabilis molybdenum nitrogenase 2, form a separate cluster (Fig. 4). This cluster, though not supported by high bootstrap values, was consistently obtained by using different methods for correction and different sequences as outgroups (data not shown). The sequence of Chroococcidiopsis sp. strain Bad Sachsa, the dominant cyanobacterial species at this rock, closely matches that of Chroococcidiopsis sp. strain PCC7203. The other sequence from Bad Sachsa (BSC-2) also stands close to Chroococcidiopsis and other related unicellular strains.
FIG. 4.
Neighbor-joining tree of deduced protein sequences of a segment of nitrogenase reductase (NifH). Sequences aligned correspond to amino acid residues 48 to 152 of Nostoc sp. strain PCC7120 (accession no. AAA22008). Sequences in black boxes were determined in this study and have been deposited in GenBank. BS, Bad Sachsa; BSC, cyanobacterium from Bad Sachsa. Bar, 10% amino acid substitutions. Numbers at the branches indicate the percentages of occurrence of the respective nodes in a bootstrap analysis of 1,000 resamplings; only values greater than 50% are shown. In addition, the lower bootstrap value at the suggested NifH2 cluster is given at the respective node. The presumably nonfunctional nitrogenase sequences of Methanococcus thermolithotrophicus (P08625) and P. azotofixans (AJ299454) served as an outgroup. The positions of the black-boxed sequences BS-A4, BS-5, BS-H1, BS-H2, BS-H4, BS-HD1, and BS-HD5 have been analyzed in another tree with the same sequences, but shortened to amino acids 48 to 135 of the Nostoc sp. strain PCC7120 sequence, and are indicated by dashed arrows.
DISCUSSION
Chroococcidiopsis has been reported to dominate in extremely adverse habitats (41), for example, on the dry inselbergs of West Africa, Australia, or Venezuela (7, 9), in Arizona deserts (8), in the solar lake at the Sinai coast (25), in hot springs in Northern Thailand (13), or in the Antarctic continent at sites where the ice melts for 2 to 3 months in the summer (40). Organisms at such sites have to cope with extreme conditions, in particular with large fluctuations in the water supply, but may also be exposed to bright sunshine (ionizing radiation) and heavy winds. Chroococcidiopsis is considered to be a living fossil, is the closest living relative to heterocystous cyanobacteria, and is able to survive only under adverse conditions where the pressure from competition is low (11). Chroococcidiopsis is perhaps the most desiccation-tolerant cyanobacterium (26), and the mechanisms underlying this tolerance have recently been studied by molecular approaches (4, 5). It has only occasionally been found in Central Europe (18). Our different visits to the Sachsenstein, indeed, showed that Chroococcidiopsis and apparently other organisms as well have adapted to the adverse conditions at such a place by their poikilohydric nature and by growing in rock fissures (chasmolithic mode of life [9]) to ensure low light intensities. The 16S rRNA gene of Chroococcidiopsis sp. strain Bad Sachsa shows only 94% identity at most to Chroococcidiopsis sequences deposited into GenBank. Thus, the Bad Sachsa isolate may represent a new species. However, since a continuum of bacterial sequences exists in soils (see, for example, reference 29), sequences alone do not allow us to separate isolates into species.
The demonstration of a frequently light-stimulated C2H2 reduction activity with the shards assayed immediately after removal from the rock indicates that cyanobacterial N2 fixation plays a role at the location. Since the size and composition of the bluish-green layer beneath the shards are variable and since the activity in single shards can hardly be assayed due to the limited sensitivity of the C2H2 reduction assay, the data could not be quantified to estimate the N2 fixation rate on the rock. The wetting experiments performed with the material collected in June 2003 show that N2 fixation, like other life activities, is strictly dependent on a sufficient supply of water. This finding is in agreement with those of earlier investigations with cyanobacteria showing that drying leads to rapid cessation of nitrogenase activity, as shown for Nostoc commune (27), and that rewetting of desiccated cyanobacteria causes a rapid recovery of N2 fixation both under laboratory culture conditions (31) and at field sites (10, 14, 21, 22).
Since the overwhelming majority of cyanobacterial cell types was Chroococcidiopsis, it is likely but not proven that this cyanobacterium is the main contributor to the C2H2 reduction activity. Different isolates of Chroococcidiopsis have been reported to fix N2, but only under microaerobic laboratory conditions (28, 33). In contrast, C2H2 reduction by the shards proceeded at similar rates in air and under reduced O2 tensions. The possibility that the activity was due to very active (perhaps heterocystous) cyanobacteria occurring in minute amounts in the bluish-green layer cannot be excluded (though we think it unlikely). A high aerobic rate of N2 fixation by unicellular cyanobacteria is also unlikely to occur at this site, since only a few such strains have been reported to fix N2 aerobically (3). Among these, a cyanobacterium related to G. membranacea was detected under the shards, but only occasionally. Thus, microaerobic sites under the shards might allow N2 fixation to proceed. This requires O2 protection mechanisms such as respiratory protection, protection by the oxyhydrogen reactions, or others.
The maximal C2H2 reduction rate determined for Chroococcidiopsis sp. strain PCC7203 was about 300 nmol/h per mg of protein and thus in the range of that observed for the heterocystous cyanobacterium A. variabilis or Anabaena cylindrica (15, 16). The extreme dependence on a narrow range of low light intensities is a peculiar feature of Chroococcidiopsis (1) (this study). If this is true for Chroococcidiopsis of the gypsum rock also, then the strict dependence on the low intensity may explain the inconsistency of the light-stimulatory effect on C2H2 reduction by shards from the gypsum rock. Shards that showed a distinct bluish-green layer differed visibly in size and color from each other. Differences in the size and thickness of the shards from flask to flask may have caused subtle changes in the light intensity to which the Chroococcidiopsis cells were exposed. Net C2H4 production by the shards may have been the result of several highly variable factors such as moisture content or even C2H4 consumption, as is known for some microorganisms (12, 17).
The hypothesis of a demand for O2 protection devices for the Chroococcidiopsis nitrogenase also arises from the nifH sequencing data utilizing more than 300 bp, more than one-third of the gene. The deduced Chroococcidiopsis NifH sequence has higher identities to microaerobically expressed nitrogenases (A. variabilis molybdenum nitrogenase 2 and the P. boryanum enzyme) than to the aerobically expressed nitrogenases of the unicellular Synechococcus, Gloeothece, and Synechocystis strains or to A. variabilis molybdenum nitrogenase 1 (Fig. 4). Although the phylogenetic tree is not supported by high bootstrap values, we suggest a separate molybdenum nitrogenase 2 cluster formed by sequences from Dermocarpa, Chroococcidiopsis, P. boryanum, A. variabilis nifH2, Xenococcus, Myxosarcina, Phormidium, and Pseudanabaena. The relevance of this cluster is corroborated by the physiological descriptions of these cyanobacteria given by the Pasteur Culture Collection(http://www.pasteur.fr/recherche/banques/PCC/search.htm).Accordingly, Myxosarcina strain ATCC 29377 (PCC7312), Xenococcus strain ATCC 29373 (PCC7305), Leptolyngbya boryana (P. boryanum), Pseudanabaena strain PCC7403, and Phormidium strain ATCC 29409 (Leptolyngbya strain PCC7375) have been reported to fix N2 anaerobically only. This requires reduction of the O2 level to less than 1% for nitrogenase expression, a condition first described for the filamentous, nonheterocystous organism P. boryanum (34), and also a prerequisite for C2H2 reduction by Chroococcidiopsis strains to proceed. In conclusion, all the evidence suggests that Chroococcidiopsis possesses the genes coding for the second molybdenum nitrogenase. The outcome for one organism, however, is problematic: Cyanothece sp. strain ATCC 51124 has been reported to perform N2 fixation aerobically (3), but its nitrogenase clusters with molybdenum nitrogenase 2 (Fig. 4).
Apart from a report on the isolation of strains from aquatic samples and salt marshes in North Carolina (21), the occurrence of non-molybdenum nitrogenases in microorganisms has been reported only for laboratory cultures. For us, the unusual Sachsenstein cliff, consisting of about 90% gypsum and with almost no humus formation, was a candidate site for the occurrence of an alternative nitrogenase. However, only bacteria with the conventional nifH gene were isolated and enriched from the gypsum shards, and the isolates were related to sulfate-reducing bacteria (to be expected at such a CaSO4-rich rock), to Clostridium pasteurianum, or to P. azotofixans. All these might have a selective advantage under the adverse conditions on the rock by virtue of their capability to form spores.
Acknowledgments
We thank Dieter Mollenhauer, Biebergemünd, Germany, for help with the morphological determinations of the cyanobacteria and M. Geoffrey Yates of the former ARC Unit of Nitrogen Fixation of the University of Sussex, Brighton, United Kingdom, for helpful comments both on the content of this article and on the English. We are indebted to Teresa Thiel, University of Missouri, St. Louis, for providing the sequence information for the presumptive vnfH sequence of A. variabilis and to Stefanie Backhausen for expert technical assistance.
REFERENCES
- 1.Almon, A., and P. Böger. 1988. Hydrogen metabolism of the unicellular cyanobacterium Chroococcidiopsis thermalis. FEMS Microbiol. Ecol. 49:445-449. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergman, B., J. R. Gallon, A. N. Rai, and R. J. Stal. 1997. N2-fixation by non-heterocystous cyanobacteria. FEMS Microbiol. Rev. 19:139-185. [Google Scholar]
- 4.Billi, D., E. I. Friedmann, R. F. Helm, and M. Potts. 2001. Gene transfer to the desiccation-tolerant cyanobacterium Chroococcidiopsis. J. Bacteriol. 183:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billi, D., M. Grilli Caiola, L. Paolozzi, and P. Ghelardini. 1998. A method for DNA extraction from the desert cyanobacterium Chroococcidiopsis and its application to identification of ftsZ. Appl. Environ. Microbiol. 64:4053-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, P. E., and R. D. Joerger. 1990. Genetics and molecular biology of alternative nitrogen fixing systems. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41:109-125. [Google Scholar]
- 7.Büdel, B., U. Becker, G. Follmann, and K. Sterflinger. 2000. Algae, fungi and lichens on Inselbergs. Biol. Studies 146:69-90. [Google Scholar]
- 8.Büdel, B., and D. C. J. Wessels. 1991. Rock inhabiting blue-green algae/cyanobacteria from hot arid regions. Algol. Studies 64:385-398. [Google Scholar]
- 9.Büdel, W., U. Lüttge, R. Stelzer, O. Huber, and E. Median. 1994. Cyanobacteria of rocks and soils of the Orinoco lowlands and the Guayana uplands, Venezuela. Bot. Acta 107:422-431. [Google Scholar]
- 10.Davey, A., and H. J. Marchant. 1983. Seasonal variation in nitrogen fixation by Nostoc commune Vaucher at the Vestfold Hills, Antarctica. Phycologia 22:375-385. [Google Scholar]
- 11.Fewer, D., T. Friedl, and B. Büdel. 2002. Chroococcidiopsis and heterocyst-differentiating cyanobacteria are each other's closest living relatives. Mol. Phylogenet. Evol. 23:82-90. [DOI] [PubMed] [Google Scholar]
- 12.Hartmans, S., J. A. M. De Bont, and W. Harder. 1989. Microbial metabolism of short-chain unsaturated hydrocarbons. FEMS Microbiol. Rev. 63:235-264. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi, N. R., Y. Peeraponpisal, H. Nishihara, M. Ishii, Y. Igarashi, and T. Kodama. 1994. Isolation and cultivation of thermophilic cyanobacteria from hot springs in Northern Thailand. J. Ferment. Bioeng. 78:179-181. [Google Scholar]
- 14.Jones, K. 1989. Interactions between desiccation and dark nitrogen fixation in tropical Nostoc commune. New Phytol. 113:1-6. [Google Scholar]
- 15.Kentemich, T., G. Danneberg, B. Hundeshagen, and H. Bothe. 1988. Evidence for the occurrence of the alternative, vanadium-containing nitrogenase in the cyanobacterium Anabaena variabilis. FEMS Microbiol. Lett. 51:19-24. [Google Scholar]
- 16.Kentemich, T., G. Haverkamp, and H. Bothe. 1991. The expression of a third nitrogenase in the cyanobacterium Anabaena variabilis. Z. Naturforsch. Sect. C 46:217-222. [Google Scholar]
- 16a.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 17.Koene-Cottaar, F. H. M., and G. Schraa. 1998. Anaerobic reduction of ethene to ethane in an enrichment culture. FEMS Microbiol. Ecol. 25:251-256. [Google Scholar]
- 18.Komárek, J., and K. Anagnostidis. 1998. Chroococcidiopsis Geitler 1933, p. 421-426. In H. Ettl, G. Gärtner, H. Heynig, and D. Mollenhauer (ed.), Süsswasserflora von Mitteleuropa, vol. 19. Cyanoprokaryota, 1. Teil Chroococcales. Gustav Fischer, Jena, Germany. [Google Scholar]
- 19.Landwehr, M., U. Hildebrandt, P. Wilde, K. Nawrath, T. Tóth, B. Borbála, and H. Bothe. 2002. The arbuscular mycorrhizal fungus Glomus geosporum in European saline, sodic and gypsum soils. Mycorrhiza 12:199-211.- [DOI] [PubMed] [Google Scholar]
- 20.Loveless, T. M., and P. E. Bishop. 1999. Identification of genes unique to Mo-independent nitrogenase systems in diverse diazotrophs. Can. J. Microbiol. 45:312-317. [PubMed] [Google Scholar]
- 21.Loveless, T. M., J. R. Saah, and P. E. Bishop. 1999. Isolation of nitrogen-fixing bacteria containing molybdenum-independent nitrogenases from natural environments. Appl. Environ. Microbiol. 65:4223-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 22.Malam Issa, O., L. J. Stal, D. C. A. Couté, and J. Trichet. 2001. Nitrogen fixation by microbial crusts from desiccated Sahelian soils (Niger). Soil Biol. Biochem. 33:1425-1428. [Google Scholar]
- 23.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, D. Dymock, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oren, A. 2000. Salts and brine, p. 281-306. In B. A. Whitton and M. A. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 26.Potts, M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potts, M., and M. A. Bowman. 1985. Sensitivity of Nostoc commune UTEX 584 (cyanobacteria) to water stress. Arch. Microbiol. 141:51-56. [Google Scholar]
- 28.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdmann, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 29.Rösch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Scherer, S., A. Ernst, T. W. Chen, and P. Böger. 1984. Rewetting of drought-resistant blue-green algae: time course of water uptake and reappearance of respiration, photosynthesis and nitrogen fixation. Oecologia 62:418-423. [DOI] [PubMed] [Google Scholar]
- 32.Schrautemeier, B., U. Neveling, and S. Schmitz. 1995. Distinct and differentially regulated Mo-dependent nitrogen-fixing systems evolved for heterocysts and vegetative cells of Anabaena variabilis ATCC 29413; characterization of the fdxH1/2 gene regions as part of the nif1/2 cluster. Mol. Microbiol. 18:357-369. [DOI] [PubMed] [Google Scholar]
- 33.Stewart, W. D. P. 1980. Some aspects of structure and function in N2-fixing cyanobacteria. Annu. Rev. Microbiol. 34:497-536. [DOI] [PubMed] [Google Scholar]
- 34.Stewart, W. D. P., and M. Lex. 1970. Nitrogenase activity in the blue-green alga Plectonema boryanum strain 594. Arch. Microbiol. 73:250-260. [DOI] [PubMed] [Google Scholar]
- 35.Tamagnini, P., O. Troshina, F. Oxelfelt, R. Salema, and P. Lindblad. 1997. Hydrogenases in Nostoc sp. strain PCC 73102, a strain lacking a bidirectional enzyme. Appl. Environ. Microbiol. 63:1801-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiel, T. 1993. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J. Bacteriol. 175:6276-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiel, T., E. M. Lyons, J. C. Erker, and A. Ernst. 1995. A second nitrogenase in vegetative cells of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. USA 92:9358-9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van de Peer, Y., and R. de Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 40.Vincent, W. F. 2000. Cynobacterial dominance in polar regions, p. 321-340. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 41.Wynn-Williams, D. D. 2000. Cyanobacteria in deserts: life at the limit? p. 341-366. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.