Abstract
Molecular epidemiological studies suggest that microRNA polymorphisms may be associated with an increased risk of coronary heart disease (CHD). However, the results of these studies were inconsistent and inconclusive. To derive a more precise evaluation, we performed a meta-analysis focused on the associations between microRNA polymorphisms and CHD risk. PubMed, Embase, CNKI and Wanfang databases were searched. Odds ratios (ORs) with 95% confidence intervals (CIs) were applied to assess the association between microRNA-146a rs2910164, microRNA-196a2 rs11614913, microRNA-499 rs3746444 and microRNA-149 rs71428439 polymorphisms and CHD susceptibility. Heterogeneity, publication bias and sensitivity analysis were conducted to measure the robustness of our findings. A total of thirteen related studies involving 8,120 patients and 8,364 controls were analyzed. Significant associations between microRNA-146a rs2910164 polymorphism and CHD risk were observed in the total population, as well as in subgroup analysis. For microRNA-196a2 rs11614913 and microRNA-499 rs3746444, similarly increased risks were also found. In addition, no significant association was detected between microRNA-149 rs71428439 polymorphism and CHD risk. In conclusion, our meta-analyses suggest that microRNA polymorphisms may be associated with increased risk of CHD development.
Keywords: microRNA, polymorphism, coronary heart disease, meta-analysis, Pathology Section
INTRODUCTION
Coronary heart disease (CHD) has become a main cause of morbidity and mortality worldwide [1]. In 2010, approximately 7,000,000 deaths were reported globally, and in which CHD took up the largest proportion of death causes and years of life lost [2]. Traditional factors, such as hypertension, diabetes and smoking have been proven to contribute to the occurrence and progression of CHD [3–5]. However, more existed risk factors leading to CHD susceptibility need to be explored. Till now, increasing molecular epidemiological studies have revealed the important role of genetic factors in CHD, and the genetic predisposition is attracting more and more attention [6, 7].
MicroRNAs (miRNAs) are small single-stranded non-coding RNA molecules which function in the post-transcriptional regulation of gene expression [8]. Emerging evidence has indicated that the functions of miRNAs appear to be in a variety of fundamental biological processes, involving proliferation, differentiation and stress resistance [9–11]. In addition, recent studies have shown that miRNAs take part in the regulation of glucose and lipid metabolism, the proliferation of smooth muscle cells and vascular inflammation, which play important roles in the pathogenesis of CHD [12–16].
By affecting the miRNA maturation and the binding to target mRNAs, single nucleotide polymorphisms (SNPs) located in pre-microRNA (pre-miR) genes may alter the expression levels of a large number of target genes and cause the complex functional consequences [17]. Therefore, functional SNPs in miRNA genes may affect disease susceptibility. Previous studies have confirmed that four common miRNA polymorphisms (rs2910164 G>C in miR-146a, rs11614913 T>C in miR-196a2, rs3746444 A>G in miR-499 and rs71428439 A>G in miR-149) were associated with several diseases, including various cancers and autoimmune diseases [18–21]. Recently, these four SNPs were under investigation to uncover the possible genetic predisposing to CHD, but the results were inconsistent. Therefore, we conducted a meta-analysis involving all related publications to assess the association between microRNA polymorphisms and CHD risk.
RESULTS
Characteristics of studies
In total, 285 relevant publications were retrieved according to the search strategy. Firstly, we excluded 254 articles after title reviewing and duplicate screening. Then, 19 studies including 6 reviews, 12 studies not for focus polymorphisms, and 1 study without available information [22] were excluded. Finally, 12 eligible articles (13 studies) published from 2012 to 2016 were selected in the meta-analysis, including ten studies on microRNA-146a rs2910164 G>C [23–32], seven studies on microRNA-196a2 rs11614913 T>C [23, 26, 27, 29, 31, 33], six publications on microRNA-499 rs3746444 A>G [23, 25, 26, 29, 31, 33], and two studies on microRNA-149 rs71428439 A>G [29, 34], respectively. The process of study selection was shown in Figure 1. Among the retrieved articles, nine articles [23, 24, 26–30, 33, 34] were written in English and three [25, 31, 32] in Chinese. Moreover, two of the studies involved Caucasians [24, 30], and eleven of them were conducted for Asians. The distribution of genotype was consistent with HWE in all studies but one study for microRNA-146a rs2910164 [32] and two for microRNA-499 rs3746444 polymorphism [25, 31]. Detailed characteristics of included studies were shown in Table 1.
Figure 1. Flow diagram of the study selection process.
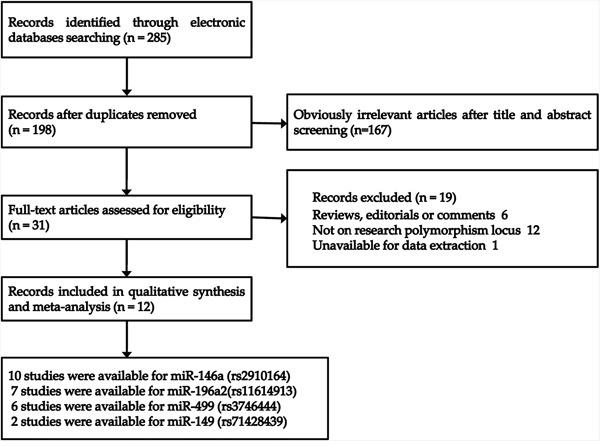
Table 1. Characteristics of case-control studies on microRNA polymorphisms and CHD risk included in the meta-analysis.
| First author | Year | Country/Region | Ethnicity | Source of controls | Case | Control | Genotype distribution | Genotyping methods | Age and sex matched | P for HWEa | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||||||||
| microRNA-146a rs2910164 G>C | CC | GC | GG | CC | GC | GG | |||||||||
| Sung JH | 2016 | Korea | Asian | Hospital | 522 | 535 | 203 | 242 | 77 | 202 | 260 | 73 | PCR-RFLP | matched | 0.460 |
| Bastami M | 2016 | Iran | Caucasian | NA | 300 | 300 | 34 | 155 | 111 | 22 | 128 | 150 | Taqman | matched | 0.454 |
| Huang SL | 2015 | China | Asian | Hospital | 722 | 721 | 266 | 308 | 143 | 237 | 348 | 132 | Taqman | matched | 0.830 |
| Xiong XD | 2014 | China | Asian | Hospital | 295 | 283 | 113 | 141 | 41 | 97 | 125 | 61 | PCR-RFLP | unmatched | 0.086 |
| Prithiksha R | 2014 | South Africa | Asian | NA | 106 | 100 | 13 | 43 | 50 | 9 | 46 | 45 | PCR-RFLP | matched | 0.569 |
| Chen CR | 2014 | China | Asian | Hospital | 919 | 889 | 187 | 463 | 269 | 153 | 435 | 301 | PCR-LDR | unmatched | 0.846 |
| Hamann L | 2014 | Germany | Caucasian | Population | 206 | 200 | 12 | 74 | 120 | 10 | 73 | 117 | PCR-HRM | unmatched | 0.748 |
| Chen L | 2013 | China | Asian | Hospital | 658 | 658 | 172 | 305 | 181 | 134 | 330 | 194 | Taqman | matched | 0.769 |
| Yang Y-a | 2012 | China | Asian | Population | 853 | 948 | 272 | 392 | 165 | 271 | 457 | 189 | Taqman | matched | 0.885 |
| Li L | 2012 | China | Asian | Hospital | 415 | 1010 | 149 | 184 | 82 | 345 | 455 | 210 | PCR-RFLP | unmatched | 0.009 |
| microRNA-196a2 rs11614913 T>C | CC | TC | TT | CC | TC | TT | |||||||||
| Sung JH | 2016 | Korea | Korean | Hospital | 522 | 535 | 107 | 236 | 179 | 108 | 274 | 153 | PCR-RFLP | matched | 0.465 |
| Huang SL | 2015 | China | Asian | Hospital | 722 | 721 | 147 | 381 | 190 | 156 | 360 | 204 | Taqman | matched | 0.905 |
| Xiong XD | 2014 | China | Asian | Hospital | 295 | 283 | 78 | 131 | 86 | 68 | 132 | 83 | PCR-RFLP | unmatched | 0.278 |
| Chen CR | 2014 | China | Asian | Hospital | 919 | 889 | 157 | 450 | 312 | 161 | 406 | 322 | PCR-LDR | unmatched | 0.097 |
| Zhi H | 2012 | China | Asian | Hospital | 916 | 584 | 155 | 470 | 291 | 98 | 278 | 208 | PCR-RFLP | matched | 0.755 |
| Yang Y-a | 2012 | China | Asian | Population | 853 | 948 | 163 | 463 | 202 | 217 | 463 | 241 | Taqman | matched | 0.853 |
| Yang Y-b | 2012 | China | Asian | Population | 1919 | 1840 | 433 | 971 | 493 | 389 | 921 | 528 | Taqman | matched | 0.734 |
| microRNA-499 rs3746444 G>A | GG | AG | AA | GG | AG | AA | |||||||||
| Sung JH | 2016 | Korea | Korean | Hospital | 522 | 535 | 9 | 155 | 358 | 13 | 168 | 354 | PCR-RFLP | matched | 0.182 |
| Xiong XD | 2014 | China | Asian | Hospital | 295 | 283 | 3 | 65 | 227 | 4 | 67 | 212 | PCR-RFLP | unmatched | 0.616 |
| Chen CR | 2014 | China | Asian | Hospital | 919 | 889 | 70 | 237 | 612 | 37 | 246 | 606 | PCR-LDR | unmatched | 0.062 |
| Chen L | 2013 | China | Asian | Hospital | 658 | 658 | 46 | 149 | 463 | 26 | 158 | 474 | Taqman | matched | 0.007 |
| Zhi H | 2012 | China | Asian | Hospital | 916 | 584 | 86 | 201 | 629 | 21 | 167 | 396 | PCR-RFLP | matched | 0.517 |
| Yang Y-a | 2012 | China | Asian | Population | 853 | 948 | 28 | 210 | 589 | 28 | 212 | 683 | Taqman | matched | 0.023 |
| microRNA-149 rs71428439 G>A | GG | AG | AA | GG | AG | AA | |||||||||
| Chen CR | 2014 | China | Asian | Hospital | 919 | 889 | 155 | 389 | 375 | 124 | 381 | 384 | PCR-LDR | unmatched | 0.062 |
| Ding SL | 2013 | China | Asian | NA | 289 | 296 | 64 | 130 | 95 | 38 | 126 | 132 | PCR-DNA sequencing | matched | 0.360 |
CHD: coronary heart disease. HWE: Hardy-Weinberg equilibrium. a HWE in control. NA: not available
Quantitative analysis
Meta-analysis for microRNA-146a rs2910164 G>C polymorphism
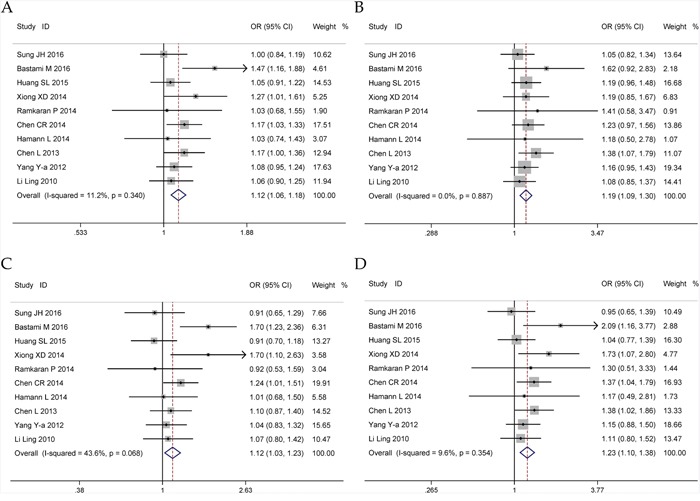
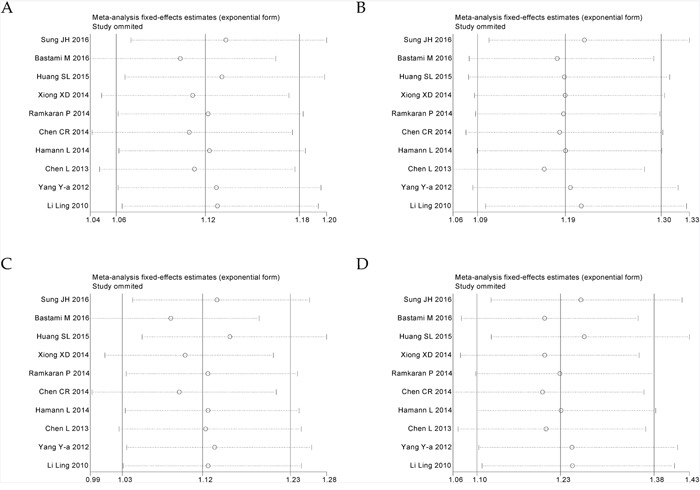
Ten eligible studies including 4,996 cases and 5,644 controls were included to assess the association between miR-146a rs2910164 polymorphism and CHD risk. The heterogeneity in all genetic models was not significant statistically (I2<0.5). So we used the fixed effect model to calculate the ORs and 95% CIs. Overall, an increased CHD risk was detected in all five genetic models (C vs. G: OR = 1.12, 95% CI = 1.06–1.18, P<0.01, I2 = 11.2%; CC vs. GG+GC: OR = 1.19, 95% CI = 1.09–1.30, P<0.01, I2 = 0%; GC + CC vs. GG: OR = 1.12, 95% CI = 1.03–1.23, P = 0.012, I2 = 43.6%; CC vs. GG: OR = 1.23, 95% CI = 1.10–1.38, P<0.01, I2 = 9.6%; GC vs. GG: OR = 1.06, 95% CI = 0.97–1.17, P = 0.211, I2 = 46.7%) (Figure 2, Table 2). Subgroup analyses of ethnicity disclosed similar results in Asians. In addition, significant associations were observed in subgroup analyses by source of controls and genotyping method (Table 2). The sensitivity analysis showed that the pooled ORs with corresponding 95%CI were not qualitatively changed by any single study in allelic, recessive, homozygous and heterozygous models, but dominant model (Figure 3). Publication bias was estimated by visual inspection of funnel plot and Egger's test, and the results revealed no asymmetrical evidence (Figure 4). The data of Egger's test supported the above results further (C vs. G: P = 0.682; CC vs. GG + GC: P = 0.283; GC + CC vs. GG: P = 0.911; CC vs. GG: P = 0.379; GC vs. GG: P = 0.877).
Figure 2. Forests for microRNA-146a rs2910164 G>C polymorphism and CHD.
A. allele model (C vs. G); B. recessive model (CC vs. GG + GC); C. dominant model (GC + CC vs. GG); D. homozygote model (CC vs. GG).
Table 2. Summary ORs and 95% CI of microRNA-146a rs2910164 polymorphisms and CHD risk.
| Locus | N* | Allele | Recessive | Dominant | Homozygote | Heterozygote | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | ||
| Total | 10 | 1.12 (1.06-1.18) <0.01 | 11.2 | 1.19 (1.09-1.30) <0.01 | 0 | 1.12 (1.03-1.23) 0.012 | 43.6 | 1.23 (1.10-1.38) <0.01 | 9.6 | 1.06 (0.97-1.17) 0.211 | 46.7 |
| Ethnicity | |||||||||||
| Asian | 8 | 1.10 (1.04-1.17) <0.01 | 0 | 1.18 (1.08-1.29) <0.01 | 0 | 1.09 (0.99-1.20) 0.083 | 22.6 | 1.20 (1.07-1.35) <0.01 | 0 | 1.03(0.93-1.14) 0.631 | 30.5 |
| Caucasian | 2 | 1.25 (0.88-1.77) 0.205 | 66.5 | 1.47 (0.92-2.35) 0.108 | 0 | 1.33 (0.80-2.21) 0.277 | 75.0 | 1.74 (1.07-2.84) 0.025 | 13.4 | 1.29 (0.79-2.11) 0.312 | 70.9 |
| Source of controls | |||||||||||
| Population | 2 | 1.08 (0.95-1.22) 0.247 | 0 | 1.17 (0.96-1.42) 0.130 | 0 | 1.04 (0.85-1.27) 0.733 | 0 | 1.15 (0.89-1.49) 0.281 | 0 | 0.98 (0.80-1.22) 0.883 | 0 |
| Hospital | 6 | 1.11 (1.04-1.19) <0.01 | 0 | 1.18 (1.07-1.31) <0.01 | 0 | 1.11 (0.99-1.24) 0.067 | 40.9 | 1.22 (1.07-1.39) <0.01 | 23.8 | 1.04 (0.93-1.17) 0.470 | 47.0 |
| Method | |||||||||||
| Taqman | 4 | 1.15 (1.03-1.29) 0.017 | 50.0 | 1.24 (1.10-1.41) <0.01 | 0 | 1.14 (0.90-1.43) 0.281 | 67.3 | 1.23 (1.05-1.44) 0.012 | 41.9 | 1.05 (0.82-1.36) 0.698 | 69.9 |
| PCR-RFLP | 4 | 1.07 (0.97-1.19) 0.177 | 0 | 1.10 (0.94-1.28) 0.227 | 0 | 1.09 (0.91-1.31) 0.347 | 45.0 | 1.16 (0.94-1.43) 0.165 | 22.9 | 1.05 (0.87-1.28) 0.598 | 44.2 |
| Age and sex matched | 6 | 1.11 (1.03-1.19) <0.01 | 35.3 | 1.21 (1.08-1.35) <0.01 | 0 | 1.08 (0.90-1.29) 0.420 | 52.7 | 1.18 (1.02-1.37) 0.024 | 25.0 | 1.00 (0.83-1.22) 0.981 | 54.1 |
| Controls in HWE | 9 | 1.13 (1.06-1.20) <0.01 | 17.0 | 1.21 (1.10-1.33) <0.01 | 0 | 1.13 (1.03-1.25) 0.012 | 49.4 | 1.25 (1.11-1.41) <0.01 | 15.7 | 1.07 (0.92-1.25) 0.396 | 52.5 |
* Numbers of comparisons. PCR-RFLP: polymerase chain reaction-based restriction fragment length polymorphism. HWE: Hardy-Weinberg equilibrium.
Figure 3. Sensitivity analyses for microRNA-146a rs2910164 G>C polymorphism and CHD.
A. allele model (C vs. G); B. recessive model (CC vs. GG + GC); C. dominant model (GC + CC vs. GG); D. homozygote model (CC vs. GG).
Figure 4. Funnel plots for microRNA-146a rs2910164 G>C polymorphism and CHD.
A. allele model (C vs. G); B. recessive model (CC vs. GG + GC); C. dominant model (GC + CC vs. GG); D. homozygote model (CC vs. GG).
Meta-analysis for microRNA-196a2 rs11614913 T>C polymorphism
Seven original studies involving 6,668 cases and 6,335 controls were analyzed for miRNA-196a2 rs11614913 T>C polymorphism and CHD susceptibility. In the overall analysis, significant associations were found in the dominant model (TC + CC vs. TT: OR = 1.08, 95%CI = 1.00–1.17, P = 0.046, I2= 27.3%) and heterozygous model (TC vs. TT: OR = 1.10, 95%CI = 1.01–1.19, P = 0.029, I2= 40%) (Figure 5, Table 3). In the stratified analysis, significant results were observed in group with population-based controls as well as genotyping method of Taqman (Table 3). Publication bias analyses were performed, and the shapes of funnel plots (Supplementary Figure 1) were consistent with the Egger's test approved (C vs. T: P = 0.262; CC vs. TT + TC: P = 0.650; TC + CC vs. TT: P = 0.226; CC vs. TT: P = 0.220; TC vs. TT: P = 0.292). However, when sensitivity analysis was performed, some changes of the pooled ORs were detected under both dominant and heterozygous models (Supplementary Figure 2).
Figure 5. Forests for microRNA-196a2 rs11614913 T>C polymorphism and CHD.
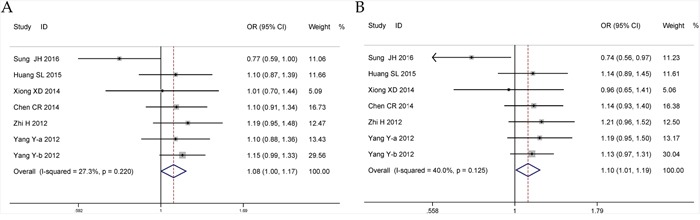
A. dominant model (TC + CC vs. TT); B. heterozygote model (TC vs. TT).
Table 3. Summary ORs and 95% CI of microRNA-196a2 rs11614913 polymorphisms and CHD risk.
| Locus | N* | Allele | Recessive | Dominant | Homozygote | Heterozygote | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | ||
| Total | 7 | 1.03 (0.98-1.09) 0.252 | 0 | 0.99 (0.90-1.08) 0.801 | 6.0 | 1.08 (1.00-1.17) 0.046 | 27.3 | 1.05 (0.95-1.16) 0.370 | 0 | 1.10 (1.01-1.19) 0.029 | 40.0 |
| Source of controls | |||||||||||
| Population | 2 | 1.03 (0.91-1.17) 0.615 | 60.3 | 0.95 (0.69-1.30) 0.736 | 81.3 | 1.13 (1.00-1.28) 0.042 | 0 | 1.05 (0.80-1.39) 0.711 | 64.8 | 1.15 (1.01-1.30) 0.032 | 0 |
| Hospital | 5 | 1.02 (0.95-1.09) 0.671 | 0 | 0.98 (0.87-1.11) 0.793 | 0 | 1.05 (0.94-1.17) 0.389 | 44.8 | 1.01 (0.88-1.17) 0.863 | 0 | 1.04 (0.87-1.24) 0.666 | 55.4 |
| Method | |||||||||||
| Taqman | 3 | 1.04 (0.97-1.11) 0.242 | 25.3 | 0.95 (0.78-1.16) 0.604 | 63.9 | 1.13 (1.01-1.25) 0.030 | 0 | 1.08 (0.94-1.23) 0.299 | 34.2 | 1.15 (1.02-1.28) 0.017 | 0 |
| PCR-RFLP | 3 | 1.01 (0.91-1.12) 0.847 | 30.4 | 1.04 (0.87-1.25) 0.661 | 0 | 0.98 (0.74-1.29) 0.867 | 68.5 | 1.02 (0.83-1.25) 0.874 | 0 | 0.95 (0.69-1.32) 0.775 | 72.4 |
| Age and sex matched | 5 | 1.03 (0.97-1.09) 0.315 | 30.4 | 0.99 (0.90-1.09) 0.825 | 28.9 | 1.07 (0.93-1.22) 0.342 | 50.4 | 1.05 (0.94-1.18) 0.387 | 18.0 | 1.08 (0.93-1.26) 0.316 | 57.3 |
* Numbers of comparisons. PCR-RFLP: polymerase chain reaction-based restriction fragment length polymorphism.
Meta-analysis for microRNA-499 rs3746444 A>G polymorphism
Six relevant studies comprising 4,163 patients and 3,897 controls were included in the meta-analysis for miRNA-499 rs3746444 A>G polymorphism and CHD risk. The pooled analyses indicated that this polymorphism was associated with an increased risk of CHD in three genetic models (G vs. A: OR = 1.11, 95% CI = 1.02–1.20, P = 0.015, I2 = 17.8%; GG vs. AA + AG: OR = 1.55, 95% CI = 1.07–2.27, P = 0.022, I2= 58.1%; GG vs. AA: OR = 1.54, 95% CI = 1.08–2.20, P = 0.017, I2= 52.6%) (Figure 6, Table 4). Subsequent subgroup analyses revealed similar results in the hospital-based control group, genotyping method of Taqman group as well as age and sex matched group (Table 4). No significant publication bias was found, indicating that the meta-analysis results are reliable (G vs. A: P = 0.092; GG vs. AA + AG: P = 0.156; AG + GG vs. AA: P = 0.182; GG vs. AA: P = 0.198; AG vs. AA: P = 0.821) (Supplementary Figure 3). However, further sensitivity analysis revealed that omission of each study made some significant differences on the findings (Supplementary Figure 4).
Figure 6. Forests for microRNA-499 rs3746444 A>G polymorphism and CHD.
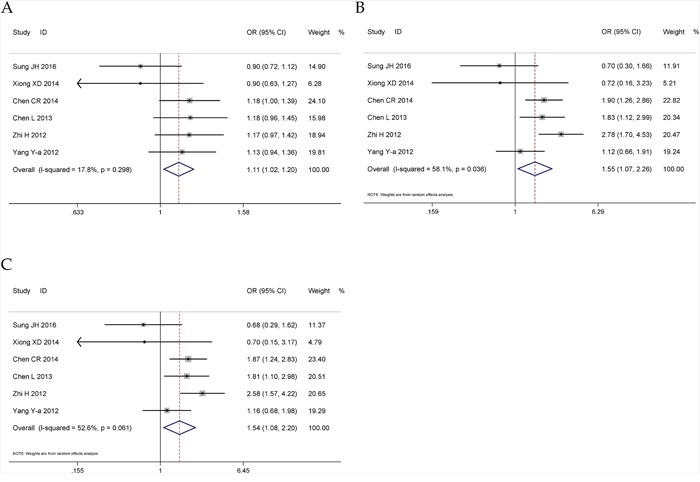
A. allele model (G vs. A); B. recessive model (GG vs. AA + AG); C. homozygote model (GG vs. AA).
Table 4. Summary ORs and 95% CI of microRNA-499 rs3746444 polymorphisms and CHD risk.
| Locus | N* | Allele | Recessive | Dominant | Homozygote | Heterozygote | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | ||
| Total | 6 | 1.11 (1.02-1.20) 0.015 | 17.8 | 1.55 (1.07-2.27) 0.022 | 58.1 | 1.03 (0.94-1.13) 0.545 | 0 | 1.54 (1.08-2.20) 0.017 | 52.6 | 0.95 (0.85-1.05) 0.275 | 21.8 |
| Source of controls | |||||||||||
| Population | 1 | 1.13 (0.94-1.36) 0.199 | NA | 1.12 (0.66-1.91) 0.676 | NA | 1.15 (0.93-1.42) 0.193 | NA | 1.16 (0.68-1.98) 0.588 | NA | 1.15 (0.92-1.43) 0.218 | NA |
| Hospital | 5 | 1.10 (1.01-1.21) 0.039 | 33.7 | 1.68 (1.11-2.55) 0.014 | 55.9 | 1.00 (0.90-1.12) 0.990 | 0 | 1.65 (1.10-2.46) 0.015 | 52.4 | 0.90 (0.80-1.01) 0.061 | 0 |
| Method | |||||||||||
| Taqman | 2 | 1.15 (1.01-1.32) 0.042 | 0 | 1.46 (1.02-2.10) 0.038 | 42.7 | 1.12 (0.96-1.31) 0.156 | 0 | 1.48 (1.03-2.12) 0.034 | 30.1 | 1.07 (0.90-1.26) 0.446 | 0.8 |
| PCR-RFLP | 3 | 1.03 (0.90-1.17) 0.693 | 48.8 | 1.24 (0.42-3.66) 0.693 | 77.9 | 0.93 (0.79-1.08) 0.330 | 0 | 1.20 (0.42-3.39) 0.737 | 76.1 | 0.84 (0.71-0.99) 0.033 | 0 |
| Age and sex matched | 4 | 1.10 (1.00-1.22) 0.052 | 27.5 | 1.51 (0.89-2.57) 0.127 | 71.1 | 1.03 (0.92-1.15) 0.631 | 0 | 1.49 (0.91-2.45) 0.113 | 66.5 | 0.94 (0.79-1.12) 0.493 | 52.7 |
| Controls in HWE | 4 | 1.08 (0.98-1.20) 0.132 | 45.2 | 1.55 (0.86-2.79) 0.144 | 66.9 | 0.98 (0.87-1.11) 0.744 | 0 | 1.51 (0.85-2.66) 0.159 | 64.3 | 0.88 (0.77-1.00) 0.050 | 0 |
* Numbers of comparisons. PCR-RFLP: polymerase chain reaction-based restriction fragment length polymorphism. HWE: Hardy-Weinberg equilibrium. NA: not available.
Meta-analysis for microRNA-149 rs71428439 A>G polymorphism
A total of 2 studies with 1,208 cases and 1,185 controls were selected in the meta-analysis. This polymorphism was not found to be significantly associated with CHD risk in all five models (G vs. A: OR = 1.30, 95% CI = 0.94–1.79, P = 0.107, I2 = 82.1%; GG vs. AA+AG: OR = 1.50, 95% CI = 0.98–2.27, P = 0.059, I2 = 64.2%; AG + GG vs. AA: OR = 1.31, 95% CI = 0.89–1.93, P = 0.169, I2 = 75.8%; GG vs. AA: OR = 1.67, 95% CI = 0.93–3.01, P = 0.086, I2 = 78.1%; AG vs. AA: OR = 1.18, 95% CI = 0.87–1.59, P = 0.281, I2 = 55.6%).
DISCUSSION
Coronary heart disease is the most common cause of morbidity and mortality in most regions worldwide. Although we have conducted some major advances in the understanding of cardiovascular disease in more recent decades, detailed pathogenesis of CHD remain to be explored. Nowadays, the association between polymorphisms of microRNAs and CHD risk is drawing more and more attention.
In the current meta-analysis, we comprehensively investigated the associations between microRNA-146a rs2910164 G>C, microRNA-196a2 rs11614913 T>C, microRNA-499 rs3746444 A>G and microRNA-149 rs71428439 A>G polymorphisms and CHD risk according to thirteen included case-control studies, consisting of 8,120 patients and 8,364 controls. Overall, significant increased risks of CHD were observed for microRNA-146a rs2910164, microRNA-196a2 rs11614913 and microRNA-499 rs3746444, but not miRNA-149 rs71428439.
As for miRNA-146a rs2910164 G>C, this is the latest and largest meta-analysis investigated the association with CHD risk. Compared with the previous meta-analysis with four studies including 2506 subjects [35], we found that the significant association existed in recessive model, as well as no association in heterozygous model. The advantages of our analysis are as follows. First, our meta-analysis had much larger sample size: we added another six recent studies involving 8,134 subjects which were not part of the previous meta-analysis [23, 24, 27, 29, 31, 32]. Second, sensitivity analyses showed that our results were statistically robust in four genetic models. Also, no significant publication bias was detected in our meta-analysis. Third, we performed a more comprehensive subgroup analyses. Stratification by ethnicity showed an increased CHD risk for microRNA-146a rs2910164 G>C polymorphism in Asians. Furthermore, similar increased results were observed in the group with genotyping method of Taqman, rather than PCR-RFLP. It revealed that Taqman was a more useful genotyping method to improve the accuracy of experiment.
To the best of our knowledge, this is the first meta-analysis assessing the association of miRNA-196a2 rs11614913 T>C, miRNA-499 rs3746444 A>G, and miRNA-149 rs71428439 A>G polymorphisms with CHD susceptibility. Interestingly, by increasing the sample size, the results of the combined analysis revealed a significant association with CHD risk for microRNA-196a2 rs11614913, even though no association was found in each single original study. How can we explain the association of miRNA-196a2 rs11614913 with CHD susceptibility? First, the miRNA-196a2 rs11614913 polymorphism involved a T to C nucleotide substitution and situated in the 3p strand of mature miRNA regions, which might affect both pre-miRNA maturation of 5p and 3p miRNAs and the interacting of target mRNAs to 3p mature miRNAs [36]. Second, it has been reported that miR-196a2 was closely associated with the regulation of annexin A1 (ANXA1) [37]. As an important modulator in atherosclerosis, ANXA1 can inhibit not only the monocyte adhesion to endothelium but also the expression of inflammatory enzymes, such as inducible cyclooxygenase 2 (COX-2) and phospholipase A2 [38, 39]. Additionally, the predicted targets of miR-196a2 included hundreds of genes ( http://www.targetscan.org). There also existed the possibility that other targets of miR-196a2 might play some roles in the development of CHD, despite it was unknown by far.
Our meta-analysis had several limitations. First of all, the ethnicity of most subjects was Asian in the current study and this restricted the general application of the results to other populations. Second, only articles published in English or Chinese were selected, potentially causing a language bias. Third, in the sensitivity analysis for miRNA-196a2 rs11614913 T>C and miRNA-499 rs3746444 A>G, we found that omission of each study made some significant differences on the results. Although it may be explained by the small number of studies included, the caution should be indicated when interpreting the association of these two miRNA polymorphisms with CHD. Third, the heterogeneity existed in our meta-analysis for miRNA-499 rs3746444 A>G and microRNA-149 rs71428439 A>G. For rs3746444, although subgroup and sensitivity analyses were performed, unfortunately, we have not found the sources of heterogeneity. Also, as for rs71428439, only two included studies were too small to analyze the sources of heterogeneity. Fourth, CHD is both multi-factorial disease influenced by genetic and environmental factors. However, in our current meta-analysis, the inter-gene and gene-environment interactions could not be conducted owing to the data deficiency. Last but not the least, genetic epidemiological studies show different genetic variants can predispose to different subtypes of CHD [40–42]. So subtypes of CHD, such as myocardial infarction, acute coronary syndrome and stable angina should be further analyzed. Unfortunately, we could not assess the difference among these subtypes of CHD due to insufficient statistical data in the literature.
In conclusion, the current meta-analysis demonstrated that three functional polymorphisms of microRNA-146a rs2910164 G>C, microRNA-196a2 rs11614913 T>C and microRNA-499 rs3746444 A>G might take important part in the development of CHD. Considering the limitations in the current meta-analysis, our results should be interpreted with caution. More eligible studies with rigorous design are needed to confirm the association of above polymorphisms in miRNA and CHD risk in the future.
MATERIALS AND METHODS
Search strategy
We searched four electronic databases (Pubmed, Embase, CNKI and Wanfang) for articles written in English or Chinese published prior to August 31, 2016. The following medical subject heading terms were used: (microRNA OR miRNA) AND (myocardial infarction OR ischemic heart disease OR ischaemic heart disease OR coronary heart disease OR coronary artery disease OR coronary syndrome OR coronary stenosis OR coronary disease OR cardiovascular disease OR CAD OR CHD OR MI) AND (genotype OR gene OR allele OR polymorphism OR variant OR SNP).
Study selection
All selected studies had to meet the following criteria: (1) published studies based on case-control design assessing the association of rs2910164 G>C in miR-146a, rs11614913 T>C in miR-196a2, rs3746444 A>G in miR-499 and rs71428439 A>G in miR-149 with CHD risk; (2) availability of allele or genotype frequency for calculating odds radio (OR) and their 95% confidence interval (CI). Studies were excluded if they investigated the progression, severity, phenotype modification, response to treatment, survival or family based studies. Moreover, meeting abstracts, case reports, editorials, review articles and non-English and non-Chinese articles were also excluded. For duplicate publications, the one with more complete design or larger sample size was finally selected.
Data extraction
The two of the authors independently extracted the data from each relevant study including the first author, publication year, study country/region, ethnicity of participants (such as Asian or Caucasian), sources of controls, genotyping method, case-control matched status, HWE status of controls and number of genotypes in CHD cases and controls. Disagreements were reconciled through group discussion. The Hardy-Weinberg equilibrium (HWE) was calculated based on the genotypes of the controls.
Statistical analysis
Heterogeneity among studies was examined with the I2 statistic and I2>50% indicates significant heterogeneity between the studies. Based on the test of heterogeneity, a pooled OR was calculated by using fixed or random effect model, along with the 95% CI to measure the strength of the genetic association. For the microRNA-146a rs2910164 G>C polymorphism, the pooled ORs were obtained for the allele contrast (C vs. G), recessive model (CC vs. GG+GC), dominant model (GC+CC vs. GG), homozygous (co-dominant) model (CC vs. GG) and heterozygous (co-dominant) model (GC vs. GG). Similar genetic models were also assessed for microRNA-196a2 rs11614913 T>C, microRNA-499 rs3746444 A>G and microRNA-149 rs71428439 A>G variants. Subgroup analyses of ethnicity, source of controls, genotyping methods, case-control matched status and HWE status of controls were also submitted to statistical testing. In order to evaluate the stability of the results, sensitivity analysis was used, which meant omitting one study at a time, and then compared to show whether a significant difference existed between the former and the latter results. Publication bias was examined by the visual inspection of funnel plot, and Egger's regression test. Data were analyzed and processed using Stata 12.0 (Stata Corporation, College Station, TX, USA). P<0.05 was considered statistically significant.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Footnotes
CONFLICTS OF INTEREST
No conflicts of interest were disclosed.
GRANT SUPPORT
This work was supported by the National Natural Science Foundation of China (Grant No. 81302127, 81400950) and the Joint Specialized Research Fund for the Doctoral Program of Higher Education in China (Grant No. 20132104120018).
REFERENCES
- 1.Newton JN, Briggs AD, Murray CJ, Dicker D, Foreman KJ, Wang H, Naghavi M, Forouzanfar MH, Ohno SL, Barber RM, Vos T, Stanaway JD, Schmidt JC, Hughes AJ, Fay DF, Ecob R, et al. Changes in health in England, with analysis by English regions and areas of deprivation, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:2257–2274. doi: 10.1016/S0140-6736(15)00195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Opadijo OG, Akande AA, Jimoh AK. Prevalence of coronary heart disease risk factors in Nigerians with systemic hypertension. African journal of medicine and medical sciences. 2004;33:121–125. [PubMed] [Google Scholar]
- 4.Fharm E, Cederholm J, Eliasson B, Gudbjornsdottir S, Rolandsson O. Time trends in absolute and modifiable coronary heart disease risk in patients with Type 2 diabetes in the Swedish National Diabetes Register (NDR) 2003-2008. Diabetic medicine. 2012;29:198–206. doi: 10.1111/j.1464-5491.2011.03425.x. [DOI] [PubMed] [Google Scholar]
- 5.Cullen P, Schulte H, Assmann G. Smoking, lipoproteins and coronary heart disease risk. Data from the Munster Heart Study (PROCAM). European heart journal. 1998;19:1632–1641. doi: 10.1053/euhj.1998.1086. [DOI] [PubMed] [Google Scholar]
- 6.Han X, Gui L, Liu B, Wang J, Li Y, Dai X, Li J, Yang B, Qiu G, Feng J, Zhang X, Wu T, He M. Associations of the uric acid related genetic variants in SLC2A9 and ABCG2 loci with coronary heart disease risk. BMC genetics. 2015;16:4. doi: 10.1186/s12863-015-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, Boer JM, Barsova RM, Favorova O, Goel A, Muller M, Feskens EJ, Consortium PC. TGFB1 genetic polymorphisms and coronary heart disease risk: a meta-analysis. BMC medical genetics. 2012;13:39. doi: 10.1186/1471-2350-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.O'Driscoll L. The emerging world of microRNAs. Anticancer research. 2006;26:4271–4278. [PubMed] [Google Scholar]
- 11.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 12.Madrigal-Matute J, Rotllan N, Aranda JF, Fernandez-Hernando C. MicroRNAs and atherosclerosis. Current atherosclerosis reports. 2013;15:322. doi: 10.1007/s11883-013-0322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santovito D, Mezzetti A, Cipollone F. MicroRNAs and atherosclerosis: new actors for an old movie. Nutrition, metabolism, and cardiovascular diseases. 2012;22:937–943. doi: 10.1016/j.numecd.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. The New England journal of medicine. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 15.Canoui-Poitrine F, Luc G, Mallat Z, Machez E, Bingham A, Ferrieres J, Ruidavets JB, Montaye M, Yarnell J, Haas B, Arveiler D, Morange P, Kee F, Evans A, Amouyel P, Ducimetiere P, et al. Systemic chemokine levels, coronary heart disease, and ischemic stroke events: the PRIME study. Neurology. 2011;77:1165–1173. doi: 10.1212/WNL.0b013e31822dc7c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers KC, Rye KA, Tabet F. MicroRNAs in the onset and development of cardiovascular disease. Clinical science. 2014;126:183–194. doi: 10.1042/CS20130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nature reviews Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, Jin M, Zhang M, Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PloS one. 2012;7:e30585. doi: 10.1371/journal.pone.0030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Tu Y, Liu L, Qi J, He L. MicroRNA-499 rs3746444 polymorphism and autoimmune diseases risk: a meta-analysis. Molecular diagnosis & therapy. 2014;18:237–242. doi: 10.1007/s40291-013-0073-0. [DOI] [PubMed] [Google Scholar]
- 21.Ma F, Zhang P, Lin D, Yu D, Yuan P, Wang J, Fan Y, Xu B. There is no association between microRNA gene polymorphisms and risk of triple negative breast cancer in a Chinese Han population. PloS one. 2013;8:e60195. doi: 10.1371/journal.pone.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buraczynska M, Zukowski P, Wacinski P, Ksiazek K, Zaluska W. Polymorphism in microRNA-196a2 contributes to the risk of cardiovascular disease in type 2 diabetes patients. Journal of diabetes and its complications. 2014;28:617–620. doi: 10.1016/j.jdiacomp.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Sung JH, Kim SH, Yang WI, Kim WJ, Moon JY, Kim IJ, Cha DH, Cho SY, Kim JO, Kim KA, Kim OJ, Lim SW, Kim NK. miRNA polymorphisms (miR146a, miR149, miR196a2 and miR499) are associated with the risk of coronary artery disease. Molecular medicine reports. 2016;14:2328–2342. doi: 10.3892/mmr.2016.5495. [DOI] [PubMed] [Google Scholar]
- 24.Bastami M, Ghaderian SM, Omrani MD, Mirfakhraie R, Vakili H, Parsa SA, Nariman-Saleh-Fam Z, Masotti A. MiRNA-Related Polymorphisms in miR-146a and TCF21 Are Associated with Increased Susceptibility to Coronary Artery Disease in an Iranian Population. Genetic testing and molecular biomarkers. 2016;20:241–248. doi: 10.1089/gtmb.2015.0253. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Wu YT. Association of genetic polymorphisms in microRNAs precursor with the risk and prognosis of coronary heart disease. Journal of Xi'an Jiaotong University (Medical Sciences) 2013;34:495–499. [Google Scholar]
- 26.Xiong XD, Cho M, Cai XP, Cheng J, Jing X, Cen JM, Liu X, Yang XL, Suh Y. A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutation research. 2014;761:15–20. doi: 10.1016/j.mrfmmm.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Huang S, Lv Z, Deng Q, Li L, Yang B, Feng J, Wu T, Zhang X, Cheng J. A Genetic Variant in Pre-miR-146a (rs2910164 C>G) Is Associated with the Decreased Risk of Acute Coronary Syndrome in a Chinese Population. The Tohoku journal of experimental medicine. 2015;237:227–233. doi: 10.1620/tjem.237.227. [DOI] [PubMed] [Google Scholar]
- 28.Ramkaran P, Khan S, Phulukdaree A, Moodley D, Chuturgoon AA. miR-146a polymorphism influences levels of miR-146a, IRAK-1, and TRAF-6 in young patients with coronary artery disease. Cell biochemistry and biophysics. 2014;68:259–266. doi: 10.1007/s12013-013-9704-7. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Hong H, Chen L, Shi X, Chen Y, Weng Q. Association of microRNA polymorphisms with the risk of myocardial infarction in a Chinese population. The Tohoku journal of experimental medicine. 2014;233:89–94. doi: 10.1620/tjem.233.89. [DOI] [PubMed] [Google Scholar]
- 30.Hamann L, Glaeser C, Schulz S, Gross M, Franke A, Nothlings U, Schumann RR. A micro RNA-146a polymorphism is associated with coronary restenosis. International journal of immunogenetics. 2014;41:393–396. doi: 10.1111/iji.12136. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y. PhD Dissertation, Peking Union Medical College; China: 2012. Common genetic variations in pre-miRNAs and the risk of coronary heart disease in a Chinese Han population. [Google Scholar]
- 32.Li L. MsD Dissertation, Central South University; China: 2010. Association of miRNA-146a polymorphism with risk of cardiovascular disease and ischemia stroke and the mechanisms. [Google Scholar]
- 33.Zhi H, Wang L, Ma G, Ye X, Yu X, Zhu Y, Zhang Y, Zhang J, Wang B. Polymorphisms of miRNAs genes are associated with the risk and prognosis of coronary artery disease. Clinical research in cardiology. 2012;101:289–296. doi: 10.1007/s00392-011-0391-3. [DOI] [PubMed] [Google Scholar]
- 34.Ding SL, Wang JX, Jiao JQ, Tu X, Wang Q, Liu F, Li Q, Gao J, Zhou QY, Gu DF, Li PF. A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the Puma protein in apoptosis. The Journal of biological chemistry. 2013;288:26865–26877. doi: 10.1074/jbc.M112.440453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao MH, Xiao Y, Zhang QS, Luo HQ, Luo J, Zhao J, Li GY, Zeng J, Li JM. Meta-Analysis of miR-146a Polymorphisms Association with Coronary Artery Diseases and Ischemic Stroke. International journal of molecular sciences. 2015;16:14305–14317. doi: 10.3390/ijms160714305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, Broaddus RR, Rashid A, Albarracin CT. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 38.Parente L, Solito E. Annexin 1: more than an anti-phospholipase protein. Inflammation research. 2004;53:125–132. doi: 10.1007/s00011-003-1235-z. [DOI] [PubMed] [Google Scholar]
- 39.Perretti M, D'Acquisto F. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nature reviews Immunology. 2009;9:62–70. doi: 10.1038/nri2470. [DOI] [PubMed] [Google Scholar]
- 40.Barbato E, Bartunek J, Mangiacapra F, Sciarretta S, Stanzione R, Delrue L, Cotugno M, Marchitti S, Iaccarino G, Sirico G, Di Castro S, Evangelista A, Lambrechts D, Sinnaeve P, De Bruyne B, Van De Werf F, et al. Influence of rs5065 atrial natriuretic peptide gene variant on coronary artery disease. Journal of the American College of Cardiology. 2012;59:1763–1770. doi: 10.1016/j.jacc.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Mokretar K, Velinov H, Postadzhiyan A, Apostolova M. Association of Polymorphisms in Endothelial Nitric Oxide Synthesis and Renin-Angiotensin-Aldosterone System with Developing of Coronary Artery Disease in Bulgarian Patients. Genetic testing and molecular biomarkers. 2015. [DOI] [PMC free article] [PubMed]
- 42.Jiang H, Wang R, Liu Y, Zhang Y, Chen ZY. BDNF Val66Met polymorphism is associated with unstable angina. Clinica chimica acta. 2009;400:3–7. doi: 10.1016/j.cca.2008.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


