Abstract
Here we compare the physiological state of Escherichia coli exposed to tellurite or selenite by using the noninvasive technique of phosphorus-31 nuclear magnetic resonance (NMR) spectroscopy. We studied glucose-fed Escherichia coli HB101 cells containing either a normal pUC8 plasmid with no tellurite resistance determinants present or the pTWT100 plasmid which contains the resistance determinants tehAB. No differences could be observed in intracellular ATP levels, the presence or absence of a transmembrane pH gradient, or the levels of phosphorylated glycolytic intermediates when resistant cells were studied by 31P NMR in the presence or absence of tellurite. In the sensitive strain, we observed that the transmembrane pH gradient was dissipated and intracellular ATP levels were rapidly depleted upon exposure to tellurite. Only the level of phosphorylated glycolytic intermediates remained the same as observed with resistant cells. Upon exposure to selenite, no differences could be observed by 31P NMR between resistant and sensitive strains, suggesting that the routes for selenite and tellurite reduction within the cells differ significantly, since only tellurite is able to collapse the transmembrane pH gradient and lower ATP levels in sensitive cells. The presence of the resistance determinant tehAB, by an as yet unidentified detoxification event, protects the cells from uncoupling by tellurite.
Tellurium (Te) and selenium (Se) are quite similar in that they are found in the same chemical group as oxygen (O), sulfur (S), and polonium (Po). Both of these elements are trace soil elements. Te and Se differ, however, in that tellurium is not considered to be an essential nutrient, whereas selenium is (26). As well, the oxidized form of Te, tellurite (TeO32−), is more toxic to most gram-negative organisms than the oxidized form of Se, selenite (SeO32−). The MIC for Escherichia coli strains lacking TeO32− resistance determinants is on the order of 1 to 2 μg/ml (29), whereas the MIC for strains lacking SeO32− resistance determinants is much higher, approximately 1,000 μg/ml (34).
Ecologically, metal compounds are disseminated in our environment through volcanic, meteorological, and anthropogenic activities. Human activity and pollution are a particular concern, as industrial effluent and mine drainage runoff create contaminated environmental niches that select for and increase the persistence of bacterial metal resistance determinants (21, 23, 24). Bacteria have developed a diverse array of strategies to counter heavy metal toxicity. These strategies include reduction or modification of the heavy metal to a less toxic species, sequestration, chelation, efflux, reduced uptake, and increased expression of cellular repair machinery (19, 23, 24, 35).
Research comparing the toxic effects of selenite and tellurite on the physiology of microorganisms was initiated 40 years ago (22), but little is known for certain about the biochemical mechanisms of toxicity (10). There is some evidence that the presence of SeO32− overwhelms intracellular antioxidant defenses, most notably the pool of reduced thiols (25). Other research has proposed that chemical oxidation events, the inactivation of proteins by the replacement of sulfhydryl groups with Te or Se, or the quenching of key respiratory chain components lies behind the toxicity of these chalcogens (12, 29, 31, 34, 35, 37). More recent studies show that significant changes in the respiratory chains of Pseudomonas pseudoalcaligenes (7) and Rhodobacter capsulatus (3) occur after exposure to TeO32− or SeO32−. It is not known whether E. coli changes its respiratory chain in the same manner.
Glutathione can reduce TeO32− and SeO32− via the Painter reaction to their elemental forms (Te0 and Se0) to form black or orange-red deposits, respectively. These deposits have been shown to be closely associated with cell membranes (32). In addition to the reduction of these oxyanions by the cellular pool of reduced thiols (36), reduction by enzymes may also be possible. In E. coli, a molybdenum-containing enzyme has been shown to have selenate reductase activity (2), and nitrate reductase has been shown to be capable of reducing both TeO32− and SeO32− (1). Examples of other organisms that reduce TeO32− via terminal reductases are numerous, particularly in the Rhodobacter spp. (16, 30, 31). NADH-dependent tellurite reductase activity has been observed in Natronococcus occultus (20) and Thermus thermophilus (4).
Overexpression of genes for the metabolism of cysteine has also been shown to mediate tellurite resistance (27). There have been a number of naturally occurring tellurite resistance determinants identified in bacteria as well (10, 29, 35). At this point, there has not been a clear biochemical connection demonstrated between the resistance determinants and the amount of Te0 and Se0 crystal formation.
Our group has concentrated its efforts on elucidating the mechanism by which the teh determinant confers tellurite resistance to E. coli. The tehAB genes are located at 32.3 min on the E. coli chromosome and provide resistance to potassium tellurite (K2TeO3) with an MIC of 128 μg/ml upon overexpression (28), well above that of 1 to 2 μg/ml observed for control strains. This determinant is specific for tellurite and shows no cross-resistance to selenite, selenate, or tellurate. The tehA gene encodes an integral membrane protein that has been shown to have efflux activity of quaternary ammonium compounds (33). The TehB protein has been shown to be capable of binding both S-adenosylmethionine and tellurite (8, 11). This suggests that the resistance mechanism of the TehAB phenotype likely involves the methylation of tellurite, an activity observed in phytopathogenic pseudomonads (5), and subsequent efflux of an organotelluro compound from the cell.
The present study involves the use of a noninvasive technique, phosphorus-31 nuclear magnetic resonance (NMR) spectroscopy, to examine the effect of tellurite and selenite on the physiological status of E. coli with or without the resistant determinant TehAB present within the cells. Using 31P NMR, we monitored intracellular ATP levels, the presence or absence of a transmembrane pH gradient, and the levels of phosphorylated glycolytic intermediates in sensitive and resistant strains under a variety of conditions. This information provides us with more of an understanding of how cellular metabolism may be affected by the presence of TeO32− and SeO32−.
MATERIALS AND METHODS
Chemicals and media.
M9 medium contained the following (per liter): 9.4 g of Na2HPO4, 1.0 g of NH4Cl, 3.0 g of KH2PO4, and 5 g of NaCl. After autoclaving, the following sterile stock solutions were added aseptically: 2.5 ml of 20% MgSO4, 20 ml of 20% glucose, 1.8 ml of 50 mM CaCl2, 25 ml of 10% case amino acids, and 2 ml of 10% vitamin B1. The final pH was 7.3. Luria broth medium consisted of 10 g of Bacto Tryptone, 5 g of yeast extract, and 5 g of NaCl per liter of distilled water (pH 7.0). Ampicillin (AMP) was added to all cultures to a concentration of 100 μg/ml. NMR buffer consisted of 100 mM PIPES [piperazine-N-N′-bis(2-ethanesulfonic acid)], 50 mM MES [2-(N-morpholino)ethanesulfonic acid], 40 mM NaCl, and 7.5 mM KCl (pH 6.5) with NaOH. The glucose stock solution for the NMR experiments was 1.0 M. Deuterated water (D2O) was obtained from CDN Isotopes. Potassium tellurite (K2TeO3) was obtained from Sigma, and a stock solution was made up to 1 mg/ml in distilled water. Sodium selenite (Na2TeO3) was obtained from Sigma, and a stock solution was made up to 10 mg/ml in distilled water.
Growth conditions.
All cultures were incubated at 37°C with 250-rpm shaking. E. coli strains HB101(pTWT100), expressing the tellurite resistance determinant tehAB (28), and HB101(pUC8), as a control, were inoculated in the morning from frozen 20% dimethyl sulfoxide cultures into 5 ml of Luria broth-AMP medium. After 8 h, the cells were transferred into 50 ml of M9-AMP medium in a 250-ml flask and grown overnight for 16 h, and the entire 50 ml was transferred to 4-liter Erlenmeyer flasks containing 1 liter of M9-AMP medium. Cell growth was monitored at 600 nm. When the cell density had reached the late log phase (optical density at 600 nm between 1.0 and 1.3), the flasks were placed on ice and cooled to 10°C with shaking so that aerobic conditions were maintained in the cultures as they were cooled. Cells were then harvested by centrifugation (7,000 × g for 10 min) and washed once with NMR buffer (see section above), and the weight of the pellet was accurately recorded. For every gram (wet weight) of cell pellet, an equal volume (in milliliters) of NMR buffer was added. The cells and buffer were then mixed together to give a concentrated cell slurry which was stored on ice until used (not longer than 8 h).
Whole-cell studies of cell metabolism by NMR.
In vivo NMR studies with Candida tropicalis provided the background to this study (14, 15). For each experiment, 2.0 ml of E. coli cell slurry was pipetted into a 15-mm NMR tube along with 4.2 ml of NMR buffer and 0.5 ml of D2O. Oxygen was bubbled through the sample at a rate of 30 ml per min. The flow rate was controlled exactly for all experiments with a Manostat flowmeter. For control experiments, 0.5 ml of 1 M glucose was added to the NMR samples just before spectra were recorded (t = 0). Ten spectra, 5-min each, were then acquired. For starved cell experiments, 0.5 ml of distilled water (instead of glucose) was added to the NMR tubes at t = 0, and data were collected as described above. For experiments requiring the addition of K2TeO3 or Na2SeO3, glucose was first added to the samples and NMR spectra were acquired for 15 min in the absence of metal ions to energize the cells. The samples were then removed from the NMR spectrometer, and either K2TeO3 or Na2SeO3 was added to give a final concentration of 0.01 or 0.1 mg/ml in the tubes. The accumulation of NMR data resumed after an interruption time of less than 2 min. Higher levels of Na2SeO3 were used to compensate for the fact that the MIC for selenite for this strain is higher than that for tellurite.
Preparation of cell extracts for NMR analysis.
Experimental conditions for NMR analysis were exactly the same as described in the previous section (NMR tube, cell slurry, and oxygen bubbling, etc.), except that the experiments were performed outside the NMR spectrometer. After exactly 30 min of incubation time, experiments were terminated by the addition of 0.7 ml of 70% perchloric acid. The acidified contents of the NMR tubes were then transferred to 50-ml plastic centrifuge tubes and subjected to three freeze-thaw cycles in liquid nitrogen to facilitate cell breakage. The samples were then centrifuged at 8,000 × g for 10 min to pellet cell debris, after which the supernatants were neutralized to pH 7.0 with the addition of solid K2CO3. The neutralized extracts were then treated with 0.3 g of Chelex resin (added directly to the tube) and recentrifuged. To each of the resulting supernatants, the following was added: 0.264 ml of EDTA, 7 μl of 1 M azide, 0.15 ml of 1.0 M Tris-HCl (pH 8.0), and 20 μl of 0.1 M methylene diphosphonate (internal NMR standard). The pH of the samples was adjusted to exactly 8.4, the samples were Millipore filtered (0.22-μM pore size), and the volumes were adjusted to exactly 8.0 ml. The extracts were stored at −20°C until analyzed by NMR.
NMR conditions.
31P NMR spectra were obtained at 25°C on a Bruker AM-400 wide-bore spectrometer operating in the Fourier-transform mode and using a 13C-31P switchable dual-tuned probe, as in previous studies (14, 15). For whole-cell experiments, samples were placed in 15-mm NMR tubes and the 5-min spectra (800 scans each) were recorded at 162 MHz by using a 9.5-μs pulse length representing a flip angle of 60°. Composite pulse 1H decoupling in a bi-level scheme with 1 W of decoupler power was employed during acquisition. The decoupler power was adjusted to minimize heating of the cells during the course of the experiments. Cell extracts were run under identical conditions, except 60,000 scans were acquired. Spectra were processed and plotted on a Silicon Graphics System, with baseline corrections performed manually. Peak areas were integrated and normalized to the internal methylene diphosphonate standard.
RESULTS
Whole-cell 31P NMR studies.
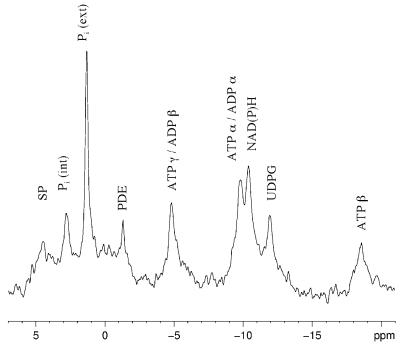
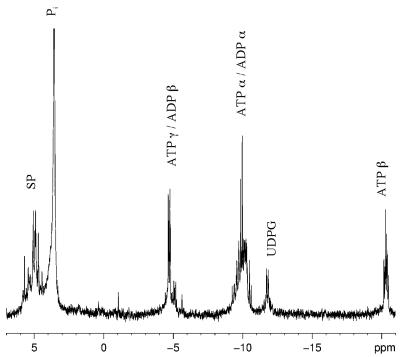
Figure 1 illustrates an in vivo phosphorus-31 NMR spectrum of E. coli HB101(pUC8), a wild-type tellurite-sensitive strain. This spectrum was acquired between 15 and 20 min after the addition of glucose, when quasi-steady-state levels of ATP had been obtained (Fig. 2). The assignment of NMR peaks was made by comparison to previously published NMR spectra (6, 14, 18). The peak in Fig. 1 labeled SP is comprised of phosphomonoesters such as glycolytic sugar phosphates as well as nucleoside monophosphates. These compounds are not well resolved for a number of reasons (the higher viscosity of the intracellular environment and the presence of paramagnetic ions, etc.). The two inorganic phosphate (Pi) peaks next to the SP peak correspond to intracellular [Pi (int)] and extracellular [Pi (ext)] pools of phosphate. The chemical shift of inorganic phosphate is sensitive to pH and can be used to estimate the intracellular pH of living cells noninvasively (9, 18). The appearance of two Pi peaks is only possible when cells have enough energy to maintain a transmembrane pH gradient.
FIG. 1.
A 5-min 31P NMR spectrum (800 scans) of tellurite-sensitive E. coli HB101(pUC8) cells 25 to 30 min after the addition of glucose. Abbreviations: SP, sugar phosphomonoesters; Pi (int) and Pi (ext), intracellular and extracellular pools of inorganic phosphate, respectively; PDE, phosphodiesters; UDPG, uridine diphosphoglucose.
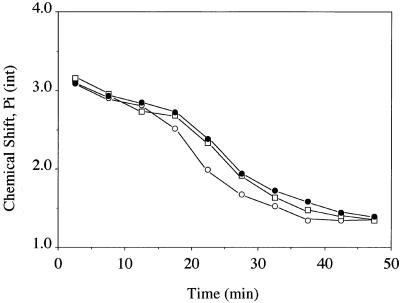
FIG. 2.
Migration of the chemical shift of the intracellular phosphate resonance in E. coli HB101(pUC8) whole-cell NMR spectra as a function of time. Each point is the average of two runs. TeO32− or SeO32− was added to the cell suspensions at the 15-min mark. Filled circles, control glucose-fed cells; open circles, tellurite-treated cells; open squares, selenite-treated cells.
Next to the pH-insensitive phosphodiester peak we see α and γ peaks of ATP (and other nucleoside triphosphates). The ATP peaks overlap with the α and β peaks of ADP (and other nucleoside diphosphates). Only the β phosphate of ATP accurately represents the intracellular nucleotide triphosphate pool. Since this peak is present, we surmise that the cells are energized.
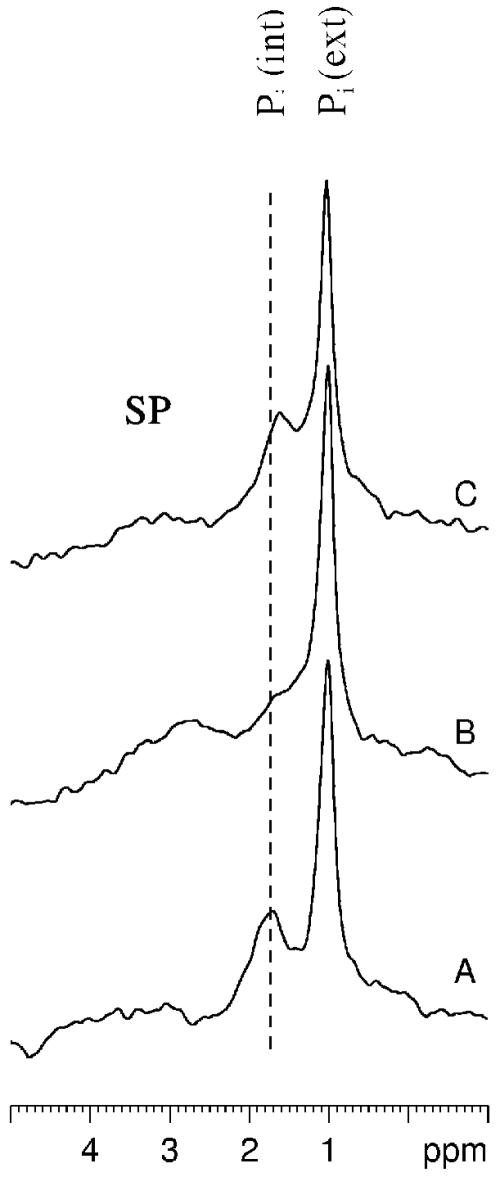
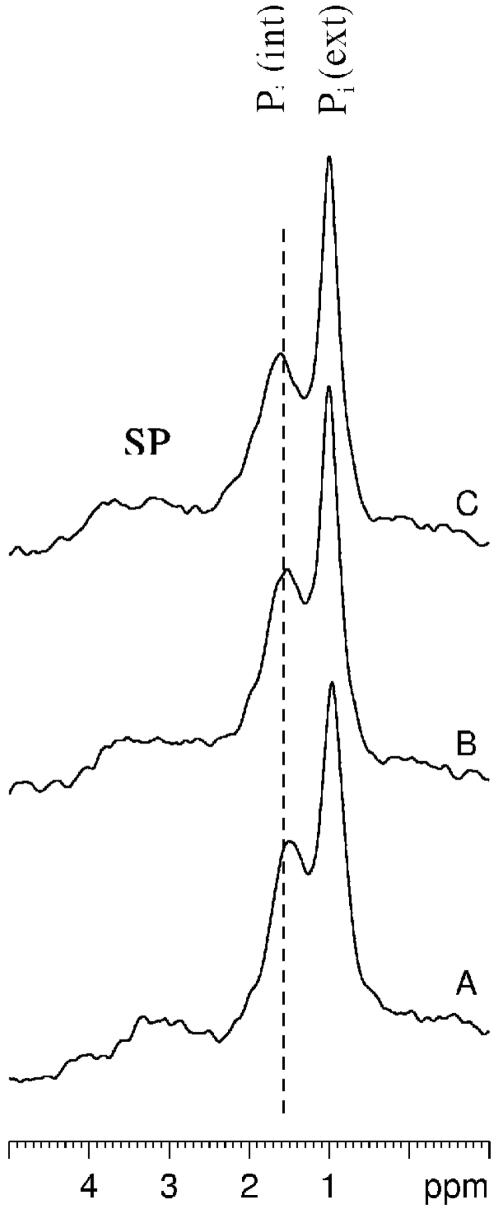
The gradual shift in the δ of the Pi (int) peak as a function of time in the E. coli tellurite-resistant strain is illustrated in Fig. 2. From a Pi titration curve generated by titrating a sample of 5 mM Pi in the presence of 130 mM KCl, 20 mM NaCl, and 1 mM MgCl2, the approximate ionic composition found within E. coli cells (17), we estimate that the intracellular pH at the start of these experiments is around 7.75, whereas by the end of the experiments, the values drop to pH 6.2, since there is not enough glucose left (as confirmed by natural abundance 13C NMR) for the cells to obtain the energy required to maintain an alkaline intracellular pH. Figure 2 shows that control and selenite-treated cells show a very similar acidification trend with time, but in tellurite-treated cells, this rate is much faster. This difference is more dramatically illustrated in Fig. 3, where phosphorus-31 NMR spectra obtained between 25 and 30 min after the start of the experiments (or 10 to 15 min after the addition of K2TeO3 or Na2SeO3) are compared for all three strains. Control (A) and selenite-treated cells (C) still maintain a transmembrane pH gradient at this point in time, but in tellurite-treated cells (B), the pH gradient has been dissipated.
FIG. 3.
Expanded SP region of whole-cell NMR spectra of tellurite-sensitive E. coli HB101(pUC8). (A) Control cells between 30 min after the addition of glucose; (B and C) tellurite- and selenite-treated cells (respectively) at the same point in time 15 min after the addition of metals but 30 min into the experiment.
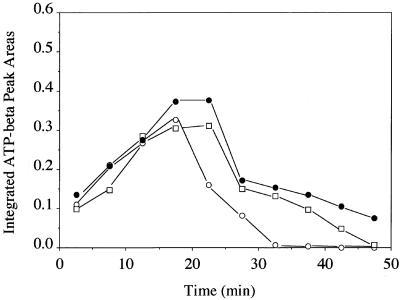
Figure 4 illustrates the changes in the integrated areas of the β-ATP peak as a function of time in control K2TeO3- and Na2SeO3-treated E. coli tellurite-resistant cells. In the control experiment, the trend is for ATP levels to increase over the first 15 min, stabilize over the next 10 min to quasi-steady-state levels, and then drop in response to diminished extracellular glucose concentrations. Selenite-treated cells exhibit a trend in ATP levels fairly similar to those of the control cells; however, after tellurite addition, the decrease in ATP levels is almost immediate and parallels the intracellular acidification observed in Fig. 2.
FIG. 4.
Integrated β-ATP peak areas in whole-cell NMR spectra of E. coli HB101(pUC8) as a function of time. Each point is the average of two runs. TeO32− or SeO32− was added to the cell suspensions at the 15-min mark. Filled circles, control glucose-fed cells; open circles, tellurite-treated cells; open squares, selenite-treated cells.
The in vivo phosphorus-31 NMR experiments described above were also performed with E. coli HB101 harboring the tellurite resistance determinant tehAB (pTWT100). In contrast to results obtained with the tellurite-sensitive strain, there were no observable differences between control, tellurite-treated, and selenite-treated tehAB cells either with respect to the chemical shift of the Pi (int) peak or the area of the β-ATP peak as a function of time. Figure 5 (when compared with Fig. 3) best illustrates the ability of the tehAB determinant to protect the E. coli from intracellular acidification in the presence of K2TeO3.
FIG. 5.
Expanded SP region of whole-cell NMR spectra of tellurite-resistant E. coli HB101(pTWT100). Experimental conditions are as described in the legend to Fig. 3.
31P NMR of cell extracts.
Since E. coli HB101(pUC8) cells exhibited a dramatic de-energization in the presence of K2TeO3, we were interested to know whether or not the metabolism of glucose had been affected in some manner by the presence of this oxyanion. The glycolytic pathway intermediates are all sugar phosphomonoesters (the SP peak in Fig. 1). To resolve the resonances from the individual components of glycolysis, we prepared cell extracts of control, tellurite-treated, and selenite-treated cells (as well as starved cells) 15 min after the addition of metal ions (or 30 min into the experiment), when the differences between resistant and sensitive strains were maximized.
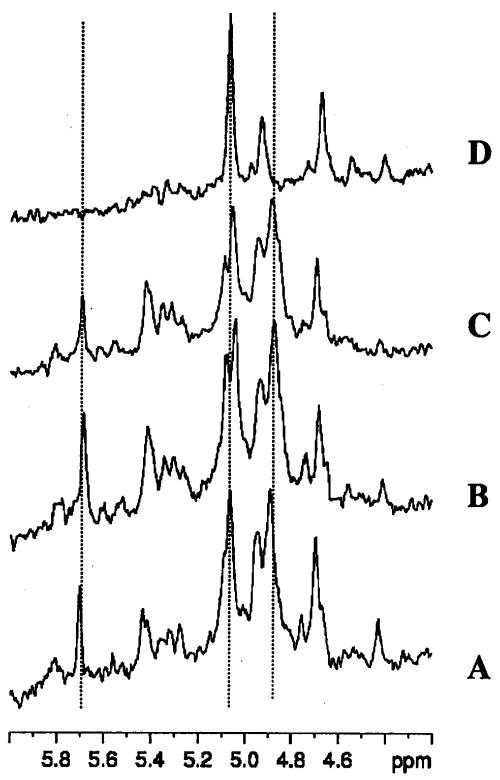
A phosphorus-31 NMR spectrum of a cell extract prepared from control (pUC8) cells is shown in Fig. 6. Here it is no longer possible to observe two inorganic phosphate peaks, since the intracellular and extracellular Pi pools have merged after lysis of the cells by perchloric acid. With extracts, though, it is possible to resolve the ATP-ADP peaks more clearly, as well as the individual peaks in the SP region. The expanded SP region (between 6.0 and 4.2 ppm) is shown in Fig. 7 for pUC8 cells. Panel A corresponds to control cells, panel B corresponds to K2TeO3-treated cells, panel C corresponds to Na2SeO3-treated cells, and panel D corresponds to starved cells. The three dotted lines transecting all four spectra correspond to the β-1 phosphate (5.7 ppm) and the α-1 and -6 phosphates (4.88 and 5.07 ppm, respectively) of fructose-1,6 bis phosphate anomers.
FIG. 6.
31P NMR spectrum (60,000 scans) of tellurite-sensitive E. coli HB101(pUC8) neutralized perchloric acid cell extracts 25 to 30 min after the addition of glucose. Abbreviations are defined in the legend to Fig. 1.
FIG. 7.
Expanded SP region of 31P NMR spectra obtained from cell extracts of E. coli HB101(pUC8). Conditions for panels A to C are described in the legend to Fig. 6. Panel D shows cells that had not received glucose and were considered starved at this point in time.
Figure 7 clearly shows that control and K2TeO3- and Na2SeO3-treated pUC8 cells have practically identical sugar phosphomonoester profiles. Had cells been unable to metabolize glucose properly in the presence of K2TeO3, one would expect to see the profile shown in panel D, where fructose-1,6 bis phosphate is no longer evident and the starvation metabolite 3-phosphoglycerate (3-PGA), which was previously observed in NMR studies with lactic acid bacteria (13), is observed.
Perchloric acid extracts of the tellurite-resistant strain E. coli HB101(pTWT100) were prepared and examined by 31P NMR spectroscopy. As with the sensitive strain, no differences in the sugar phosphomonoester profiles were observed between control and K2TeO3- and Na2SeO3-treated cells (data not shown).
DISCUSSION
Previous studies in our group have shown that the tehA gene codes for an integral membrane protein that has been shown to be capable of effluxing quaternary ammonium compounds (33). We have also shown that the tehB gene product is a protein that is capable of binding both S-adenosylmethionine and tellurite (8, 11). To date, our data suggest that the mechanism by which tehAB confers resistance to E. coli involves methylation of tellurite and subsequent efflux of this product from the cells. Tellurite resistance determinants, including tehAB, are specific for tellurite, and their expression confers no resistance to selenite (35).
31P NMR spectroscopy was used here to further our understanding of how E. coli metabolism is affected by the presence of tellurium and selenium and the route by which the tehAB resistance phenotype may occur. Earlier in vivo 31P NMR work with the bacterium Streptococcus lactis and the yeast C. tropicalis (13, 14, 15) formed the background to this study. Since E. coli, like S. lactis, has a fast metabolism (though a different pH optimum for cell growth), the experimental protocol employed stronger buffering conditions and higher concentrations of glucose than was used in the yeast studies (14, 15). With careful fine-tuning of the oxygen levels, it was possible to obtain highly reproducible data (less than 5% variation in peak areas) in duplicate runs with E. coli control, K2TeO3-treated, Na2SeO3-treated, and starved cells harvested the same day. As it took 1 h to complete an experiment, and since it was not possible to store cells longer than 8 h on ice before metabolism was compromised, we did not attempt to perform experiments in triplicate.
When K2TeO3 was added to the E. coli tellurite-sensitive strain, the NMR tubes turned black over the course of 15 min, suggesting that reduction of K2TeO3 to Te0 had occurred. In contrast, cells expressing the tehAB resistance determinant did not display the same degree of darkening during the course of the experiments. Previous studies have shown that toxicity or resistance to tellurite does not strictly depend on the formation of Te0 crystals (38); however, it was interesting that the presence of the tehAB resistance determinant did diminish the darkening of the cells.
In our 31P NMR spectra, we observed an immediate and rapid loss of the transmembrane proton gradient when tellurite-sensitive E. coli cells were treated with K2TeO3. This trend was paralleled by the depletion of ATP in the cells, suggesting that an uncoupling event is taking place. The presence of the tehAB resistance determinant protects E. coli against tellurite uncoupling, resulting in similar in vivo 31P NMR spectra, as observed with control cells. These observations suggest that our proposed methylation and efflux mechanism detoxification event is effective in diverting tellurite away from its uncoupling targets in the membrane. Unfortunately, we were unable to observe the appearance of a unique phosphorylated compound that could be attributed to a degradation by-product of tellurite metabolism.
A very important observation in our NMR study was that the presence of tellurite did not affect the sugar-phosphomonoester profile of the K2TeO3-sensitive strain. Had any of the glycolytic enzymes been inhibited by tellurite, we would have expected to see evidence of the starvation metabolite 3-PGA in our cell extracts. In a previous study, it was shown that 3-PGA levels increase when S. lactis consumes maltose, a source of carbon which is less rapidly metabolized through the glycolytic pathway than glucose (13). The fact that we did not observe 3-PGA in tellurite-treated sensitive cells, even at the point where the transmembrane pH gradient was no longer evident (Fig. 3), suggests that lack of glucose or a decreased glycolytic flux per se was not the direct cause of cellular de-energization.
The selenite biochemistry of E. coli is reasonably well understood (34). Selenite has some chemical similarities to tellurite, so the route of SeO32− and TeO32− metabolism in E. coli may be expected to show similarities. However, our 31P NMR spectra showed no differences in the degree of transmembrane pH gradient loss, ATP levels, or levels of glycolytic intermediates in selenite-metabolizing strains relative to the control situation (absence of metal ions). Both the tellurite-sensitive and -resistant strains also reduced selenite to the same degree, with bright orange-red precipitates forming during the course of the in vivo NMR experiments. Our results suggest that the routes for selenite and tellurite metabolism in E. coli differ significantly and that selenite metabolism does not involve the tehAB methylation and efflux mechanism (35).
From the data presented here, we see that exposure to tellurite but not selenite causes an uncoupling of the pH gradient across the membrane in tellurite-sensitive cells. Additionally, we find that the tellurite resistance determinant TehAB provides protection against this uncoupling. These results significantly add to our understanding of tellurite metabolism in bacteria.
Acknowledgments
This work was supported by an NSERC grant (to R.J.T.).
E.L.-V. thanks Howard Hunter for teaching her to process NMR spectra with the Silicon Graphics System and for plotting the final composite figures for this paper. Deane McIntyre is thanked for assistance with the crucial timing and running of the NMR experiments.
The work was performed at the Bio-NMR center (University of Calgary). The Bio-NMR center was recently upgraded with funds provided by the Canada Foundation for Innovation, the Alberta Science and Research Authority, the Alberta Intellectual Infrastructure Partnership Program, and the Alberta Heritage Foundation for Medical Research. Maintenance of the centre is supported by the University of Calgary and the Canadian Institutes of Health Research.
REFERENCES
- 1.Avazeri, C., R. J. Turner, J. Pommier, J. H. Weiner, G. Giordano, and A. Vermeglio. 1997. Tellurite and selenate reductase activity of nitrate reductases from Escherichia coli: correlation with tellurite resistance. Microbiology 143:1181-1189. [DOI] [PubMed] [Google Scholar]
- 2.Bébien, M., J. Kirsch, V. Méjean, and A. Verméglio. 2002. Involvement of a putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology 148:3865-3872. [DOI] [PubMed] [Google Scholar]
- 3.Boresetti, F., R. Borhgese, F. Francia, M. R. Randi, S. Fedi, and D. Zannoni. 2003. Reduction of potassium tellurite to elemental tellurium and its effect on the plasma membrane redox components of the facultative phototroph Rhodobacter capsulatus. Protoplasma 221:153-161. [DOI] [PubMed] [Google Scholar]
- 4.Chiong, M., E. González, R. Barra, and C. Vásquez. 1988. Purification and biochemical characterization of tellurite-reducing activities from Thermus thermophilus HB8. J. Bacteriol. 170:3269-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cournoyer, B., S. Watanabe, and A. Vivian. 1998. A tellurite resistance genetic determinant from phytopathogenic pseudomonads encodes a thiopurine methyltransferase: evidence of a widely-conserved family of methyltransferases. Biochem. Biophys. Acta 1397:161-168. [DOI] [PubMed] [Google Scholar]
- 6.den Hollander, J. A., K. Ugurbil, T. R. Brown, and R. G. Shulman. 1981. Phosphorus-31 nuclear magnetic resonance studies of the effect of oxygen upon glycolysis in yeast. Biochemistry 20:5871-5880. [DOI] [PubMed] [Google Scholar]
- 7.Di Tomaso, G., S. Fedi, M. Carnevali, M. Manegatti, C. Taddei, and D. Zannoni. 2002. The membrane-bound respiratory chain of Pseudomonas pseudoalcaligenes KF707 cells grown in the presence or absence of potassium tellurite. Microbiology 148:1699-1708. [DOI] [PubMed] [Google Scholar]
- 8.Dyllick-Brenzinger, M., M. Liu, T. L. Winstone, D. E. Taylor, and R. J. Turner. 2000. The role of cysteine residues in tellurite resistance mediated by the TehAB determinant. Biochem. Biophys. Res. Commun. 277:394-400. [DOI] [PubMed] [Google Scholar]
- 9.Gadian, D. G. 1992. NMR and its applications to living systems. Clarendon Press, Oxford, United Kingdom.
- 10.Guzzo, J., and M. S. Dubow. 2000. A novel selenite- and tellurite-inducible gene in Escherichia coli. Appl. Environ. Microbiol. 66:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, M., R. J. Turner, T. L. Winstone, A. Saetre, M. Dyllick-Brenzinger, G. Jickling, L. W. Tari, J. H. Weiner, and D. E. Taylor. 2000. Escherichia coli TehB requires S-adenosylmethionine as a cofactor to mediate tellurite resistance. J. Bacteriol. 182:6509-6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd-Jones, G., A. M. Osborn, D. A. Ritchie, P. Stride, J. L. Hobman, N. L. Brown, and D. A. Rouch. 1994. Accumulation and intracellular fate of tellurite in tellurite-resistant Escherichia coli: a model for the mechanism of resistance. FEMS Microbiol. Lett. 118:113-120. [DOI] [PubMed] [Google Scholar]
- 13.Lohmeier-Vogel, E. M., K. Skoog, H. Vogel, and B. Hahn-Hägerdal. 1986. Phosphorus-31 NMR studies of maltose and glucose metabolism in Streptococcus lactis. Appl. Microbiol. Biotechnol. 25:43-51. [Google Scholar]
- 14.Lohmeier-Vogel, E. M., K. Skoog, H. Vogel, and B. Hahn-Hägerdal. 1989. 31P-Nuclear magnetic resonance study of the effect of azide on xylose fermentation by Candida tropicalis. Appl. Environ. Microbiol. 55:1974-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohmeier-Vogel, E. M., B. Hahn-Hägerdal, and H. J. Vogel. 1995. Phosphorus-31 and carbon-13 nuclear magnetic resonance studies of glucose and xylose metabolism in Candida tropicalis cell suspensions. Appl. Environ. Microbiol. 61:1414-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore, M. D., and S. Kaplan. 1992. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J. Bacteriol. 174:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidhardt, F. C., et al. (ed.). 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 18.Nicolay, K., W. A. Scheffers, P. M. Bruinenberg, and R. Kaptein. 1983. Phosphorus-31 nuclear magnetic resonance studies of the intracellular pH, phosphate compartmentation and phosphate transport in yeasts. Arch. Microbiol. 133:83-89. [Google Scholar]
- 19.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 20.Pearion, C. T., and P. E. Jablonski. 1999. High level, intrinsic resistance of Natronococcus occultus to potassium tellurite. FEMS Microbiol. Lett. 174:19-23. [Google Scholar]
- 21.Roane, T. M., and S. T. Kellogg. 1996. Characterization of bacterial communities in heavy metal contaminated soils. Can. J. Microbiol. 42:593-603. [DOI] [PubMed] [Google Scholar]
- 22.Scala, J., and H. H. Williams. 1963. A comparison of selenite and tellurite toxicity in Escherichia coli. Arch. Biochem. Biophys. 101:319-324. [DOI] [PubMed] [Google Scholar]
- 23.Silver, S. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 24.Silver, S. 1998. Genes for all metals—a bacterial view of the periodic table. The 1996 Thom Award Lecture. J. Ind. Microbiol. Biotechnol. 20:1-12. [DOI] [PubMed] [Google Scholar]
- 25.Spallholz, J. E. 1994. On the nature of selenium toxicity and carcinostatic activity. Free Radic. Biol. Med. 17:45-64. [DOI] [PubMed] [Google Scholar]
- 26.Stadtman, T. C. 1996. Selenocysteine. Annu. Rev. Biochem. 65:83-100. [DOI] [PubMed] [Google Scholar]
- 27.Tantaleán, J. C., M. A. Araya, C. P. Saavedra, D. E. Fuentes, J. M. Pérez, O. L. Calderón, P. Youderian, and C. C. Vásquez. 2003. The Geobacillus stearothermophilus V iscS gene, encoding cysteine desulfurase, confers resistance to potassium tellurite in Escherichia coli K-12. J. Bacteriol. 185:5831-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor, D. E., Y. Hou, R. J. Turner, and J. H. Weiner. 1994. Location of a potassium tellurite resistance operon (tehA tehB) within the terminus of Escherichia coli K-12. J. Bacteriol. 176:2740-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor, D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111-115. [DOI] [PubMed] [Google Scholar]
- 30.Trutko, S. M., N. E. Suzina, V. I. Duda, V. K. Akimendo, and A. M. Boronin. 1998. Involvement of the respiratory chain in potassium tellurite reduction in bacteria. Dokl. Biochem. 358:13-15. [PubMed] [Google Scholar]
- 31.Trutko, S. M., V. K. Akimenko, N. E. Suzina, L. A. Anisimova, M. Shlyapnikov, B. P. Baskunov, V. I. Duda, and A. M. Boronin. 2000. Involvement of the respiratory chain of gram-negative bacteria in the reduction of tellurite. Arch. Microbiol. 173:178-186. [DOI] [PubMed] [Google Scholar]
- 32.Tucker, F. L., J. E. Walper, M. D. Appleman, and J. Donohue. 1962. Complete reduction of tellurite to pure tellurium metal by microorganisms. J. Bacteriol. 83:1313-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner, R. J., D. E. Taylor, and J. H. Weiner. 1997. Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob. Agents Chemother. 41:440-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner, R. J., J. H. Weiner, and D. E. Taylor. 1998. Selenium metabolism in Escherichia coli. BioMetals 11:223-227. [DOI] [PubMed] [Google Scholar]
- 35.Turner, R. J. 2001. Tellurite toxicity and resistance in gram-negative bacteria. Rec. Res. Dev. Microbiol. 5:69-77. [Google Scholar]
- 36.Turner, R. J., J. H. Weiner, and D. E. Taylor. 1999. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology 14:2549-2557. [DOI] [PubMed] [Google Scholar]
- 37.Turner, R. J., Y. Aharonowitz, J. H. Weiner, and D. E. Taylor. 2001. Glutathione is a target in bacterial tellurite toxicity and is protected by tellurite resistance determinants in Escherichia coli. Can. J. Microbiol. 47:33-40. [PubMed] [Google Scholar]
- 38.Yurkov, V., J. Jappe, and A. Vermeglio. 1996. Tellurite resistance and reduction by obligate aerobic photosynthetic bacteria. Appl. Environ. Microbiol. 62:4195-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]