Abstract
This work describes the purification and characterization of propionicin F, the first bacteriocin isolated from Propionibacterium freudenreichii. The bacteriocin has a bactericidal activity and is only active against strains of P. freudenreichii. Propionicin F appears to be formed through a processing pathway new to bacteriocins. The mass of the purified bacteriocin was determined by mass spectrometry, and the N-terminal amino acid sequence was determined by Edman degradation. Sequencing of pcfA, the bacteriocin structural gene, revealed that propionicin F corresponds to a 43-amino-acid peptide in the central part of a 255-amino-acid open reading frame, suggesting that mature propionicin F is excised from the probacteriocin by N- and C-terminal proteolytic modifications. DNA sequencing and Northern blot hybridizations revealed that pcfA is cotranscribed with genes encoding a putative proline peptidase and a protein from the radical S-adenosylmethionine family. A gene encoding an ABC transporter was also identified in close proximity to the bacteriocin structural gene. The potential role of these genes in propionicin F maturation and secretion is discussed.
Propionic acid bacteria (PAB) and lactic acid bacteria (LAB) have been utilized for flavor development and preservation of food products for centuries. The preservative capacity of PAB and LAB has previously been attributed to the lowering of pH by production of propionic acid, acetic acid, and lactic acid, which are the main fermentative end products produced by these organisms. However, it has become evident that PAB and LAB also produce antimicrobial substances in addition to organic acids (25). Among these are the bacteriocins, which are gene-encoded, ribosomally synthesized peptides that usually display antimicrobial activity against species closely related to the producer organism (43).
A vast number of LAB bacteriocins have been purified; some of these, including nisin, pediocin PA-1, and sakacin P, have been thoroughly characterized at the genetic and biochemical level (5, 6, 16, 21, 24, 26, 27, 29, 33, 40, 44, 45). In general, LAB bacteriocins are small, cationic peptides, with a propensity to adopt amphiphilic helical structures (36). There are, however, exceptions from this rule, including helveticin J and enterolysin A, which both are large proteins with antimicrobial activity (23, 37). The LAB bacteriocins are usually synthesized as prepeptides, containing an N-terminal secretion signal peptide of the sec or the double-glycine type. The leader sequence is cleaved off concomitant with secretion either by the general secretory machinery or, in the case of double-glycine leaders, by dedicated ABC transporters (15, 47).
In contrast to the numerous bacteriocins characterized from LAB, only three PAB bacteriocins have been identified and characterized at the amino acid and genetic level. Propionicin T1 isolated from Propionibacterium thoenii is a 65-amino-acid bacteriocin, synthesized with a 31-residue signal peptide (13). The 204-amino-acid bacteriocin SM1 from Propionibacterium jensenii also appears to be exported via the sec pathway. In contrast to the other PAB bacteriocins, SM1 is encoded by a plasmid-located gene (32). This plasmid is identical to pRG01, which is found in several PAB species (39). Finally, an antimicrobial peptide (PAMP) was recently purified from protease-activated supernatants of P. jensenii. This bacteriocin is synthesized as a proprotein of 198 amino acids with an N-terminal signal peptide of 27 amino acids, whereas the 64 C-terminal residues comprise the active bacteriocin. It is possible that the mature form of PAMP is produced via proteolytic cleavage of the proprotein by a hitherto undisclosed protease (11). The production of propionicin T1 and pro-PAMP is prevalent in P. jensenii and P. thoenii strains but has not been identified as occurring in P. freudenreichii (12). In fact, no bacteriocin has been purified from P. freudenreichii, although the antimicrobial potential of this organism has frequently been demonstrated (1, 19, 31).
Characterization of bacteriocins from P. freudenreichii is of special interest, since this species is the most important secondary starter in the production of Swiss-type cheeses. In this work we describe the molecular and genetic characterization of propionicin F, the first bacteriocin isolated from P. freudenreichii. Intriguingly, the active bacteriocin appears to be formed through an unusual processing mechanism involving extensive C- and N-terminal proteolytic modifications of a large probacteriocin.
MATERIALS AND METHODS
Bacterial strains and media.
Propionibacteria were propagated anaerobically at 30°C in sodium lactate broth (SLB) (1% [wt/vol] tryptone, 1% [wt/vol] yeast extract, 1% [wt/vol] sodium lactate, 0.25 g of K2HPO4/liter) (28) or MRS (Oxoid, Basingstoke, Hampshire, United Kingdom). Escherichia coli BL 21(DE3) (Novagen, Madison, Wis.) was grown at 37°C in Luria-Bertani medium supplemented with 100 μg of carbenicillin/ml when appropriate.
Colony assay of bacteriocin activity.
Strains of propionibacteria were spotted on SLB agar plates and grown for 5 days. A total of 5 ml of soft agar was mixed with 0.5-ml cultures of indicator bacteria in late logarithmic growth phase and then poured over the plates. After incubation for 24 h at 30°C, the plates were examined for zones of growth inhibition surrounding the colonies.
Quantitative determination of bacteriocin activity.
Bacteriocin activity was determined in microtiter plate assays (20). Each well of the microtiter plate contained 50 μl of a twofold serial dilution of the sample in SLB or MRS and 150 μl of a 100-fold-diluted culture of the indicator strain. The plates were incubated anaerobically at 30°C for 24 h, and growth inhibition was measured spectrophotometrically at 620 nm with a microtiter plate reader (Multiscan Ascent; Labsystems, Helsinki, Finland). By definition, one bacteriocin unit caused 50% growth inhibition of the standard indicator P. freudenreichii ISU-P59 compared to that for a control culture with no bacteriocin added.
Determination of bactericidal activity.
A stationary-phase culture of P. freudenreichii ISU-P59 was diluted 100 times in SLB, and aliquots were incubated with up to 40 nM purified propionicin F at 30°C. Growth and viability was monitored by optical density measurements and viable counts.
Purification of propionicin F.
The bacteriocin was purified from a 2-liter culture of P. freudenreichii LMGT 2946. The bacteria were grown in MRS at 30°C until the onset of stationary phase (96 h). The culture was centrifuged at 12,000 × g for 20 min at 4°C, and the bacteriocin was precipitated from the supernatant with 40% (wt/vol) ammonium sulfate (Applichem, Darmstadt, Germany). The pellet was dissolved in 400 ml of distilled water, and the pH was adjusted to 9.0 by the addition of solid Trisma base (Sigma, St. Louis, Mo.). This sample was applied to a 10-ml Q-Sepharose Fast-Flow anion-exchange column (Amersham Pharmacia Biotech, Uppsala, Sweden) preequilibrated with 10 mM Trisma base buffer (pH 9.0). The column was washed with 100 ml of 10 mM sodium phosphate buffer at pH 7.2 before the bacteriocin was eluted in 100 ml of 1 M NaCl. The purification was completed by three consecutive rounds of reverse-phase chromatography on an Äkta purifier system (Amersham Pharmacia Biotech). The sample was applied onto the RESOURCE RPC 1-ml column first followed by application onto a Source 5RPC ST 4.6/415 column and then a Sephasil peptide C8 5-μm ST 4.6/250 column (all columns were from Amersham Pharmacia Biotech). In each run the peptide was eluted from the column in a water-2-propanol gradient containing 0.1% trifluoroacetic acid. The fractions showing the highest specific bacteriocin activity were pooled and used in the subsequent purification step.
N-terminal amino acid sequencing.
The N-terminal amino acid sequence was determined by automated Edman degradation with a 447A automatic sequence analyzer (Applied Biosystems, Foster City, Calif.) and an on-line 120A amino acid phenylthiohydantoin analyzer as described by Cornwell et al. (8).
Mass spectrometry (MS).
The molecular mass of propionicin F was determined with a matrix-assisted laser desorption ionization-time of flight mass spectrometer (Voyager-RP DE; Applied Biosystems) in the linear positive-ion mode. The total acceleration voltage was 25 kV. The voltage on the first grid and the delay time between ion production and extraction were adapted to the mass of the sample. A total of 100 single scans were accumulated for each spectrum. The matrix, α-cyano-4-hydroxycinnamic acid (C-2020; Sigma), was dissolved at a concentration of 15 mg/ml in a mixture of 1:1 acetonitrile-0.1% aqueous trifluoroacetic acid. Afterwards, 0.5 μl of sample and 1.5 μl of matrix were mixed on the sample plate and air dried. All data were calibrated by using an external calibration standard mixture (Applied Biosystems).
Quantification of purified propionicin F.
The concentration of purified propionicin F was determined spectrophotometrically at 280 nm with a molar extinction coefficient deduced from the amino acid sequence (14).
DNA manipulation.
The molecular biological techniques used in this study were described by Sambrook et al. (41). Restriction enzymes and T4 DNA ligase were purchased from New England BioLabs, Inc. (Beverly, Mass.) and Fermentas (Vilnius, Lithuania). Taq and Pfx DNA polymerases were obtained from QIAGEN (Hilden, Germany) and Invitrogen (Paisley, United Kingdom), respectively. Plasmid DNA was purified by use of a Qiaprep Miniprep spin column (QIAGEN). Isolation of total DNA from P. freudenreichii was done using Advamaxbeads (Advanced Genetic Technologies Corp., Gaithersburg, Md.). All products were used according to the manufacturers' instructions.
DNA sequencing and sequence analysis.
On basis of the N-terminal sequence of propionicin F, degenerate primers pFf1 (5′TGG TTC TAY CAG GGN ATG AA 3′) and pFr1 (5′GAT RTT NGC NAC NCC NCC GAT 3′) were designed and used in PCR amplification of a 63-bp fragment. Further sequence was obtained by a primer walking strategy. First, total DNA of P. freudenreichii LMGT 2946 was digested with BamHI, EcoRI, EcoRV, HindIII, and SmaI. The DNA fragments obtained from each of the digests were ligated to pBluescript II SK+ (Stratagene, La Jolla, Calif.) digested with the same enzyme. Each of the ligation mixtures was used as a template in PCR amplification with combinations of the vector-specific primer T7 and bacteriocin-specific primers. Amplified DNA fragments were purified by use of agarose gel electrophoresis and Qiaquick PCR purification columns (QIAGEN). The purified fragments were sequenced using a BigDye version 3.1 terminator cycle sequencing ready reaction kit and a model 3100 genetic analyzer according to procedures provided by the supplier (Applied Biosystems).
Heterologous expression of propionicin F in E. coli.
The propionicin F encoding part of pcfA was cloned and expressed in E. coli BL21(DE3) as a thioredoxin fusion protein by use of an LIC pET32/Xa vector system (Novagen) according to the manufacturer's protocol. The thioredoxin-propionicin F fusion protein was purified from a 200-ml culture. The culture was induced at an A600 of 0.6 with 1 mM isopropyl-β-d-thiogalactopyranoside and incubated for 3 h before the cells were harvested by centrifugation. The cells were suspended in 20 ml of 1× binding buffer (Novagen) and subsequently lysed by use of a French press homogenizer. Inclusion bodies were recovered as described by Novagen. Cell debris was removed by centrifugation, and the supernatant was applied onto a His · Bind column (Novagen). The purified fusion protein eluted in 1× elution buffer (Novagen) was washed and concentrated using an AmiconUltra-15 centrifugal filter device (10-kDa cutoff) (Millipore, Bedford, Mass.). Approximately 10 μg of fusion protein was processed in a 100-μl reaction mixture with 1 unit of factor Xa at 21°C for 4 h. The sample was analyzed by MS, and bacteriocin activity was measured by microtiter plate assay using the most sensitive strain, P. freudenreichii ISU-P104, as indicator.
RNA isolation and Northern hybridization analysis.
Samples of P. freudenreichii LMGT 2946 grown in MRS broth were harvested at time intervals. The cells were put on ice and suspended in diethylpyrocarbonate-treated STE buffer (100 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0), washed once, suspended in 0.25 ml of diethylpyrocarbonate-treated STE buffer, and mixed with 0.75 ml of RTL buffer (QIAGEN) containing 10 mM β-mercaptoethanol. The cells were mechanically disrupted with Rnase-free glass beads (BIO1; Savant, Vista, Calif.) and a Fast Prep FP120 apparatus (BIO101) at full speed for 20 sec. The disrupted cell fractions were centrifuged. The supernatants were transferred into new tubes, mixed with 0.5 ml of 96% ethanol, and subsequently applied onto RNEasySpin columns (QIAGEN). Nucleic acids were removed by DNase treatment, and the samples were processed according to RNEasy protocol (QIAGEN). For Northern analysis (41), samples containing 10 μg of RNA were fractionated by agarose gel electrophoresis and transferred to a Hybond-N membrane (Amersham Pharmacia Biotech). A pcfA-specific probe covering the nucleotides encoding Trp102 to Val255 of PcfA was labeled with [α-32P]dCTP using a Rediprime labeling kit (Amersham Pharmacia Biotech) and subsequently hybridized to the membrane at 65°C over night. The Northern blot was developed for 10 min in storage phosphor screen cassettes (Molecular Dynamics, Sunnyvale, Calif.) and subsequently analyzed on a Typhoon 8600 PhosphorImager (Molecular Dynamics). Band sizes were determined by comparison to a size marker RNA ladder (Promega, Madison, Wis.).
Nucleotide sequence accession number.
The DNA sequence described here has been deposited in the GenBank database under accession no. AY587566.
RESULTS
Characterization of propionicin F.
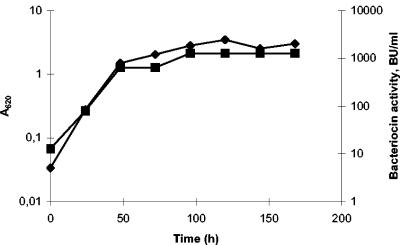
As part of a continued effort to study the antimicrobial potential of PAB, a collection of P. freudenreichii strains were screened for inhibitory activity against a selection of indicator strains (19). Two strains, P. freudenreichii LMGT 2946 and P. freudenreichii LMGT 2956, were found to inhibit the growth of P. freudenreichii ISU-P59 on agar plates. When the strains were anaerobically grown in MRS broth at 30°C, the antimicrobial substance was constitutively secreted, with the maximum activity level reached in stationary phase (Fig. 1).
FIG. 1.
Growth kinetics and bacteriocin production of P. freudenreichii LMGT 2946. Antimicrobial activity was determined from ammonium sulfate-precipitated cell-free culture supernatant by use of P. freudenreichii ISU-P59 as an indicator. Symbols: ♦, A620; ▪, AU/ml.
The antimicrobial substance could be precipitated from culture supernatants of strains LMGT 2946 and LMGT 2956 with ammonium sulfate. Furthermore, heat treatment (85°C for 10 min) of the samples did not affect the activity, but extensive proteinase K treatment reduced the antimicrobial activity by 90%.
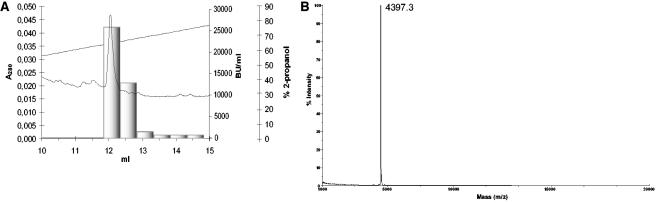
These findings led to the assumption that the specific antimicrobial activity of strains LMGT 2946 and LMGT 2956 was proteinaceous. When ammonium sulfate precipitation, ion exchange chromatography, and reverse-phase chromatography were subsequently used, a bacteriocin designated propionicin F was purified from the culture supernatant of strain LMGT 2946 (Table 1 and Fig. 2). When a fraction of purified propionicin F was analyzed by MS, a single peak corresponding to a molecular mass of 4,397 m/z (±1 Da) was observed (Fig. 2A). The same fraction was subjected to sequencing by Edman degradation, and the N-terminal amino acid sequence WFYQGMNIAIYANIGGVANIIGY was obtained.
TABLE 1.
Purification of propionicin from P. freudenreichii LMGT 2946
| Purification step | Volume (ml) | A280 | Antimicrobial activity (AU/ml) | Specific activity (AU/ml/A280) | Severalfold increase in specific activity |
|---|---|---|---|---|---|
| Culture supernatant | 2,000 | 20.1 | 1,256 | 62.5 | 1 |
| Ammonium sulphate precipitate | 400 | 12.0 | 2,560 | 2.1 × 102 | 3.4 |
| Anion-exchange chromatography | 100 | 1.0 | 2,560 | 2.5 × 103 | 40.0 |
| Reverse-phase chromatography (RESOURCE-I) | 5 | 1.2 | 10,000 | 8.3 × 103 | 132.8 |
| Reverse-phase chromatography (RESOURCE-II) | 2 | 0.09 | 12,800 | 1.4 × 105 | 2,240.0 |
| Reverse-phase chromatography (Sephasil-C8) | 1 | 0.016 | 12,500 | 7.8 × 105 | 12,480.0 |
FIG. 2.
(A) Chromatogram and activity measurement of the final reversed phase purification of propionicin F. (B) MS analysis of purified propionicin F.
Identification of the propionicin F-encoding gene.
Two degenerate primers were designed on basis of the amino acid sequence. When these primers were used in a PCR with genomic DNA from strain LMGT 2946, the expected 63-bp fragment was successfully amplified. The DNA sequence of this fragment was subsequently used to devise a new set of primers that were applied as part of a primer walking strategy to obtain flanking sequences. Using this technique, approximately 10 kb of DNA surrounding the initial 63-bp fragment were sequenced (GenBank accession no. AY587566). The DNA fragment encoding the previously identified 23-amino-acid sequence was found as part of a 765-bp open reading frame (ORF) (pcfA). The N-terminal amino acid sequence corresponded to residues 102 to 125 of the 255-amino-acid putative protein PcfA. As determined on the basis of the experimentally determined molecular mass of 4,397 ± 1 Da, the mature propionicin F bacteriocin was predicted to include residues Trp102 to Pro145 of PcfA. The calculated mass of this peptide is 4,379 ± 1 Da. The discrepancy between this mass and the mass obtained by the MS analysis can probably be ascribed to oxidation of the only methionine residue in propionicin F into methionine sulfoxide.
To confirm the antimicrobial nature of propionicin F, the peptide was cloned and expressed as a thioredoxin fusion protein in E. coli. Inclusion bodies of the fusion protein were purified and subjected to processing by factor Xa. The treatment with factor Xa resulted in low but significant levels of bacteriocin activity. MS analysis of these fractions resulted in a peak corresponding to a mass of 4,396 ± 1 Da (data not shown), confirming the presence of propionicin F.
Although a putative ribosome binding site could be found, no promoter −10 or −35 sequences were identified in the upstream region of pcfA. No obvious leader peptide sequence could be identified either. The results of secondary structure analysis using TMpred, DAS, and TopPred algorithms (9, 18, 46) suggested that PcfA contains one transmembrane segment, spanning residues 127 to 142. Notably, this particular region comprises the C-terminal part of mature propionicin F. PcfA did not show any similarity to protein or translated DNA sequences in public databases.
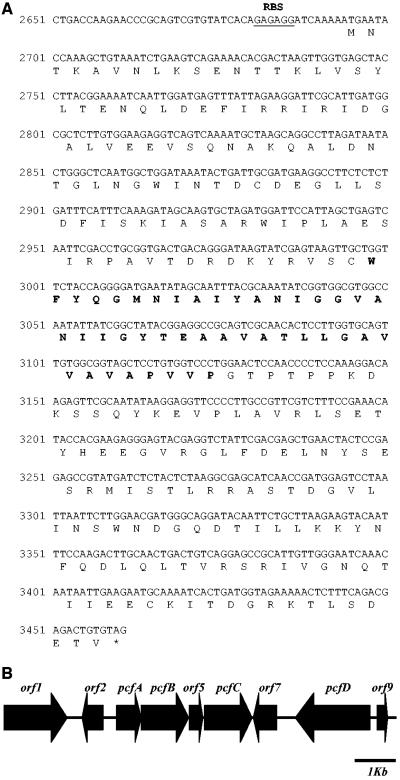
Including the bacteriocin-encoding gene, nine ORFs were identified in the 10-kb DNA sequence (Fig. 3; Table 2). The ORF pcfB, which is located directly downstream of pcfA, encodes a protein with homology to members of the radical S-adenosylmethionine (SAM) family. Enzymes of this family participate in biosynthetic pathways, where they perform various reactions, including methylation, radical formation, anaerobic oxidation, and protein ring formation (42). Further downstream an ORF (pcfC) encoding a protein with homology to membrane-associated and secreted proteases was identified (7, 38). Another ORF, pcfD, showed high similarity to ABC transporter superfamily genes (17). Database searches could not identify any homologues to orf5, orf7, or orf9. Strikingly, the GC content of the genes summarized above was approximately 50%, while P. freudenreichii DNA normally has a GC content of 65% (10). Comparison of the pcfA coding sequence to sequences in the codon usage table database (34) revealed that this gene contains codons that are untypical for P. freudenreichii. Interestingly, the low-GC-content gene cluster is preceded by an ORF (orf2) encoding a protein with homology to proteins in the TnpA family of transposases. These findings suggest that the propionicin F gene cluster could have been acquired by horizontal gene transfer.
FIG. 3.
(A) Sequence of nucleotides 2650 to 3461 of the propionicin F locus (GenBank accession no. AY587566), corresponding to the pcfA gene with translation of propropionicin F. Bold characters denote the amino acid sequence of mature propionicin F. The putative ribosome binding site (RBS) of pcfA is underlined. (B) Schematic representation of the genetic organization of the propionicin F locus. The bacteriocin structural gene pcfA was identified by reverse genetics on the basis of the amino acid sequence from purified propionicin F. Eight additional ORFs were identified in the surrounding 10-kb region; three of these (pcfB, pcfC, and pcfD) have a proposed function connected to production of propionicin F (Table 2).
TABLE 2.
Summary of proposed gene functions of the identified ORFs in the propionicin F gene cluster and homology to proteins in public databases
| ORF (gene) | Size of product (no. of amino acids) | Function | Homologya | Accession no. |
|---|---|---|---|---|
| ORF 1 | 469 | Putative ABC sugar transporter | Membrane transport protein, Bordetella pertussis (29.7% identity) | CAE43127b |
| Sugar transport proteins, ABC transporters of the major facilitator superfamily (MFS) | pfam00083b | |||
| ORF 2 | 212 | Transposase | Transposase, Arthrobacter nicotinovorans (IS1473) (23.4% identity) | AAK64271 |
| Transposase and inactivated derivatives | COG3415b | |||
| ORF 3 (pcfA) | 255 | Prebacteriocin | ||
| ORF 4 (pcfB) | 349 | Hypothetical protein (RSc2181) Ralstonia solanacearum (19.3% identity) | CAD15888b | |
| Radical SAM superfamily | COG0535b | |||
| ORF 5 | 127 | |||
| ORF 6 (pcfC) | 452 | Protease, putatively membrane associated or secreted | Predicted hydrolase or acyltransferase (Mdeg1145), Microbulbifer degradans (19.2% identity) | ZP00065771 |
| ORF 7 | 169 | Alpha/beta hydrolase fold | pfam00561 | |
| ORF 8 (pcfD) | 523 | ABC transporter | ABC transporter (p503), Staphylococcus epidermidis (21.2% identity) | NP863206b |
| ABC-type multidrug transport system, ATPase and permease components | COG1132b | |||
| ORF 9 | 125 |
Only the best matches are shown.
Accession number of sequence deposited directly in the National Center for Biotechnology Information database.
Distribution of the pcfA gene in PAB.
To investigate the distribution of the pcfA gene in various PAB, a set of primers specific to the pcfA gene was synthesized. By the use of these primers, 20 PAB strains were screened by PCR for the presence of the pcfA gene (Table 3). Successful amplification was obtained only with DNA from P. freudenreichii LMGT 2946 and P. freudenreichii LMGT 2956. This confirms the identity of the antimicrobial activity produced by these strains as propionicin F.
TABLE 3.
Antimicrobial activity of propionicin F and distribution of the pcfa genea
| Indicator species | Straina | Presence of pcfAb | Sensitivity to propionicin F (MIC [nM]) |
|---|---|---|---|
| P. acidipropionici | ATCC 4965 | − | >320 |
| P. acidipropionici | ATCC 4875 | − | >320 |
| P. freudenreichii ssp. freudenreichii | ATCC 6207T | − | 0.7 |
| P. freudenreichii ssp. shermanii | ATCC 9614T (DSM 4902) | − | 5.0 |
| P. freudenreichii | ATCC 9616 | − | 20 |
| P. freudenreichii | IFO12426 | − | 2.5 |
| P. freudenreichii | LMGT 2842 | − | 5.0 |
| P. freudenreichii | LMGT 3001 | − | 5.0 |
| P. freudenreichii | LMGT 2946 | + | 80, >640c |
| P. freudenreichii | LMGT 2956 | + | 80, >640c |
| P. freudenreichii | ISU-P24 | − | 0.7 |
| P. freudenreichii | ISU-P59 | − | 2.5 |
| P. freudenreichii | ISU-P83 | − | 5.0 |
| P. freudenreichii | ISU-P98 | − | 1.3 |
| P. freudenreichii | ISU-P104 | − | 0.2 |
| P. jensenii | ATCC 4868T | − | >320 |
| P. jensenii | ATCC 14072 | − | >320 |
| P. thoenii | ATCC 4872 | − | >320 |
| P. thoenii | ATCC 4874 | − | >320 |
Abbreviations: ATCC, American Type Culture Collection (Rockville, Md.); DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen; NCDO, National Collection of Food Bacteria (Reading, United Kingdom); ISU, Iowa State University, Ames, Iowa, United States; IFO, the Institute for Fermentation (Osaka, Japan); LMGT, our strain collection; superscript T, type strain.
+, amplification of the expected 350-bp pcfA gene fragment; −, negative PCR amplification result.
MIC values correspond to strains LMGT 2946 and LMGT 2956 precultivated in SLB and MRS, respectively.
Propionicin F activity.
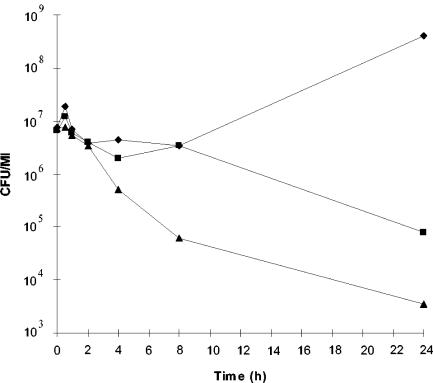
The sensitivity of various PAB strains to propionicin F was determined. Only strains of P. freudenreichii were sensitive to the bacteriocin (Table 3). P. freudenreichii ISU-P104 proved to be the most sensitive strain, with a propionicin F MIC of 0.2 nM. Treatment of the standard indicator strain P. freudenreichii ISU-P59 with propionicin F resulted in inhibition of growth after 60 min and a more than 3-log10 reduction in viable cell counts after 24 h (Fig. 4). In contrast to the ISU-P59 strain, the producer strains were resistant to the bacteriocin. However, during the optimization of propionicin F production, it was discovered that both P. freudenreichii LMGT 2946 and P. freudenreichii LMGT 2956 produced little or no bacteriocin when grown in SLB. We therefore investigated whether cultivation of these strains in SLB would also alter the sensitivity. Indeed, strains LMGT 2946 and LMGT 2956 grown in SLB medium were sensitive to 80 nM propionicin F. In comparison, the sensitivity of P. freudenreichii ISU-P59 was not altered by a change of growth medium. The resistance phenotype of the LMGT 2946 and LMGT 2956 strains was restored upon transfer from SLB to MRS. This strongly suggests that a propionicin F-specific immunity factor is coproduced with the bacteriocin.
FIG. 4.
Viable counts of P. freudenreichii ISU-P59 treated with propionicin F. ▪, 0 nM propionicin F (control); ♦, 10 nM propionicin F; ▴, 40 nM propionicin F.
Transcriptional analysis of pcfA.
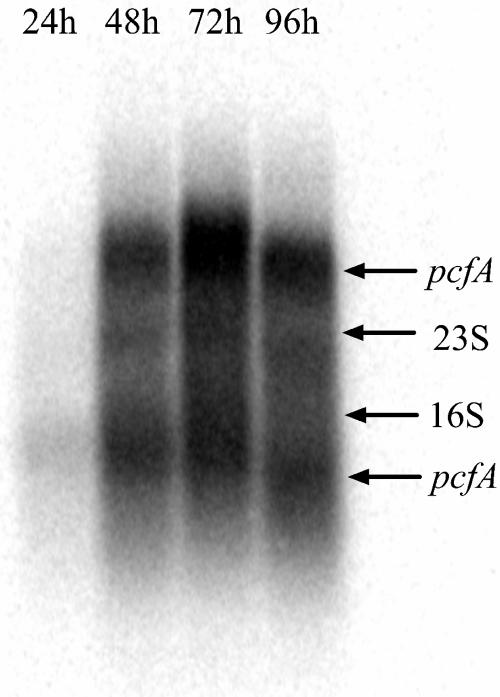
A synthetic radiolabeled probe, covering the propionicin F-encoding region of pcfA, was used to determine the size of the pcfA transcript. Northern blotting revealed the presence of two strong signals of approximately 3.2 and 1.5 kb (Fig. 5). The large transcript indicates that pcfA is cotranscribed with other genes, most probably pcfB, orf5, and pcfC. Furthermore, pcfA displayed a high transcription level during exponential growth, but expression declined when the cells reached stationary phase. This transcription pattern is consistent with the production profile of propionicin F (Fig. 1).
FIG. 5.
Analysis of the pcfA expression profile in propionicin F-producing cells. Total RNA was isolated from P. freudenreichii LMGT 2946 at indicated time points. Approximately 10 μg of RNA from each sample was used in the blot that was hybridized to a pcfA-specific probe.
DISCUSSION
Propionicin F is the first bacteriocin characterized from P. freudenreichii. This bacteriocin is a small, unmodified, and heat-stable peptide that is inhibitory only to strains belonging to the same species as the producer. These properties are common to the majority of bacteriocins from gram-positive bacteria. However, propionicin F also displays some unique features. In contrast to all previously characterized LAB and PAB bacteriocins, propionicin F is negatively charged, with a pI of 4.0. Furthermore, the maturation of propionicin F appears to proceed through unique processing steps. The mature bacteriocin encompasses 43 amino acids in the central region of a putative 255-residue probacteriocin, encoded by the pcfA gene. Examination of the pcfA DNA sequence did not reveal any potential alternative start codons with corresponding ribosome binding sites in the region immediately upstream of the propionicin F-encoding part. No N-terminal leader signal sequence could be identified either. The data thus indicate that maturation of propionicin F involves proteolytic processing, in which the 101 N-terminal and the 111 C-terminal residues of the probacteriocin are removed. The probacteriocins of PAMP from P. jensenii and cytolysin from Enterococcus faecalis undergo two steps of N-terminal proteolytic processing that result in release of the mature bacteriocins (4, 11). However, there have been no previous reports of probacteriocins that are processed at both the N and C termini. Interestingly, pcfC, which appears to be cotranscribed with pcfA, encodes a protein with high similarity to the tripeptidyl aminopeptidase (TAP) from Streptomyces lividans (7). TAP is a member of the S33 peptidase family, a subgroup of serine proteases with amino proline peptidase activity (38). Multiple alignment of PcfC to members of the S33 peptidase family revealed that PcfC contains the Ser-His-Asp catalytic triad pattern characteristic of the S33 peptidases, indicating that PcfC is a member of this family (data not shown). Since the C-terminal position of propionicin F contains a proline, a proline peptidase is required. The presence of such a peptidase in the bacteriocin gene cluster strongly suggests that it performs the C-terminal processing of propionicin F.
The pcfB gene located directly downstream of pcfA encodes a protein with homology to members of the radical SAM superfamily. These proteins catalyze a broad range of reactions, including isomerizations, ring formations, and anaerobic oxidations (22). Notably, albA in the subtilosin A bacteriocin gene cluster encodes a radical SAM protein (35, 42, 49). AlbA is required for the maturation of subtilosin A, possibly by acting as an oxidoreductase that aids the formation of intramolecular thioether linkages (30) or in the condensation process that leads to the ring formation (48). Propionicin F is not a cyclic molecule, and the bacteriocin does not contain any modified residues. However, propropionicin F contains a cysteine residue located adjacent to the N-terminal cleavage site in the probacteriocin. Notably, some radical SAM enzymes, such as AtsB in Pseudomonas aeruginosa, have been shown to catalyze the formation of unstable radical cysteine and serine residues in substrate polypeptides (2). Hence, it is tempting to speculate that PcfB is involved in releasing the N terminus of propionicin F by generating such a radical on the cys101 residue of the probacteriocin.
DNA sequencing of the propionicin F gene cluster also revealed the presence of an ORF, pcfD, encoding a protein with strong homology to various ABC transporters, including the SunT bacteriocin and members of the HlyB family. Such transporters are responsible for secretion of peptide bacteriocins and large proteins such as proteases and RTX toxins (3, 15). Therefore, it is possible that PcfD is specifically involved in transport of the propionicin F.
In LAB, immunity genes are usually coregulated with the bacteriocin and are often located in the vicinity of the bacteriocin-encoding gene (36). There are some results which indicate that bacteriocin-producing PAB also have specific immunity factors (13). Results presented here add further support to this idea. In P. freudenreichii LMGT 2946 and LMGT 2956, the bacteriocin-positive phenotype was consistently associated with immunity. Equally, a bacteriocin-negative phenotype was associated with bacteriocin sensitivity. Hence, it is likely that the propionicin F regulon also contains an immunity gene, but an assigned immunity function has not yet been identified.
In summary, this work presents the molecular characterization of the propionicin F bacteriocin and its corresponding gene. Maturation of propionicin F appears to require a type of processing new to bacteriocins. We suggest that genes located in the vicinity of the propionicin F structural gene participate in this process. The development of genetic tools for P. freudenreichii is still in its infancy. Hopefully, in the near future it will be possible to conduct detailed investigations of propionicin F biosynthesis and immunity.
Acknowledgments
We thank D. Mantzilas and S. Kjæraas (University of Oslo) for performing MS analysis and amino acid sequencing, respectively.
D. A. Brede was funded by the Nordic Industrial Fund grant P98089 and a grant from the Norwegian Research Council. T. Faye was funded by a grant from the Norwegian Research Council.
REFERENCES
- 1.al-Zoreky, N., J. W. Ayres, and W. E. Sandine. 1991. Antimicrobial activity of Microgard against food spoilage and pathogenic microorganisms. J. Dairy Sci. 74:758-763. [DOI] [PubMed] [Google Scholar]
- 2.Beil, S., H. Kehrli, P. James, W. Staudenmann, A. M. Cook, T. Leisinger, and M. A. Kertesz. 1995. Purification and characterization of the arylsulfatase synthesized by Pseudomonas aeruginosa PAO during growth in sulfate-free medium and cloning of the arylsulfatase gene (atsA). Eur. J. Biochem. 229:385-394. [DOI] [PubMed] [Google Scholar]
- 3.Blight, M. A., and I. B. Holland. 1990. Structure and function of haemolysin B,P-glycoprotein and other members of a novel family of membrane translocatorsa. Mol. Microbiol. 4:873-880. [DOI] [PubMed] [Google Scholar]
- 4.Booth, M. C., C. P. Bogie, H. G. Sahl, R. J. Siezen, K. L. Hatter, and M. S. Gilmore. 1996. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol. Microbiol. 21:1175-1184. [DOI] [PubMed] [Google Scholar]
- 5.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 6.Brurberg, M. B., I. F. Nes, and V. G. Eijsink. 1997. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol. Microbiol. 26:347-360. [DOI] [PubMed] [Google Scholar]
- 7.Butler, M. J., C. Binnie, M. A. DiZonno, P. Krygsman, G. A. Soltes, G. Soostmeyer, E. Walczyk, and L. T. Malek. 1995. Cloning and characterization of a gene encoding a secreted tripeptidyl aminopeptidase from Streptomyces lividans 66. Appl. Environ. Microbiol. 61:3145-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornwell, G. G., III, K. Sletten, B. Johansson, and P. Westermark. 1988. Evidence that the amyloid fibril protein in senile systemic amyloidosis is derived from normal prealbumin. Biochem. Biophys. Res. Commun. 154:648-653. [DOI] [PubMed] [Google Scholar]
- 9.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 10.Cummings, C. S., and J. L. Johnson. 1986. Genus I. Propionibacterium Orla-Jensen 1909, p. 1346-1353. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. William and Wilkins, Baltimore, Md. [Google Scholar]
- 11.Faye, T., D. A. Brede, T. Langsrud, I. F. Nes, and H. Holo. 2002. An antimicrobial peptide is produced by extracellular processing of a protein from Propionibacterium jensenii. J. Bacteriol. 184:3649-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faye, T., D. A. Brede, T. Langsrud, I. F. Nes, and H. Holo. 2004. The prevalence of genes encoding the antimicrobial peptides propionicin T1 and protease-activated peptide (PAMP) and their expression in classical Propionibacteria. Appl. Environ. Microbiol. 70:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faye, T., T. Langsrud, I. F. Nes, and H. Holo. 2000. Biochemical and genetic characterization of propionicin T1, a new bacteriocin from Propionibacterium thoenii. Appl. Environ. Microbiol. 66:4230-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 15.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, J. T., A. L. Chopko, and P. D. van Wassenaar. 1992. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Arch. Biochem. Biophys. 295:5-12. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, C. F., I. D. Hiles, G. P. Salmond, D. R. Gill, J. A. Downie, I. J. Evans, I. B. Holland, L. Gray, S. D. Buckel, A. W. Bell, et al. 1986. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323:448-450. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann, K. S., W. 1993. TMBASE—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 19.Holo, H., T. Faye, D. A. Brede, T. Nilsen, I. Ødegård, T. Langsrud, J. Brendehaug, and I. F. Nes. 2002. Bacteriocins of propionic acid bacteria-review. Lait 82:59-68. [Google Scholar]
- 20.Holo, H., O. Nilssen, and I. F. Nes. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huhne, K., L. Axelsson, A. Holck, and L. Krockel. 1996. Analysis of the sakacin P gene cluster from Lactobacillus sake Lb674 and its expression in sakacin-negative Lb. sake strains. Microbiology 142:1437-1448. [DOI] [PubMed] [Google Scholar]
- 22.Jarrett, J. T. 2003. The generation of 5′-deoxyadenosyl radicals by adenosylmethionine-dependent radical enzymes. Curr. Opin. Chem. Biol. 7:174-182. [DOI] [PubMed] [Google Scholar]
- 23.Joerger, M. C., and T. R. Klaenhammer. 1990. Cloning, expression, and nucleotide sequence of the Lactobacillus helveticus 481 gene encoding the bacteriocin helveticin J. J. Bacteriol. 172:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaletta, C., and K. D. Entian. 1989. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J. Bacteriol. 171:1597-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 26.Kuipers, O. P., M. M. Beerthuyzen, P. G. de Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 27.Kuipers, O. P., H. S. Rollema, W. M. de Vos, and R. J. Siezen. 1993. Biosynthesis and secretion of a precursor of nisin Z by Lactococcus lactis, directed by the leader peptide of the homologous lantibiotic subtilin from Bacillus subtilis. FEBS Lett. 330:23-27. [DOI] [PubMed] [Google Scholar]
- 28.Malik, A. C., G. W. Reinbold, and E. R. Vedamuthu. 1968. An evaluation of the taxonomy of Propionibacterium. Can. J. Microbiol. 14:1185-1191. [DOI] [PubMed] [Google Scholar]
- 29.Marugg, J. D., C. F. Gonzalez, B. S. Kunka, A. M. Ledeboer, M. J. Pucci, M. Y. Toonen, S. A. Walker, L. C. Zoetmulder, and P. A. Vandenbergh. 1992. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1.0. Appl. Environ. Microbiol. 58:2360-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marx, R., T. Stein, K. D. Entian, and S. J. Glaser. 2001. Structure of the Bacillus subtilis peptide antibiotic subtilosin A determined by 1H-NMR and matrix assisted laser desorption/ionization time-of-flight mass spectrometry. J. Protein Chem. 20:501-506. [DOI] [PubMed] [Google Scholar]
- 31.Miescher, S. 1999. Antimicrobial and autolytic systems of dairy propionibacteria. Ph.D.a dissertation. ETH, Zürich, Switzerland.
- 32.Miescher, S., M. P. Stierli, M. Teuber, and L. Meile. 2000. Propionicin SM1, a bacteriocin from Propionibacterium jensenii DF1: isolation and characterization of the protein and its gene. Syst. Appl. Microbiol. 23:174-184. [DOI] [PubMed] [Google Scholar]
- 33.Miller, K. W., R. Schamber, O. Osmanagaoglu, and B. Ray. 1998. Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl. Environ. Microbiol. 64:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakano, M. M., G. Zheng, and P. Zuber. 2000. Dual control of sbo-alb operon expression by the Spo0 and ResDE systems of signal transduction under anaerobic conditions in Bacillus subtilis. J. Bacteriol. 182:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 37.Nilsen, T., I. F. Nes, and H. Holo. 2003. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 69:2975-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawlings, N. D., and A. J. Barrett. 1994. Families of serine peptidases. Methods Enzymol. 244:19-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehberger, T. G., and B. A. Glatz. 1990. Characterization of Propionibacterium plasmids. Appl. Environ. Microbiol. 56:864-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Risoen, P. A., M. B. Brurberg, V. G. Eijsink, and I. F. Nes. 2000. Functional analysis of promoters involved in quorum sensing-based regulation of bacteriocin production in Lactobacillus. Mol. Microbiol. 37:619-628. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional charac-terization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tagg, J. R., A. S. Dajani, and L. W. Wannamaker. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tichaczek, P. S., R. F. Vogel, and W. P. Hammes. 1994. Cloning and sequencing of sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH 673. Microbiology 140:361-367. [DOI] [PubMed] [Google Scholar]
- 45.Venema, K., J. Kok, J. D. Marugg, M. Y. Toonen, A. M. Ledeboer, G. Venema, and M. L. Chikindas. 1995. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol. Microbiol. 17:515-522. [DOI] [PubMed] [Google Scholar]
- 46.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 47.Woboro, R. W., M. J. van Belkum, M. Sailer, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1995. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J. Bacteriol. 177:3143-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng, G., R. Hehn, and P. Zuber. 2000. Mutational analysis of the sbo-alb locus of Bacillus subtilis: identification of genes required for subtilosin production and immunity. J. Bacteriol. 182:3266-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, G., L. Z. Yan, J. C. Vederas, and P. Zuber. 1999. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol. 181:7346-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]