Abstract
Genogroup I noroviruses from five genetic clusters and genogroup II noroviruses from eight genetic clusters were detected in stool extracts using degenerate primers and single-tube, real-time reverse transcription-PCR (RT-PCR) with SYBR Green detection. Two degenerate primer sets, designated MON 431-433 and MON 432-434, were designed from consensus sequences from the major clusters of norovirus based on the RNA-dependent RNA polymerase region of the norovirus genome. Viruses were extracted from stool samples within 20 min using a viral RNA extraction kit. Real-time RT-PCR for noroviruses generated semiquantitative results by means of the cycle threshold data and dilution endpoint standard curves. Presumptive product verification was achieved by evaluation of first-derivative melt graphs. Multiple clusters of noroviruses were identified simultaneously in a multiplex fashion by virtue of slight differences in melting temperature. The detection of 13 different genetic clusters suggests that the MON primers may serve as universal primers for most, if not all, of the noroviruses in a multiplex assay. Our technique provides a framework for broad application of real-time RT-PCR in clinical, environmental, and food testing laboratories for a wide range of noroviruses.
In spite of over 30 years of intensive research on the noroviruses, illnesses continue relatively unabated. Recent outbreaks on cruise ships (5) and in hospitals (3), schools (14), nursing homes (11), and day care centers (7) highlight the importance of the noroviruses as enteric pathogens. Each year, an estimated 23 million cases of norovirus illness occur in the United States, of which 40% are believed to be food-related (12), making the noroviruses the most common cause of acute gastroenteritis (6). Although the economic costs of these outbreaks have not been measured, the financial impact is likely of staggering proportions. Rapid spread of viruses and their persistence in some environments have made the control of outbreaks difficult and, at times, seemingly impossible. Among the limitations experienced in curtailing outbreaks is the failure of rapid diagnosis. Such failures are, in large part, attributable to the complexity of norovirus assays. Since the viruses cannot be propagated in cell culture, molecular methods have been the assays of choice for their detection.
The noroviruses are a broad group of enteric pathogens with high sequence diversity. Molecular analysis for noroviruses has been hampered by the need for multiple PCR primers to detect the many genetic variants. These variants include five genetic clusters within the genogroup I (GI) noroviruses and at least eight clusters within GII. Even within the most conserved regions of the norovirus genome, some sequence divergence exists. Consequently, multiple primers and multiple reverse transcription (RT)-PCRs have been required to test for the noroviruses. Recently, a paper describing the use of a broadly reactive degenerate primer set was published for the real-time RT-PCR detection of GI and GII noroviruses from stool samples using TaqMan probes (9). These degenerate primers were obtained from the ORF1-ORF2 junction of the norovirus genome, and the probe was designed to bind to a sequence within ORF1. In the present study, we developed an alternate procedure using degenerate primers from the RNA-dependent RNA polymerase region encoded by ORF1 of the norovirus genome and with SYBR Green detection in place of TaqMan probes. SYBR Green is a fluorescent dye which intercalates into double-stranded amplicons, so fluorescence does not depend on probe binding to widely divergent sequences as it does for TaqMan probes. First-derivative melt curves, made possible by the use of SYBR Green, were evaluated as a means to verify products. In a recent study, members of our laboratory demonstrated the use of SYBR Green real-time RT-PCR and specific primers to detect and semiquantitate the 8FIIa strain of classical GI, cluster 1 (GI/1) Norwalk virus (15), which is a commonly used laboratory strain. Here we expand that study by providing analytical methods to detect 13 different genetic clusters of GI and GII noroviruses.
MATERIALS AND METHODS
Viruses.
Genogroup I and II noroviruses were obtained from clinical specimens submitted for virus identification to the Centers for Disease Control and Prevention. Viruses within these stool samples were classified as GI genetic clusters 1 to 5 and GII clusters 1 to 8 based on nucleotide sequence analyses (1) performed on PCR products from region A or region B or both, as previously described (4). The prototypical GI virus is Norwalk virus (10), while the GII prototype is Snow Mountain virus. The 8FIIa strain of Norwalk virus, obtained from a volunteer study (8, 15), was used to optimize real-time RT-PCR conditions.
Processing of stool samples.
Approximately 50 to 80 μg of stool samples containing GI and GII noroviruses was weighed, diluted 1:10 in nuclease-free H2O, and vortexed for 30 s. Samples were clarified by centrifugation at 6,800 × g for 10 min at room temperature. Viral RNA was extracted from 140 μl of the supernatant with the QIAamp Viral RNA kit (QIAGEN, Valencia, Calif.) according to the column centrifugation procedure described by the manufacturer. In essence, 140 μl of each 10% stool suspension was denatured, adsorbed to a silica column, washed, and then eluted to give a final volume of 60 μl. Purification required <20 min to perform. The RNA extracts were stored on ice, and 1 μl or less was assayed by real-time RT-PCR on the day of preparation. To determine the feasibility of the virus extraction and assay methods for the detection of noroviruses in field samples, 24 stool samples containing noroviruses were supplied by the Delaware Public Health Laboratory, the Florida Department of Health, and the Rhode Island Department of Health. Samples were extracted with the QIAamp Viral RNA kit, as described above, and 1 μl was assayed for norovirus by real-time RT-PCR.
Real-time RT-PCR.
Real-time RT-PCR was performed using a SYBR Green Quantitative RT-PCR kit (catalog no. QR0100; Sigma Chemical Co., St. Louis, Mo.) consisting of 2× SYBR Green Taq ReadyMix (with jump-start Taq DNA polymerase, SYBR Green I fluorescent dye, a 0.4 mM concentration of each deoxynucleoside triphosphate, and 6 mM MgCl2), DuraScript reverse transcriptase (20 U/μl), and 10× PCR buffer. The RT-PCR master mixes were made by combining 12.5 μl of 2× SYBR Green Taq ReadyMix, 1 μl of DuraScript reverse transcriptase (after a 1:10 dilution of the DuraScript reverse transcriptase was carried out with 1× PCR buffer), 0.5 μl of RNase inhibitor (5 U; Invitrogen, Carlsbad, Calif.), degenerate primers, and nuclease-free H2O to give a total volume of 24 μl for each reaction tube. Degenerate, multiplex primers for the GI (MON 432 and 434) and GII (MON 431 and 433) noroviruses (Table 1) were added to final concentrations of 640 nM each. In preliminary studies with Norwalk virus, primer concentrations of 160 to 1280 nM each were evaluated, and 640 nM gave optimal results without excessive primer dimer formation. All four degenerate primers (MON 431 to 434) were combined in a single reaction tube for the detection of either GI or GII noroviruses. The master mixes were maintained on ice at all times prior to real-time RT-PCR.
TABLE 1.
Primer sets evaluated for real-time RT-PCR
| Norovirus genogroup | Primer designation | Sequence (description)a | Amplicon size (bp) |
|---|---|---|---|
| I | MON 432 | 5′ TGG ACI CGY GGI CCY AAY CA 3′ (RNA sense) | |
| MON 434 | 5′ GAA SCG CAT CCA RCG GAA CAT 3′ (cDNA sense) | 213 | |
| II | MON 431 | 5′ TGG ACI AGR GGI CCY AAY CA 3′ (RNA sense) | |
| MON 433 | 5′ GGA YCT CAT CCA YCT GAA CAT 3′ (cDNA sense) | 213 |
Ambiguity code: I, inosine; R, purine (A/G); Y, pyrimidine (C/T); S, strong (C/G).
To chilled 25-μl Smart Cycler tubes (Cepheid, Sunnyvale, Calif), 24 μl of cold master mix was added followed by 1 μl of purified RNA from stool. The mix was forced into the bottom of each tube by centrifugation, as recommended by the manufacturer, for 5 s, and the tubes were immediately placed into a Smart Cycler block (Cepheid). Cycling parameters were optimized, including the concentration of primer used; the times and temperatures for the annealing, extension, and denaturation steps; and the temperature of the optical read step. The optical read step was evaluated to ensure that the read would be performed at the highest temperature possible to eliminate detection of primer dimer or spurious products.
Optics graphs, showing the fluorescence intensity of each reaction plotted against PCR cycles, were obtained for each run. Optics graphs indicate the cycle threshold (Ct), which is the number of cycles required for the fluorescence to cross a preassigned fluorescence threshold. A threshold of 30 fluorescence units, the default value for the Cepheid Smart Cycler, was used throughout this study. Samples containing the highest initial levels of virus have the lowest Ct, and those with the lowest levels of virus have the highest Ct. PCR was performed for 40 cycles; therefore, the Ct has an upper limit of 40 cycles. Melt graphs and first-derivative melt graphs were obtained immediately after completion of the PCR using the software and graphics programs provided with the Smart Cycler and were produced by plotting the fluorescence intensity against temperature as the temperature was increased from 60 to 95°C at 0.2°C/s.
Generation of standard curves.
Dilution endpoint standard curves (15) were determined and compared for GI and GII noroviruses by performing real-time RT-PCR with 10-fold dilutions of stool extract followed by the assay of 1 μl of 100 to 10−5 dilutions of each extract. The Ct value obtained from the assay of each dilution was used to plot a standard curve by assigning a value of 1 RT-PCR unit (RT-PCRU) to the highest dilution showing a positive Ct and 10, 100, and 1,000 RT-PCRU sequentially to the lower dilutions. Data were used to plot standard curves. In cases where stool samples contained low levels of virus, twofold dilutions were made. The highest dilution giving a positive result was assigned a value of 1 RT-PCRU, lower dilutions were sequentially multiplied by 2, and the RT-PCRU values were used to plot standard curves. Samples were considered norovirus positive when the Ct values were <40 and the melting temperature (Tm) of the amplicon was between 78 and 85°C, as determined by the first-derivative melt curves. Stool extracts were evaluated for the presence of real-time RT-PCR inhibitors by evaluation of the dilution endpoint standard curves. Inhibitors were considered present if the Ct for 1 μl of undiluted extract was appreciably off the curve (line) formed from the real-time RT-PCR of higher dilutions.
RESULTS
Optimization of real-time RT-PCR.
Optimal conditions for the real-time RT-PCR of the noroviruses were determined using GI/1 virus stock and all four MON primers combined. Optimal conditions were determined to be 640 nM of each primer; RT at 50°C for 30 min; Taq activation at 95°C for 15 min; and 40 cycles of PCR with denaturation at 95°C for 1 min, annealing at 50°C for 1 min, extension at 60°C for 1 min, and an optical read step at 77°C for 6 s, using the Sigma SYBR Green RT-PCR kit. A first-derivative melt curve, programmed to run immediately after the final PCR cycle, provided verification of amplicon purity. In preliminary studies, a QIAGEN SYBR Green RT-PCR kit gave only a weak fluorescence signal, even when optimized (data not shown).
Primer dimer or spuriously produced products were frequently observed when testing control stool samples without norovirus. This was likely due to the high concentration of primer used (640 nM), the high degeneracy of the primers, and the low annealing and extension temperatures employed. Adjusting the temperature of the optical read to 77°C, just below the Tm of the desired amplicon but above the Tm of the nonspecific fluorescent signal, allowed the Ct values to be used in a semiquantitative fashion. Positive results were obtained with negative controls (NC) from spuriously produced products or primer dimers and were readily recognized as negative by the absence of product within the normal melting temperatures for norovirus amplicon on first-derivative melt graphs. In such cases, the Ct values were discounted.
Stool analyses.
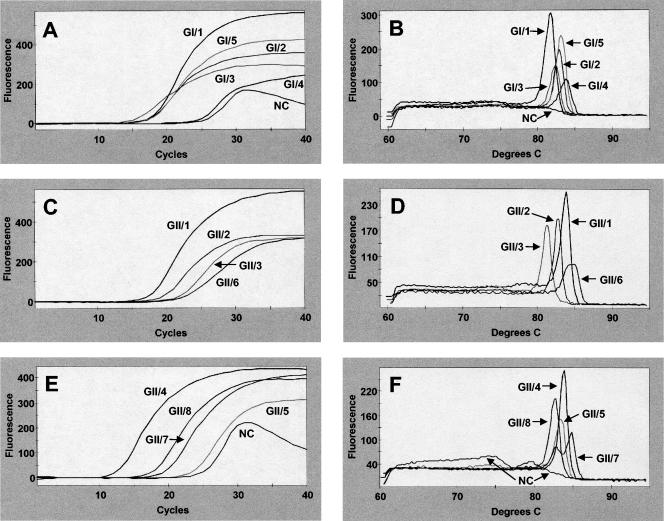
The primers MON 431 to 434 amplified all of the GI and GII noroviruses (Fig. 1). All five GI noroviruses amplified with high fluorescence intensity as seen on the optics graphs (Fig. 1A). Corresponding first-derivative melt graphs (Fig. 1B) show differences in Tm for the various cluster viruses ranging from 81.84 to 83.79°C (Table 2). Optics graphs for the eight GII cluster viruses (Fig. 1C and E) and their corresponding first-derivative melt graphs (Fig. 1D and F) show clear-cut amplification. Melting temperatures for the GII cluster viruses range from 81.18 to 84.90°C (Table 2). Because the Tms of the amplicons varied within each genogroup, the Tms cannot be used to identify the genogroup in which the virus resides. The NC, shown in Fig. 1A and E, give a positive Ct value because of the presence of some spurious fluorescence. The more prominent peak for the NC in Fig. 1F represents primer dimer, while the smaller peak is spuriously produced amplicon. These peaks are easily discounted as true amplicon by viewing the first-derivative melt curve. No peak was present for the NC around the Tms of the desired norovirus amplicons (Fig. 1B and F). Gel electrophoresis confirmed the presence of a 213-bp amplicon for each of the 13 cluster noroviruses but only primer dimer for the negative control (data not shown). In each case where virus was present, the melt graph shows preference for amplicon production over primer dimer or spurious products, as evidenced by a stable baseline between 60 and 77°C on first-derivative melt graphs (Fig. 1B, D, and F).
FIG. 1.
Results of real-time RT-PCR amplification of genogroup I (A and B) and II (C to F) noroviruses and NC (F) from stool extracts. Optics graphs (A, C, and E) show amplicon production, and first-derivative melt graphs (B, D, and F) show corresponding melting temperatures for each of 13 norovirus clusters.
TABLE 2.
Noroviruses amplified by real-time RT-PCR of stool extracts and accompanying Tms of the amplicons, slopes of the standard curves, correlation coefficients (R2) of the standard curves, and the presence of inhibitors in 1 μl of undiluted extract
| Genogroup | Clustera | Representative virus | Tm (°C) | Slope | R2 | Inhibitorsb |
|---|---|---|---|---|---|---|
| I | 1 | Norwalk | 81.84 | −0.344 | 0.999 | No |
| 2 | Southampton | 83.00 | −0.447 | 0.997 | No | |
| 3 | Desert Shield | 82.39 | −0.357 | 0.984 | Yes | |
| 4 | Chiba | 83.79 | −0.803 | 0.974 | No | |
| 5 | Musgrove | 83.17 | −0.388 | 0.981 | No | |
| II | 1 | Hawaii | 83.82 | −0.390 | 0.970 | Yes |
| 2 | Snow Mountain | 82.67 | −0.428 | 0.991 | No | |
| 3 | Toronto | 81.18 | −0.459 | 1.000 | No | |
| 4 | Lordsdale | 83.96 | −0.400 | 0.968 | Yes | |
| 5 | Hillingdon | 83.34 | −0.364 | 0.998 | Yes | |
| 6 | Seacroft | 84.90 | −0.669 | 1.000 | Yes | |
| 7 | Leeds | 84.90 | −0.590 | 1.000 | Yes | |
| 8 | Amsterdam | 82.81 | −0.337 | 0.995 | No |
Genetic clusters are based on criteria described by Ando et al. (1).
A “yes” indicates that the real-time RT-PCR reaction was partially inhibited by the use of 1 μl of stool extract in the real-time RT-PCR reaction mix.
Optical read temperatures.
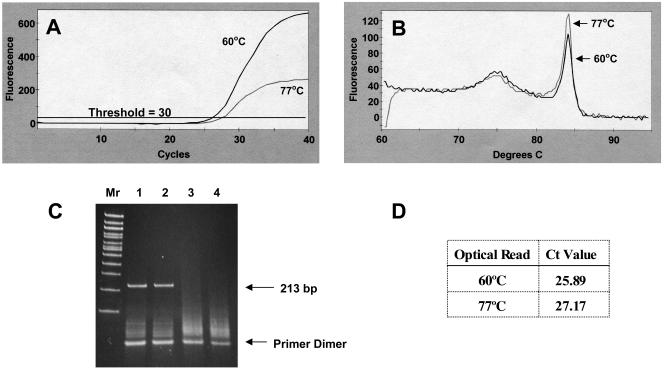
A comparison of results obtained with optical reads at 60 and 77°C is shown for stool extract containing a low norovirus titer (Fig. 2A). First-derivative melt curves (Fig. 2B) show essentially the same products with a characteristic early peak associated with the formation of primer dimer and a later peak representing authentic amplicon. Primer dimer peaks are usually observed in negative controls (Fig. 1F). The Ct value obtained from an optical read taken at 60°C is lower than that taken at 77°C (Fig. 2D), indicating the presence of more fluorescence because of primer dimer presence at 60°C (Fig. 2A). Although PCR amplifications produce the same products regardless of the temperature of the optical read (Fig. 2B and C), the 77°C optical read avoids the detection of fluorescence from primer dimer, resulting in more accurate Ct values. The presence of authentic product at 213 bp and lower-molecular-mass primer dimer can be seen in an electrophoretic gel (Fig. 2C). Negative controls show the presence of only primer dimer (Fig. 2C, lanes 3 and 4). Since dilution endpoint standard curves require real-time RT-PCR of highly diluted samples, often resulting in increases in the formation of primer dimer, it is crucial in quantitative studies to employ an elevated optical read temperature when using SYBR Green. High correlation coefficients for the dilution endpoint standard curves (see below) for the various norovirus clusters are attributable to the use of an optical read temperature above the melting temperature of potential primer dimer.
FIG. 2.
Comparison of (A) optics graphs, (B) first-derivative melt curves, (C) electrophoretic gel, and (D) Ct values for low-titer norovirus RNA amplification by real-time RT-PCR with optical reads performed at 60 and 77°C. (C) Electrophoretic gel with 100-bp ladder (Mr). Lanes: 1, amplicon produced with an optical read at 60°C; 2, amplicon with an optical read at 77°C; 3 and 4, negative controls with optical reads performed at 60 and 77°C, respectively. The 213-bp amplicon represents authentic norovirus amplicon, and the lower-mass products represent primer dimers. The optical read temperatures and corresponding Ct values derived from panel A are shown in the table in panel D.
Dilution endpoint standard curves.
Standard curves were obtained for the GI and GII noroviruses. The slopes and correlation coefficients (R2) are shown in Table 2. Some inhibition in the anticipated amplicon was experienced for some samples when 1 μl of direct extract was tested (Table 2), even though the RNA had been purified with the QiaAmp Viral RNA kit. This inhibition did not jeopardize virus detection but did reduce the amount of amplicon expected. In cases where inhibition occurred, these points were omitted when determining the standard curve. For quantitative analyses, dilutions of 1:10 and higher should be assayed. The high R2 values for the standard curves (Table 2) indicate high assay precision. Slopes obtained from the standard curves were not sufficiently different to distinguish between the genogroups.
Field samples.
Stool samples containing noroviruses were obtained from state testing laboratories in Delaware, Florida, and Rhode Island and were extracted and assayed as described above. Of the 24 specimens tested, 100% gave positive results (Table 3). First-derivative melt curve data produced single peaks with melting temperatures within the range for noroviruses (Table 3).
TABLE 3.
Ct values and Tms for noroviruses detected by real-time RT-PCR of stool samples obtained from health department laboratories
| Sample origin | Ct | Tm (°C) |
|---|---|---|
| Delaware | 21.99 | 84.04 |
| 26.10 | 83.88 | |
| 24.17 | 83.34 | |
| 26.23 | 83.94 | |
| 21.36 | 83.99 | |
| 26.09 | 83.90 | |
| 21.66 | 83.90 | |
| 20.14 | 83.67 | |
| 26.11 | 83.81 | |
| 21.46 | 84.21 | |
| 23.33 | 83.92 | |
| 18.10 | 83.80 | |
| 25.83 | 84.08 | |
| 22.16 | 83.60 | |
| Florida | 21.72 | 83.00 |
| Rhode Island | 22.74 | 85.12 |
| 18.13 | 84.13 | |
| 26.42 | 83.09 | |
| 25.58 | 83.23 | |
| 20.44 | 82.96 | |
| 23.73 | 83.18 | |
| 17.42 | 83.38 | |
| 20.26 | 83.54 | |
| 16.62 | 83.35 |
Stool samples containing mixed noroviruses.
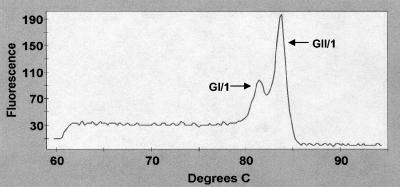
Stool samples containing a mixture of GI and GII noroviruses were tested by real-time RT-PCR and occasionally produced multiple peaks on first-derivative melt graphs. Figure 3 shows one example where two stool samples, one containing GI/1 and the other containing GII/1 noroviruses, were combined followed by viral RNA extraction and assay. These peaks allow the use of melt graphs in a multiplex context, rather than the conventional method of multiplexing, which involves different amplicon sizes.
FIG. 3.
Detection of two noroviruses in stool samples by first-derivative melt curves. GI/1 and GII/1 noroviruses, representing Norwalk and Hawaii viruses, respectively, are simultaneously detected in a single stool extract.
DISCUSSION
Noroviruses have been divided into two genogroups, each of which contains multiple genetic clusters. The difficulty in assaying viruses from all these clusters stems from the wide divergence in nucleotide sequences. The norovirus assay by RT-PCR has been tedious because of the need for multiple primer sets to identify viruses from the various genetic clusters. We demonstrated the utility of real-time RT-PCR for the detection of five GI cluster and eight GII cluster noroviruses by using SYBR Green fluorescence and highly degenerate primers. These clusters represent the vast majority of the noroviruses circulating in the world today (4). To our knowledge, no other real-time RT-PCR technique has been universally successful in detecting such a wide variety of noroviruses.
In a recent paper, Kageyama et al. (9) reported the development and use of degenerate primers designed around the capsid sequences of norovirus to detect viruses in stool samples obtained from various clinical settings. Fluorescence detection was achieved using TaqMan probes. It is unclear which clusters could be detected with their probes. We chose SYBR Green over TaqMan probes because of the high diversity of the norovirus genome and the knowledge that SYBR Green would intercalate into the norovirus amplicon regardless of sequence. TaqMan probes are oligonucleotides designed to bind to complementary target sequences present within the PCR amplicon. Any mismatching, due to nucleotide diversity within the amplicon, could interfere with probe binding and assay sensitivity.
A recently published method for the multiplex detection of GI and GII noroviruses, human astroviruses, and enteroviruses using SYBR Green was reported successful in amplifying each virus (2); however, the method was shown to detect only one GI and one GII cluster norovirus, namely the GI/2 (Southampton) and GII/4 (Lordsdale) viruses. Another study used SYBR Green to detect noroviruses, but it did not evaluate the detection of different genetic clusters (13). The very low degeneracy or lack of degeneracy for these norovirus primers, or for the previously mentioned TaqMan probes, make it doubtful that these methods would detect noroviruses from a broad array of genetic clusters.
Our method utilized one-step, hot-start RT-PCR with thermostable DNA polymerase. One-step protocols simplify the method and reduce the likelihood of cross-contamination of samples or inactivation of newly transcribed RNA by RNases during subsequent transfer to PCR tubes. The incorporation of RNase inhibitor into the reaction mixture provides protection for naked viral RNA until cDNA copies are made. Amplification of the GI and GII noroviruses by real-time RT-PCR required a four-step PCR cycle which included an optical read step set at a temperature above the melting temperature of primer dimers. Ordinarily, real-time RT-PCR is more rapid than standard RT-PCR; however, the highly degenerate MON primers required more extended periods for denaturation, annealing, and extension, comparable to those used in standard RT-PCR. In addition, the annealing and extension temperatures were intentionally kept low, due to the high degeneracy of the primers and the need for primer-template binding where some mismatching occurred. The low assay stringency used in this study would foster the amplification of close relatives of these noroviruses and may facilitate the detection of viruses assigned to other clusters as they are discovered.
Although applicable for real-time detection methods, the use of SYBR Green has been limited because it intercalates not only into the desired amplicon but into primer dimer and spuriously produced products as well. Our optics graphs demonstrated the presence of fluorescent product not only from amplicon, but also from primer dimer formation. In fact, real-time RT-PCRs where no virus was present still produced some fluorescence from primer dimer (Fig. 1F, NC). The melting curves in Fig. 1 and 2 showed that the primer dimers or other artifactual amplicons had Tms below those of the norovirus amplicons. Consequently, a brief (6-s) optical read step at 77°C was incorporated after the normal 60°C extension step for each cycle of PCR. This ensured that primer dimer and spurious products with Tms less than that of the amplicon were fully melted before the optical read, so that SYBR Green fluorescence would only be attributable to the double-stranded amplicon. Assay specificity was enhanced by performing the optical read at this elevated temperature. When viral RNA is limited and the formation of primer dimer is likely (Fig. 2), it is essential to use an elevated optical read temperature to prevent the detection of fluorescence from primer dimer.
First-derivative melt curves are beneficial in assessing the presence of multiple noroviruses in a sample; asymmetry of this peak could indicate the presence of samples contaminated with multiple noroviruses. Stool samples containing two noroviruses with appreciably different Tms gave two distinct peaks on first-derivative melt curves (Fig. 3). The ability to simultaneously discriminate the presence of noroviruses from two or more clusters will depend on the melting temperatures of the individual viruses. Sufficient differences are required to produce separate peaks, as was the case with GI/1 and GII/1 noroviruses (Fig. 3). Multiple amplicons were apparently detected in the GII/7 stool sample (Fig. 1F), suggesting the presence of two noroviruses in some clinically derived specimens; however, the dilution endpoint standard curve for this sample gave a perfect R2 value of 1.000. It is possible that one of the two viruses was present in very limited amounts not detectable by real-time RT-PCR of diluted samples. In epidemiological investigations, multiple peaks may signify a general source of contamination, such as molluscan shellfish contaminated with municipal sewage. In such cases, a consumer could simultaneously become infected with more than one virus. Both GI and GII noroviruses were recently detected in stool samples from 10 of 80 (12.5%) individuals involved in outbreaks of acute gastroenteritis (9). We tested stool samples obtained from health department laboratories for norovirus, and all produced amplicon with a single peak on first-derivative melt graphs.
Universal primers combined with real-time RT-PCR facilitated the rapid detection of a wide range of noroviruses. It is anticipated that these methods will foster the development of improved diagnostic capabilities in clinical settings as well as enhanced monitoring capabilities for food, environmental, and regulatory laboratories.
Acknowledgments
We thank Suzanne Beard and Leslie Hadley, Centers for Disease Control and Prevention, for providing the collection of representative norovirus samples. We also thank Jong-Ho Jean (Delaware Public Health Laboratory), Deanna Simmons (Rhode Island Department of Health), and Lillian Stark (Florida Department of Health) for providing stool samples containing noroviruses.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 2.Beuret, C. 2004. Simultaneous detection of enteric viruses by multiplex real-time RT-PCR. J. Virol. Methods 115:1-8. [DOI] [PubMed] [Google Scholar]
- 3.Browne, A., and A. Dalby. 2003. Major incidents. Norwalk on the wild side. Health Serv. J. 113:26-27. [PubMed] [Google Scholar]
- 4.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 5.Flemmer, M., and E. C. Oldfield III. 2003. The agony and the ecstasy. Am. J. Gastroenterol. 98:2098-2099. [DOI] [PubMed] [Google Scholar]
- 6.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181(Suppl. 2):S254-S261. [DOI] [PubMed] [Google Scholar]
- 7.Gotz, H., K. Ekdahl, J. Lindback, B. de Jong, K. Hedlund, and J. Giesecke. 2001. Clinical spectrum and transmission characteristics of infection with Norwalk-like virus: findings from a large community outbreak in Sweden. Clin. Infect. Dis. 33:622-628. [DOI] [PubMed] [Google Scholar]
- 8.Graham, D. Y., X. Jiang, T. Tanaka, A. R. Opekun, H. P. Madore, and M. K. Estes. 1994. Norwalk virus infection of volunteers: new insights based on improved assays. J. Infect. Dis. 170:34-43. [DOI] [PubMed] [Google Scholar]
- 9.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapikian, A. Z., R. G. Wyatt, R. Dolin, T. S. Thornhill, A. R. Kalica, and R. M. Chanock. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 10:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marx, A., D. K. Shay, J. S. Noel, C. Brage, J. S. Bresee, S. Lipsky, S. S. Monroe, T. Anso, C. D. Humphrey, E. R. Alexander, and R. I. Glass. 1999. An outbreak of acute gastroenteritis in a geriatric long-term-care facility: combined application of epidemiological and molecular diagnostic methods. Infect. Control Hosp. Epidemiol. 20:306-311. [DOI] [PubMed] [Google Scholar]
- 12.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Chapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and deaths in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, I., R. Gunson, and W. F. Carman. 2002. Norwalk like virus by light cycler PCR. J. Clin. Virol. 25:231-232. [DOI] [PubMed] [Google Scholar]
- 14.Moe, C. L., W. A. Christmas, L. J. Echols, and S. E. Miller. 2001. Outbreaks of acute gastroenteritis associated with Norwalk-like viruses in campus settings. J. Am. Coll. Health 50:57-66. [DOI] [PubMed] [Google Scholar]
- 15.Richards, G. P., M. A. Watson, and D. H. Kingsley. 2004. A SYBR Green, real-time RT-PCR method to detect and quantitate Norwalk virus in stools. J. Virol. Methods 116:63-70. [DOI] [PubMed] [Google Scholar]