Abstract
Proteasomes are energy-dependent proteases that are central to the quality control and regulated turnover of proteins in eukaryotic cells. Dissection of this proteolytic pathway in archaea, however, has been hampered by the lack of substrates that are easily detected in whole cells. In the present study, we developed a convenient reporter system by functional expression of a green fluorescent protein variant with C-terminal fusions in the haloarchaeon Haloferax volcanii. The levels of this reporter protein correlated with whole-cell fluorescence that was readily detected in culture. Accumulation of the reporter protein was dependent on the sequence of the C-terminal amino acid fusion, as well as the presence of an irreversible, proteasome-specific inhibitor (clasto-lactacystin β-lactone). This inhibitor was highly specific for H. volcanii 20S proteasomes, with a Ki of ∼40 nM. In contrast, phenylmethanesulfonyl fluoride did not influence the levels of fluorescent reporter protein or inhibit 20S proteasomes. Together, these findings provide a powerful tool for the elucidation of protein substrate recognition motifs and the identification of new genes which may be involved in the proteasome pathway of archaea.
The tree of life is divided into three main evolutionary lineages, the domains Archaea, Bacteria, and Eucarya (31). Despite the fact that archaea are prokaryotes, their information machinery (DNA replication, transcription, and translation) and protein turnover and quality control systems (proteasomes and chaperones) are more closely related to their eucaryal counterparts than to their bacterial counterparts. In fact, archaeal homologs of eucaryal 26S proteasomes have greatly assisted in understanding basic mechanisms of energy-dependent proteolysis (2, 20). The elaborate ubiquitin-proteasome system of eukaryotes (15) and the tmRNA-ssrA system of bacteria (30), however, are not conserved in archaea, and the mechanisms of targeting proteins for destruction remain enigmatic.
Genome sequencing has provided insights into the central role that proteasomes are likely to play in archaeal physiology. The proteolytic 20S core is universally distributed in Archaea and is postulated to be the major cytosolic protease responsible for protein quality control and regulated proteolysis in this domain (19). Global analysis of Halobacterium sp. strain NRC-1 (1) revealed little correlation between the levels of mRNA and protein, suggesting that posttranscriptional control mechanisms are of central importance. Unfortunately, linking the archaeal proteasome to posttranscriptional control and other in vivo functions has been hampered by the lack of substrate proteins that are readily detected in whole cells.
Green fluorescent protein (GFP) and its variants are often used as reporters for investigating proteolysis. The advantages of using these proteins as substrates are multifaceted and include ease of detection and little, if any, perturbation to the overall structure upon addition of N- or C-terminal amino acid residues (14). Recently, GFP was used to assay the activity of a reconstituted proteasome system consisting of the Methanocaldococcus jannaschii proteasome-activating nucleotidase (PAN) and the Thermoplasma acidophilum 20S proteasome (3, 4, 22). Energy-dependent proteolysis of GFP was dependent on the presence of C-terminal residues identical to the Escherichia coli ssrA tag (AANDENYALAA) (30). Although ssrA homologs are not predicted based on archaeal genomics, the in vitro study raised the possibility that the amino acid sequence of the ssrA tag has structural similarity to motifs of proteins with short half-lives in archaeal cells. The absence of a suitable GFP expression system, however, precluded examination of this in vivo.
Here we describe the generation and characterization of a GFP-based reporter system that enabled quantification of proteasome-dependent proteolysis in archaeal cells. The haloarchaeon Haloferax volcanii was chosen as the host based on its ease of culture, established genetic exchange system, and synthesis of two 20S proteasome isoforms (17, 29) and PAN proteins (25). Although the concentration of inorganic ions (2.5 to 6 M) in the cytosol of haloarchaea can limit the synthesis of nonhalophilic proteins in active forms (21, 23), purified GFP is resistant to many denaturants (e.g., 6 M urea) (27), suggesting that it or derivatives of it may be suitable for expression. A soluble modified derivative of GFP (Phe99Ser, Met153Thr, and Val163Ala) (8) with a red-shifted mutation (Ser65Thr) (smRS-GFP) (9) was found to be ideal for use as a fluorescent reporter protein to examine the proteolytic activity of 20S proteasomes in H. volcanii.
MATERIALS AND METHODS
Materials.
Organic and inorganic analytical grade chemicals were obtained from Fisher Scientific (Atlanta, Ga.) and Sigma Chemical Co. (St. Louis, Mo.) unless otherwise indicated. clasto-Lactacystin β-lactone was obtained from Boston Biochem (Cambridge, Mass.). Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs (Beverly, Mass.). Desalted oligonucleotides were obtained from Operon Technologies, Inc. (Alameda, Calif.) and Integrated DNA Technologies (Coralville, Iowa).
Strains, media, and plasmids.
The strains, oligonucleotide primers, template DNA, and plasmids used are summarized in Table 1. H. volcanii DS70 and E. coli BL21(DE3) were used as hosts for synthesis of recombinant fluorescent protein derivatives. E. coli strains were grown in Luria-Bertani medium, and H. volcanii strains were grown in ATCC 974 medium at 37°C and 200 rpm. Media were supplemented with 100 mg of ampicillin per liter, 50 mg of kanamycin per liter, and/or 0.1 mg of novobiocin per liter as needed. The fidelity of all PCR-amplified products was confirmed by DNA sequencing by using the dideoxy termination method (26) with Perkin-Elmer/Applied Biosystems and LICOR automated DNA sequencers (DNA Sequencing Facilities, Interdisiplinary Center for Biotechnology Research and Department of Microbiology and Cell Science, University of Florida).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Phenotype, genotype, description, and/or PCR primers | Source or reference(s) |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | F−ompT [lon]hsdSB(rB− mB−) (E. coli B strain) with DE3, a λ prophage carrying the T7 RNA polymerase gene | Novagen |
| DH5α | F−recA1 endA1 hsdR17(rk− mk+) supE44 thi-1 gyrA relA1 | New England Biolabs |
| GM2163 | F−ara-14 leuB6 flntA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 rpsL 136 dam 13::Tn9 xylA5 mtl-1 thi-1 mcrB1 hsdR2 | New England Biolabs |
| H. volcanii DS70 | Cured of plasmid pHV2 | 28 |
| Plasmids | ||
| pBAP5010 | Apr Nvr; H. volcanii-E. coli shuttle expression vector | 16, 17 |
| pEGFP | Apr; GenBank accession no. U76561 | Clontech |
| pSMGFP | Apr; GenBank accession no. U70495 | 9 |
| pSMRSGFP | Apr; GenBank accession no. U70496 | 9 |
| pSMBFP | Apr; GenBank accession no. U70497 | 9 |
| pGFP-11 | Apr; pET22b-derived plasmid expressing smGFP-ssrA | A. Horwich 27 |
| pET22b | Apr; E. coli expression vector | Novagen |
| pET28b | Kmr; E. coli expression vector | Novagen |
| pJAM202 | Apr Nvr; H. volcanii-E. coli shuttle expression plasmid with psmB-his6 gene; β subunit of 20S proteasomes expressed with C-terminal His tag in H. volcanii | 17 |
| pJAM801 | Kmr; 0.72-kb fragment generated by PCR from pEGFP ligated into pET28b by using NdeI and blunt-ended BlpI (5′-TTTGATTCATATGGTGAGCAAGGGCG-3′ and 5′-TTACTTGTACAGCTCGTCCATGCCGAG-3′; NdeI site in boldface type); EGFP with N-terminal His tag expressed in E. coli | This study |
| pJAM1011 | Apr Nvr; 0.97-kp XbaI-to-BspEI fragment of pJAM801 blunt end ligated with a 9.52-kb KpnI-to-BamHI fragment of pBAP5010; EGFP with N-terminal His tag expressed in H. volcanii | This study |
| pJAM657 | Apr Nvr; 0.74-kb BamHI-to-SacI fragment of pSMBFP blunt end ligated with a 9.94-kb NdeI-to-BlpI fragment of pJAM202; smBFP expressed in H. volcanii | This study |
| pJAM1019 | Apr Nvr; like pJAM657, except pSMGFP replaced pSMBFP; smGFP expressed in H. volcanii | This study |
| pJAM1020 | Apr Nvr; like pJAM657, except pSMRSGFP replaced pSMBFP; smRSGFP expressed in H. volcanii | This study |
| pJAM1022 | Apr; 356-bp BalI-to-MunI fragment of pJAM1020 ligated with a 4-kb BalI-to-MunI fragment of pGFP-11; smRS-GFP-ssrA expressed in E. coli | This study |
| pJAM1023 | Apr Nvr; 910-bp XbaI-to-BlpI fragment of pJAM1022 blunt end ligated with a 9.94-kb NdeI-to-BlpI fragment of pJAM202; smRS-GFP-ssrA expressed in H. volcanii | This study |
| pJAM1024 | Apr; 191-bp fragment generated by PCR from pJAM1022 ligated into pJAM1022 by using MfeI and BamHI (primer 1 [5′-TATCAACAAAATACTCCAATTGGCGATG-3′] and primer 2 [5′-CCGGATCCTTAGTCGTCTAAAGCGTAGT-3′] [MfeI and BamHI sites in boldface type]; smRS-GFP-ssrDD expressed in E. coli | This study |
| pJAM1025 | Apr Nvr; like pJAM1023, except pJAM1024 replaced pJAM1022; smRS-GFP-ssrDD expressed in H. volcanii | This study |
| pJAM1026 | Apr; like pJAM1024, except 5′-GGATCCTTACTCCTCTAAAGCGTAGTT-3′-replaced primer 2 (BamHI site in boldface type); smRS-GFP-ssrEE expressed in E. coli | This study |
| pJAM1027 | Apr; like pJAM1024, except 5′-GGATCCTTAGAGGAGTAAAGCGTAGTT-3′ replaced primer 2 (BamHI site in boldface type); smRS-GFP-ssrLL expressed in E. coli | This study |
| pJAM1028 | Apr; like pJAM1024, except 5′-CGGATCCTTACTTCTTTAAAGCGTAGT-3′ replaced primer 2 (BamHI site in boldface type); smRS-GFP-ssrKK expressed in E. coli | This study |
| pJAM1029 | Apr; like pJAM1024, except 5′-CGGATCCTTAGTCTCATAAAGCGTAGT-3′ replaced primer 2 (BamHI site in boldface type); smRS-GFP-ssr9 expressed in E. coli | This study |
| pJAM1030 | Apr Nvr; 858-bp XbaI-to-BamHI fragment of pJAM1026 blunt end ligated with a 9.94-kb NdeI-to-BlpI fragment of pJAM202; smRS-GFP-ssrEE expressed in H. volcanii | This study |
| pJAM1031 | Apr Nvr; like pJAM1030, except pJAM1027 replaced pJAM1026; smRS-GFP-ssrLL expressed in H. volcanii | This study |
| pJAM1032 | Apr Nvr; like pJAM1030, except pJAM1028 replaced pJAM1026; smRS-GFP-ssrKK expressed in H. volcanii | This study |
| pJAM1033 | Apr Nvr; like pJAM1030, except pJAM1029 replaced pJAM1026; smRS-GFP-ssr9 expressed in H. volcanii | This study |
| pJAM1035 | Apr; 0.85-kb fragment generated by PCR from pJAM1022 ligated into pJAM1022 by using XbaI and BamHI (primer 3 [5′-GGAGACCACAACGGTTTCCCTCTAGAAA-3′] and primer 4 [5′-AGCGTAGGATCCGTCGTTTTAGGCTTT-3′] [XbaI and BamHI sites in boldface type]); smRS-GFP-ssr1 expressed in E. coli | This study |
| pJAM1037 | Apr; like pJAM1035, except 5′-AGCGTAGGATCCTTAGTTGGCGGCTTT-3′ replaced primer 4 (BamHI site in boldface type); smRS-GFP-ssr3 expressed in E. coli | This study |
| pJAM1038 | Apr; like pJAM1035, except 5′-TAAGGATCCTTATTCGTCGTTGGCGGC-3′; replaced primer 4 (BamHI site in boldface type); smRS-GFP-ssr? expressed in E. coli | This study |
| pJAM1039 | Apr; 0.85-kb fragment generated by PCR from pJAM1022 ligated into pJAM1022 by using XbaI and BamHI (primer 5 [5′-CGCGGAGACCACAACGGTTCCCTCTAGAAA-3′] and primer 6 [5′-AGCGGATCCTTAGTTTTCGTCGTTGGCGGC-3′] [XbaI and BamHI sites in boldface type]); smRS-GFP-ssr6 expressed in E. coli | This study |
| pJAM1040 | Apr; like pJAM1039, except 5′-TGCGGATCCTTAGTAGTTTTCGTCGTTGGCGGC-3′ replaced primer 6 (BamHI site in boldface type); smRS-GFP-ssr7 expressed in E. coli | This study |
| pJAM1041 | Apr; like pJAM1039, except 5′-TTAGGATCCTTACGCGTAGTTTTCGTCGTTGGCGGC-3′ replaced primer 6 (BamHI site in boldface type); smRS-GFP-ssr8 expressed in E. coli | This study |
| pJAM1044 | Apr Nvr; 858-bp XbaI-to-BamHI fragment of pJAM1035 blunt end ligated with a 9.94-kb NdeI-to-BlpI fragment of pJAM202; smRS-GFP-ssr1 expressed in H. volcanii | This study |
| pJAM1046 | Apr Nvr; like pJAM1044, except pJAM1037 replaced pJAM1035; smRS-GFP-ssr3 expressed in H. volcanii | This study |
| pJAM1048 | Apr Nvr; like pJAM1044, except pJAM1038 replaced pJAM1035; smRS-GFP-ssr? expressed in H. volcanii | This study |
| pJAM1049 | Apr Nvr; like pJAM1044, except pJAM1039 replaced pJAM1035; smRS-GFP-ssr6 expressed in H. volcanii | This study |
| pJAM1050 | Apr Nvr; like pJAM1044, except pJAM1040 replaced pJAM1035; smRS-GFP-ssr7 expressed in H. volcanii | This study |
| pJAM1051 | Apr Nvr; like pJAM1044, except pJAM1041 replaced pJAM1035; smRS-GFP-ssr8 expressed in H. volcanii | This study |
PCR was used to introduce restriction enzyme sites for directional cloning of enhanced GFP (EGFP) (GFP with Phe64Leu and Ser65Thr mutations) into plasmid pET28b and synthesis of His6-tagged EGFP in E. coli. A 0.97-kb DNA fragment of this plasmid (pJAM801) was subcloned into shuttle expression vector pBAP5010 for synthesis of His6-EGFP in H. volcanii. Plasmids expressing soluble modified blue-, green-, and red-shifted fluorescent proteins in H. volcanii were generated by blunt-end ligation of DNA fragments from plasmids pSMBFP, pSMGFP, and pSMRSGFP into the pBAP5010-derived plasmid pJAM202. To introduce C-terminal modifications into smRS-GFP, an internal 356-bp DNA fragment from the smRS-GFP gene was ligated into pGFP-11 by using BalI and MunI. The resulting pET-based plasmid (pJAM1022) encoded smRS-GFP with a C-terminal ssrA tag (AANDENYALAA). To create smRS-GFP-ssrA variants in pET22b, PCR was used to modify the ssrA sequence, and the PCR products were ligated with pJAM1022 fragments as indicated in Table 1. Genes encoding these smRS-GFP-ssrA variants were cloned into plasmid pJAM202 as indicated in Table 1 for expression in H. volcanii. The Halobacterium cutirubrum rRNA P2 promoter and T7 terminator were used for transcription of the cloned gene in H. volcanii. For all expression plasmids, the presence and orientation of the gene with respect to the promoter and terminator were determined by restriction endonuclease cleavage, PCR, and/or DNA sequencing.
DNA purification and transformation.
Plasmids were isolated with a QIAGEN miniprep kit (QIAGEN Inc., Valencia, Calif.). DNA fragments were eluted from 0.8% (wt/vol) SeaKem GTG agarose (FMC Bioproducts, Rockland, Maine) gels with 1× TAE buffer (40 mM Tris-acetate, 2 mM EDTA; pH 8.5) by QIAquick gel extraction (QIAGEN). H. volcanii DS70 cells were transformed with plasmid DNA isolated from E. coli GM2163 as described by Cline et al. (7).
Protein techniques and immunoblotting.
Protein concentrations were determined by using the Bradford reagent with bovine serum albumin (Bio-Rad, Hercules, Calif.) as the standard. For immunoblotting, proteins were separated by reducing and denaturing 12% polyacrylamide gel electrophoresis (PAGE) with β-mercaptoethanol and sodium dodecyl sulfate (SDS) (18). The molecular mass standards included phosphorylase b (97.4 kDa), serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa) (Bio-Rad). After separation, proteins were transferred to Hybond-P membranes (Amersham Biosciences, Piscataway, N.J.) by using 10 mM 2-N-morpholino) ethanesulfonic acid (MES) at pH 6.0 with 10% (vol/vol) methanol (20 V, 4°C, 16 h). Antigens were detected by using Living Colors Aequorea victoria peptide rabbit immunoglobulin anti-GFP antibody as the primary antibody, which is suitable for use with all GFP variants (Clontech Laboratories, Inc., Palo Alto, Calif.). Horseradish peroxidase-conjugated anti-rabbit immunoglobulin (H+L) antibody raised in goats (Southern Biotechnology Associates, Inc., Birmingham, Ala.) was used as the secondary antibody. Horseradish peroxidase activity was detected by ECL Plus chemiluminescence (Amersham Biosciences) with a VersaDoc 1000 imaging system (Bio-Rad). For preparation of samples with and without clasto-lactacystin β-lactone, 5-ml cultures of H. volcanii(pJAM1023) were grown to an optical density at 600 nm (OD600) of 0.23. Dimethyl sulfoxide (DMSO) (0.01%) with and without the proteasome inhibitor (30 μM) was added, and cells were grown for an additional 9.5 h to OD600 of 0.37 and 0.80, respectively. Thus, cells were exposed to the inhibitor during exponential growth, when synthesis of new 20S proteasome proteins occurs (25). Cells were harvested and resuspended in SDS-PAGE loading dye (50 μl). Each boiled sample (20 μl) was used for immunoblot analysis. For all other immunoblots, samples were prepared from H. volcanii cultures (60 ml) grown in triplicate to OD600 of 0.8 to 1.0. Cells were harvested by centrifugation (10,000 × g, 20 min, 4°C), resuspended in 0.5 ml of distilled H2O, lysed by sonication, and centrifuged again (14,000 × g, 20 min, 4°C). Samples (10 to 50 μg) were analyzed within the linear range of detection as determined by using the His6-EGFP (10.7, 21.4, 42.8, 64.2, and 85.6 ng) purified from recombinant E. coli(pJAM801) by Ni2+-Sepharose chromatography as previously described (17). Cell lysate (10 μg) of H. volcanii(pJAM1020) expressing smRS-GFP was also included on all blots for comparison to the smRS-GFP variants.
Fluorescence measurements for GFP variants in whole cells.
Colonies of H. volcanii were viewed and photographed on culture plates by using a Leica fluorescence stereomicroscope operated with and without GFP cube band pass filters (excitation 480/40 nm and emission 510 nm; Microoptics of Florida, Davie, Fla.) and a Nikon Coolpix 4500 camera. For whole-cell fluorescence, cells were viewed at a magnification of ×100 by using differential interference contrast microscopy with a Nikon OptiPhot-2 microscope with a Nikon B-1E filter combination and were photographed with a Kodak DC290 zoom camera. Fluorescence of liquid cultures was monitored by using an Aminco-Bowman series 2 luminescence spectrometer (Spectronic Instruments, Rochester, N.Y.). The excitation and emission wavelengths were as follows: 488 and 509 nm for EGFP and smRS-GFP; 395 and 509 nm for soluble modified GFP (smGFP) (GFP with Phe99Ser, Met153Thr, and Val163Ala mutations); and 380 and 440 nm for soluble modified blue fluorescent protein (smBFP), which was smGFP with a Tyr66His mutation. One unit was defined as the fluorescence intensity of fluorescein at a concentration of 0.025 nM.
Purification of EGFP and 20S proteasomes from H. volcanii.
One-liter cultures of H. volcanii DS70(pJAM1011) and DS70(pJAM202) were grown in 2.8-liter Fernbach flasks at 42°C and 200 rpm to the late stationary phase (OD600, 3.5). Cells (9 g [wet weight]) were harvested by centrifugation (10,000 × g, 20 min, 4°C) and resuspended in 20 ml of 20 mM Tris buffer (pH 7.2) supplemented with 2 M NaCl (buffer A). Cells were passed through a chilled French pressure cell at 20,000 lb/in2. Cell debris was removed by centrifugation (16,000 × g, 20 min, 4°C). The filtrate obtained with a 45-μm-pore-size filter (Nalge Nunc International, Rochester, N.Y.) was applied to a 5-ml Ni2+-Sepharose column (HiTrap chelating; Amersham Biosciences) equilibrated with buffer A and washed with a step gradient consisting of 5 and 60 mM imidazole in buffer A. Fractions containing His-tagged proteins were eluted in buffer A with 500 mM imidazole. Proteasomes were purified further by application onto a 5-ml Bio-scale hydroxyapatite type I column (Bio-Rad) equilibrated in 10 mM sodium phosphate buffer (pH 7.2) with 2 M NaCl (buffer B). The column was washed with 15 ml of buffer B and developed with a 30-ml linear sodium phosphate gradient (10 to 500 mM sodium phosphate [pH 7.2] with 2 M NaCl). Protein fractions (300 μg per ml) with peptide-hydrolyzing activity which eluted at ∼350 mM sodium phosphate were pooled. A sample (0.5 ml per run) was applied to a Superose-6 HR 10/30 gel filtration column (Amersham Biosciences) equilibrated in buffer A. Peptide-hydrolyzing fractions containing 600-kDa 20S proteasomes, which were determined to be homogeneous with Coomassie blue R-250-stained SDS-PAGE gels, were pooled and used for kinetic and inhibitor analysis. To determine the percentage of inhibition of 20S proteasomes by clasto-lactacystin β-lactone in cell culture, H. volcanii DS70(pJAM202) was grown in a 5-ml culture to an OD600 of 0.28. DMSO (0.01%) with and without a proteasome inhibitor (15 μM) was added, and cells were harvested after an additional 9.5 h of incubation (OD600, 1.58 and 2.0, respectively). His-tagged 20S proteasomes were purified from these cultures by Ni2+-Sepharose chromatography (as described above) and assayed for peptide-hydrolyzing activity.
Peptide-hydrolyzing activity.
Chymotrypsin-like peptide-hydrolyzing activity was assayed in buffer A with 20 mM N-Suc-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin unless otherwise indicated. Release of 7-amino-4-methylcoumarin was monitored by determining the increase in fluorescence over time by using excitation and emission wavelengths of 350 and 460 nm with an Aminco Fluoro-colorimeter (Spectronic Instruments). Assays were performed at the approximate temperature (60°C), pH (pH 7.2), and NaCl concentration (2 M) optima determined previously for H. volcanii 20S proteasomes (29).
RNA isolation and analysis.
Cultures (2 ml) of strains DS70, DS70(pJAM1020), DS70(pJAM1023), and DS70(pJAM1025) were grown to an OD600 of 0.8. Total RNA was isolated with an RNeasy mini kit for bacteria (QIAGEN), with the following modifications. Cells were centrifuged for 1 min at 18,000 × g and immediately resuspended in 100 μl of RNase-free deionized water. Total RNA was treated with DNase I (Sigma) and NucAway spin columns. RNA quality was confirmed by using ethidium bromide-stained 0.8% (wt/vol) agarose gels. The RNA concentration was determined by measuring the absorbance at 260 nm. Total RNA (1.5 μg) was used as a template for One-Step reverse transcriptase PCR (RT-PCR) (QIAGEN) with oligonucleotides specific for the coding region of smGFP (5′-GGAGAAGAACTTTTCACTGGAGT-3′ and 5′-ATGTTCATCCATGCCATGTGTAATC-3′). cDNA products (0.7 kb) were detected by agarose gel electrophoresis (as described above). Control reaction mixtures, incubated on ice (instead of 50°C) for the RT step, were included to ensure that cDNA products were not due to contamination of total RNA with recombinant plasmid DNA.
RESULTS AND DISCUSSION
Functional synthesis of an archaeal GFP reporter protein.
A variety of GFP variants were explored with the goal of generating of a fluorescent reporter protein for functional expression in H. volcanii. Initially, EGFP was examined based on amino acid modifications (Phe64Leu and Ser65Thr) which allowed enhanced fluorescence compared to that of GFP. Quantitative immunoblotting of a recombinant strain which expressed the EGFP gene [H. volcanii DS70(pJAM1011)] revealed levels of fluorescent protein that were ∼0.1% of the total cell protein (Fig. 1); however, whole-cell fluorescence was not enhanced. An N-terminal polyhistidine tag allowed rapid purification of EGFP in high-salt (2 M NaCl) buffer from recombinant H. volcanii. The partially pure EGFP exhibited significant fluorescence (data not shown), suggesting that it folded in the high-ionic-strength cytosol of H. volcanii. Thus, the low levels of EGFP may have limited its detection by fluorescence in whole cells.
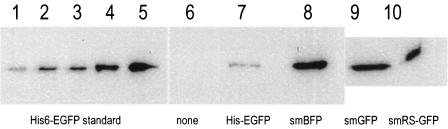
FIG. 1.
Optimization of fluorescent protein expression in recombinant H. volcanii. The levels of fluorescent protein were estimated by quantitative immunoblotting as described in Materials and Methods. His6-EGFP (18, 36, 54, 90, and 180 ng) was purified from recombinant E. coli and used as a standard (lanes 1 to 5). This protein was compared to cell lysates (15 to 16 μg) of H. volcanii DS70 (lane 6), DS70(pJAM1011) (lane 7), DS70(pJAM657) (lane 8), DS70(pJAM1019) (lane 9), and DS70(pJAM1020) (lane 10). The recombinant protein expressed in each strain is indicated below the immunoblot.
To enhance the levels of fluorescent protein, genes encoding soluble modified derivatives of GFP (smGFP, smRS-GFP, and smBFP) were expressed in H. volcanii. The soluble modifications (Phe99Ser, M153Thr, and Val163Ala) were derived from the UV-optimized cycle 3 variant which has reduced surface hydrophobicity, increased thermostability, and a lower tendency to aggregate during folding compared to GFP (13). The soluble modifications were expected to increase the stability and/or folding of GFP in the high-ionic-strength cytosol (2 to 3 M) of H. volcanii. The genes encoding the fluorescent protein variants also included silent mutations which removed mRNA processing sites and optimized codons for expression in plants (9). The red shift in excitation and emission wavelengths was selected to minimize autofluorescence of the host cell.
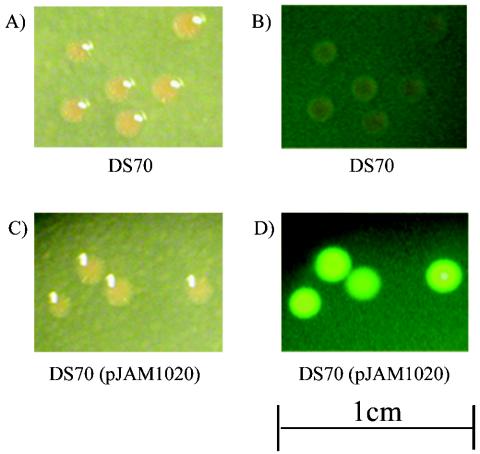
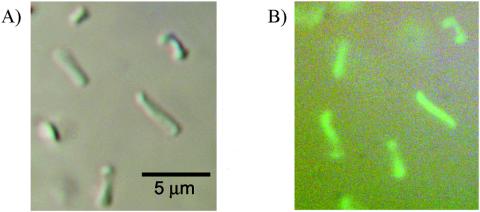
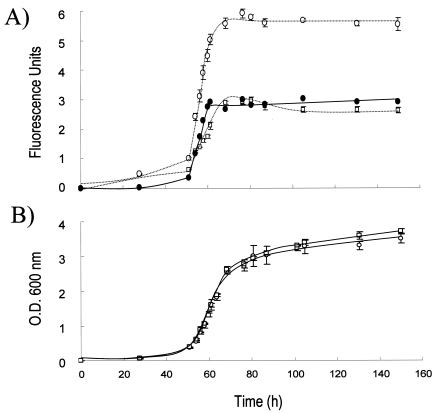
Based on quantitative immunoblotting, all three soluble modified derivatives of GFP (smGFP, smBFP, and smRS-GFP) were expressed at relatively high levels (>1% of the total cell protein) in recombinant H. volcanii (Fig. 1). Immunoblotting of the 10,000-×-g fraction of cell lysate revealed no significant accumulation of the soluble modified derivatives of GFP as inclusion bodies (data not shown). Furthermore, recombinant cells which produced smRS-GFP [strain DS70(pJAM1020)] exhibited significant increases in fluorescence compared with the wild type when cells were grown in liquid and plate cultures (Fig. 2, 3, and 4). The fluorescence attributed to smRS-GFP was uniformly distributed throughout the colonies on plates (Fig. 2) and in whole cells (Fig. 3). The smRS-GFP fluorescence paralleled growth in liquid cultures, with an exponential increase during the log phase and a plateau during the stationary phase (Fig. 4). The persistence of smRS-GFP fluorescence in the stationary phase suggested that this reporter protein was relatively stable in whole cells.
FIG. 2.
Colonies of H. volcanii expressing smRS-GFP exhibit fluorescence. H. volcanii DS70 and DS70(pJAM1020), which expresses smRS-GFP, were grown to an OD600 of 1, and 0.1 ml of a 10−6 dilution was spread onto ATCC 974 solid medium. Cells were incubated for 5 days at 37°C, and colonies were visualized by bright-field (A and C) and fluorescence (B and D) microscopy.
FIG. 3.
H. volcanii expressing smRS-GFP displays uniform cell fluorescence. Liquid cultures of H. volcanii DS70 and DS70(pJAM1020), which expresses smRS-GFP, were grown to an OD600 of 1. Cultures were transferred (10 μl) to glass slides with coverslips, and whole cells were visualized by differential interference contrast microscopy with and without fluorescence.
FIG. 4.
Liquid cultures of H. volcanii expressing smRS-GFP exhibit fluorescence that parallels cell growth. The fluorescence (A) and growth (B) of H. volcanii DS70(pJAM1020) expressing smRS-GFP (○) were compared to the fluorescence and growth of parent strain DS70 (□) in liquid culture. Fluorescence was measured at an excitation wavelength of 488 nm and an emission wavelength of 510 nm in triplicate cultures. Fluorescence attributed to smRS-GFP expression (•) was calculated by determining the difference between DS70(pJAM1020) and DS70 cell cultures.
C-terminal modifications reduce the level of smRS-GFP in vivo.
C- and N-terminal tags are often fused to GFP to define structural elements or motifs that target proteins for degradation (6). The apparent stability of smRS-GFP in H. volcanii suggested that a similar approach could be used to determine what residues and/or motifs destabilize proteins in vivo. Thus, a variety of amino acids were fused to the C terminus of smRS-GFP (Table 2). Included in the modifications was addition of the 11-residue sequence AANDENYALAA (the ssrA tag). Although archaea do not encode ssrA homologs, the 11-residue amino acid sequence had previously been shown to target GFP for degradation by reconstituted archaeal 20S proteasome and PAN complexes purified from recombinant E. coli (3). The modifications (Table 2) also included C-terminal deletions and alteration of the C-terminal Ala-10 and Ala-11 residues of the ssrA tag to acidic (Glu and Asp), basic (Lys), and hydrophobic (Leu) residues. In addition, a random sequence was included which had four contiguous charged residues (Asp-Asp-Lys-Asp) in place of the two charged residues of the ssrA tag (Asp-Glu). All of these C-terminal tags were predicted to be on the surface of smRS-GFP and to have little if any impact on the overall structure or fluorescence of the intact protein, based on circular dichroism of GFP versus GFP-ssrA (14) and the GFP crystal structure (24).
TABLE 2.
Modification of the C terminus of smRS-GFP influences whole-cell fluorescence and the level of fluorescent protein in recombinant H. volcanii
| Expression plasmid | smRS-GFP C terminusa | Amino acid tag (abbreviation)b | Relative protein level (%)c | Whole-cell fluorescenced |
|---|---|---|---|---|
| pJAM1023 | ...GITHGMDELYK | AANDENYALAA (ssrA) | UD | − |
| pJAM1030 | ...GITHGMDELYK | AANDENYALEE (ssrEE) | 6.6 ± 1.3 | + |
| pJAM1025 | ...GITHGMDELYK | AANDENYALDD (ssrDD) | 5.5 ± 0.6 | + |
| pJAM1031 | ...GITHGMDELYK | AANDENYALLL (ssrLL) | <0.5 | − |
| pJAM1032 | ...GITHGMDELYK | AANDENYALKK (ssrKK) | <0.5 | − |
| pJAM1048 | ...GITHGMDELYK | AANDDKDLSNN (ssr?) | 28.9 ± 5.5 | ++ |
| pJAM1033 | ...GITHGMDELYK | AANDENYAL (ssr9) | <0.5 | − |
| pJAM1051 | ...GITHGMDELYK | AANDENYA (ssr8) | <0.5 | − |
| pJAM1050 | ...GITHGMDELYK | AANDENY (ssr7) | <0.5 | − |
| pJAM1049 | ...GITHGMDELYK | AANDEN (ssr6) | <0.5 | − |
| pJAM1046 | ...GITHGMDELYK | AAN (ssr3) | <0.5 | − |
| pJAM1044 | ...GITHGMDELYK | A (ssr1) | 8.1 ± 1.9 | + |
| pJAM1020 | ...GITHGMDELYK | 100 ± 4.4 | ++++ |
C-terminal 11 residues of smRS-GFP.
Amino acid residues added to the C terminus of smRS-GFP. The abbreviations used to indicate the type of modification are in parentheses.
Protein level based on quantitative immunoblotting and expressed as a percentage relative to smRS-GFP. Compared to proteins that were present at levels that were less than 0.5% of the smRS-GFP level, the ssrA-tagged fusion protein was undetectable (UD).
Relative level of fluorescence as observed by fluorescence microscopy of colonies, where ++++ is the highest level and − is undetectable.
The majority of amino acid residues which were fused to the C terminus of smRS-GFP, including the ssrA tag, completely abolished whole-cell fluorescence (Table 2). In fact, only a few of the tags examined resulted in fluorescent colonies, and the levels of fluorescence of these colonies were low to moderate compared to those of the colonies with unmodified smRS-GFP. To determine whether the observed reductions in fluorescence reflected changes in the levels of the reporter protein, quantitative immunoblotting was performed. Based on this analysis, the levels of reporter protein were proportional to whole-cell fluorescence. Most of the C-terminal fusions, including the ssrA-tagged protein, were reduced at least 200-fold compared to the untagged smRS-GFP (Table 2). The exceptions included the addition of AANDDKDLSNN, whose length was comparable to that of the ssrA tag but which had four contiguous charged residues. This modification resulted in a >50-fold increase in protein levels compared to smRS-GFP-ssrA. Likewise, deletion of Ala-2 to Ala-11 of the ssrA tag enhanced the levels of this fusion protein more than 16-fold. Replacement of the C-terminal Ala residues (Ala-10 and Ala-11) of ssrA with acidic residues (Glu or Asp) also enhanced the protein levels more than 10-fold.
Some general trends in the type of C-terminal modification and the level of reporter protein were apparent. Most prominent was the finding that in all cases addition of a hydrophobic residue (i.e., Ala or Leu) to the C terminus dramatically reduced the level of reporter protein, even when the length of the fusion varied. In fact, addition of a single Ala to smRS-GFP resulted in a >10-fold decrease in the level of protein. The type of residue added to the C terminus, however, was not the sole determinant of reporter protein levels, as exemplified by fusion proteins with Asn as the terminal residue. These Asn fusion proteins varied from <0.5 to 30% of protein relative to smRS-GFP. Furthermore, the length of the tag was not directly correlated with the reporter protein levels (e.g., addition of the 11-residue AANDDKDLSNN tag had less impact on smRS-GFP levels than addition of a single Ala).
Although the ssrA system that targets proteins for degradation by the Clp protease in Bacteria is not conserved in Archaea, there are some similarities in C-terminal amino acid sequences that reduce or enhance the GFP reporter protein levels in these two domains. For example, in E. coli, modification of the Ala-10 and Ala-11 residues of ssrA to acidic residues increases the Km of ClpXP by a factor of at least 100 (i.e., no degradation is observed at a saturating protein substrate concentration) (12). Likewise, in H. volcanii, alteration of the terminal Ala residues to acidic residues enhanced protein levels 10-fold. However, in contrast to E. coli, the levels of these acidic ssrA derivatives were significantly reduced compared to the levels of the unmodified smRS-GFP reporter protein in H. volcanii.
C-terminal modifications do not affect the level of smRS-GFP-specific mRNA in H. volcanii.
Based on the results presented above, a variety of C-terminal fusions were found to reduce the level of smRS-GFP in H. volcanii. All of the expression plasmids used for this analysis were designed to have identical DNA sequences and spacing for the ribosomal binding site, transcriptional promoter, terminator, and the majority of the open reading frame encoding smRS-GFP. Thus, the C-terminal modifications to smRS-GFP were expected to have little impact on mRNA levels. To confirm this, the levels of smRS-GFP-specific mRNA were analyzed by RT-PCR by using cells expressing smRS-GFP, smRS-GFP-ssrA, and the smRS-GFP-ssrA derivative with Ala10Asp and Ala11Asp mutations (i.e., smRS-GFP-ssrDD). Comparable levels of smRS-GFP-specific mRNA were detected in all three recombinant strains, while none of this transcript was detected in parent strain DS70 (data not shown). Thus, mRNA levels did not limit the synthesis of the smRS-GFP variants in these recombinant H. volcanii strains.
clasto-Lactacystin β-lactone as an in vivo inhibitor of H. volcanii 20S proteasomes.
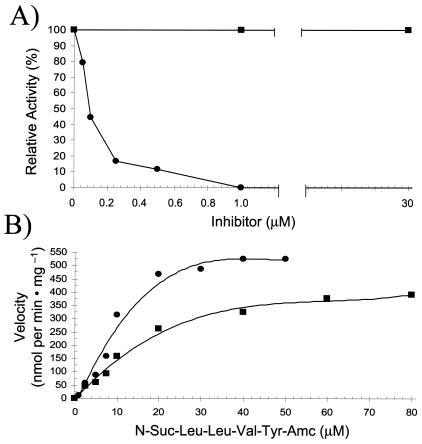
clasto-Lactacystin β-lactone has been shown to permeate eukaryotic cell membranes and inhibit 20S proteasomes by covalent modification of the active site threonine hydroxyl (10, 11). This molecule is considered by many to be the “gold standard” for proteasome inhibitor specificity and is used routinely to investigate proteasome function in eukaryotic cells (5). To determine whether H. volcanii 20S proteasomes are inhibited by this compound, two approaches were taken. First, the peptide-hydrolyzing activity of purified 20S proteasomes was assayed in the presence of clasto-lactacystin β-lactone or the general serine protease inhibitor phenylmethanesulfonyl fluoride (PMSF) (Fig. 5A). Compared to PMSF, which had little if any influence on the peptide-hydrolyzing activity of the halophilic 20S proteasomes, the lactacystin derivative was highly specific, with a Ki of ∼40 nM (Fig. 5B). clasto-Lactacystin β-lactone increased the Km for N-Suc-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin from 9.85 μM to an apparent Km of 22.2 μM without altering the Vmax (550 nmol of 7-amino-4-methylcoumarin released per min per mg of protein). Based on these results, it appears that the lactacystin derivative competes effectively for the active site of the halophilic 20S proteasomes. Thus, the influence of clasto-lactacystin β-lactone on proteasome activity was examined in cell culture. For this analysis, H. volcanii strain DS70(pJAM202), which expressed the β subunit of 20S proteasomes with a His tag, was grown in the presence and absence of clasto-lactacystin β-lactone. The specific activity of the 20S proteasomes was significantly reduced when purified from cells grown in the absence of this inhibitor compared with the specific activity in the presence of this inhibitor (204 versus 175 nmol of 7-amino-4-methylcoumarin released per min per mg of protein). The reason for the incomplete inhibition of 20S proteasomes in vivo remains to be determined. Based on previous work (17), expression of the β-His protein from plasmid pJAM202 enhances the levels of β-protein twofold compared with the wild type. Thus, the levels of 20S proteasomes may be higher in DS70(pJAM202). However, it is more likely that clasto-lactacystin β-lactone is not fully soluble at the concentration and composition of salts required for growth of H. volcanii. The latter possibility is supported by the 30 to 40% decrease in inhibition of the halophilic 20S proteasome activity in vitro when clasto-lactacystin β-lactone was preincubated for 30 min at 37°C in ATCC 974 medium salts (∼2.6 M NaCl, 530 mM MgCl2, 30 mM KSO4, and 1.2 mM CaCl2) prior to the assay (data not shown). It is also possible that clasto-lactacystin β-lactone does not permeate H. volcanii cells as readily as it permeates eukaryotic cells due to differences in cell wall structure. The cell wall of H. volcanii includes a glycoprotein S-layer and cell membrane of isoprenoid ether lipids not found in eukaryotes.
FIG. 5.
H. volcanii 20S proteasomes are inhibited by clasto-lactacystin β-lactone. (A) Purified 20S proteasomes (1 μg/ml; ∼1.67 nM) were preincubated with various concentrations of clasto-lactacystin β-lactone (•) and PMSF (▪) for 30 min at 37°C in 20 mM Tris-HCl buffer (pH 7.2) supplemented with 2 M NaCl and with 0.0075% (vol/vol) DMSO and 0.01% (vol/vol) ethanol, respectively. Peptide hydrolysis was assayed as described in Materials and Methods by using 20 mM substrate with an additional 0.004% DMSO. Even at a PMSF concentration of 100 μM, 92% of the activity remained. (B) Purified proteasomes (0.5 μg/ml; ∼0.84 nM) were assayed with various concentrations of substrate in the absence (•) and in the presence (▪) of 50 nM clasto-lactacystin β-lactone. Amc, 7-amino-4-methylcoumarin.
Inhibition of 20S proteasomes enhances the level of smRS-GFP-ssrA protein in H. volcanii.
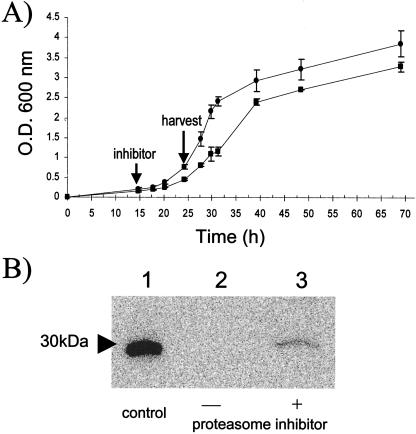
One of the C-terminal smRS-GFP derivatives, the ssrA-tagged fluorescent protein, was chosen as a reporter to investigate whether 20S proteasomes are involved in modulating the levels of this protein in the cell. The rationale for this choice was that an ssrA-tagged derivative of GFP is degraded by 20S proteasomes reconstituted with PAN in vitro (4). Thus, H. volcanii cells expressing smRS-GFP-ssrA were grown in the presence and absence of clasto-lactacystin β-lactone and analyzed for accumulation of the reporter protein. Interestingly, addition of the proteasome inhibitor significantly reduced the growth rate (from a doubling time of ∼4 h to 6 h), suggesting that 20S proteasomes are needed for normal growth of H. volcanii (Fig. 6A). Immunoanalysis revealed that the reporter protein (smRS-GFP-ssrA) accumulated when cells were grown in the presence of clasto-lactacystin β-lactone (Fig. 6B). In contrast, the general serine protease inhibitor PMSF (30 μM) had no influence on cell growth or reporter protein levels (data not shown). Although the ssrA-tagged smRS-GFP did not accumulate to levels comparable to those of the untagged smRS-GFP in the presence of clasto-lactacystin β-lactone, H. volcanii 20S proteasomes were not completely inactivated by this inhibitor in cell culture (see above).
FIG. 6.
Inhibition of H. volcanii 20S proteasomes enhances the level of smRS-GFP-ssrA in cell culture. (A) H. volcanii DS70(pJAM1023) was grown in the absence (•) and presence (▪) of clasto-lactacystin β-lactone (30 μM). An arrow indicates the time (15 h) when the inhibitor was added to the cell culture. (B) Immunoblot with anti-GFP antibody of E. coli purified His6-EGFP (42.8 ng) (lane 1) and log-phase H. volcanii DS70(pJAM1023) expressing smRS-GFP-ssrA without (lane 2) and with (lane 3) clasto-lactacystin β-lactone.
Conclusions.
This study revealed that a soluble, modified, red-shifted derivative of GFP (smRS-GFP) is synthesized and readily detected in recombinant H. volcanii. The fluorescence of smRS-GFP was uniformly distributed in whole cells and paralleled the reporter protein levels, as determined by immunoblotting. Addition of amino acid residues in various sequences and with various lengths to the C terminus of smRS-GFP had a differential effect on the levels of this reporter protein. Proteasomes were found to be responsible, at least in part, for modulating the levels of the ssrA-tagged GFP derivative in vivo.
Recombinant smRS-GFP and C-terminal variant reporter proteins have tremendous potential in a variety of applications in the haloarchaea. For example, chromosomal mutations that stabilize C-terminus-tagged smRS-GFP derivatives, regulated promoters, and chemical libraries of 20S proteasome inhibitors can be rapidly screened in H. volcanii by using fluorescent plate assays. The localization of H. volcanii proteins can be determined by fusion to smRS-GFP and imaging via whole-cell fluorescence. Although the current host strain necessitates the use of immunoblotting for quantitative analysis of smRS-GFP levels, the analysis is bolstered by the rapid and qualitative analysis of GFP levels by whole-cell fluorescence. Efforts aimed at reducing the autofluorescence of H. volcanii should result in greater versatility of this reporter protein in the future (e.g., sensitive quantification of smRS-GFP levels via fluorescence and dual-fluorescent-label studies).
Acknowledgments
We thank Francis Davis and Jack Shelton for DNA sequencing. We also thank Henry Aldrich, Donna Williams , and Paul Shirk for assistance with fluorescent microscopy and photography.
This work was supported in part by NIH grant GM57498 and by the Florida Agricultural Experiment Station.
Footnotes
Florida Agricultural Experiment Station Journal Series no. R-10375.
REFERENCES
- 1.Baliga, N. S., M. Pan, Y. A. Goo, E. C. Yi, D. R. Goodlett, K. Dimitrov, P. Shannon, R. Aebersold, W. V. Ng, and L. Hood. 2002. Coordinate regulation of energy transduction modules in Halobacterium sp. analyzed by a global systems approach. Proc. Natl. Acad. Sci. USA 99:14913-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumeister, W., J. Walz, F. Zühl, and E. Seemüller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367-380. [DOI] [PubMed] [Google Scholar]
- 3.Benaroudj, N., and A. L. Goldberg. 2000. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat. Cell Biol. 2:833-839. [DOI] [PubMed] [Google Scholar]
- 4.Benaroudj, N., P. Zwickl, E. Seemuller, W. Baumeister, and A. L. Goldberg. 2003. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell 11:69-78. [DOI] [PubMed] [Google Scholar]
- 5.Bogyo, M., and E. W. Wang. 2002. Proteasome inhibitors: complex tools for a complex enzyme. Curr. Top. Microbiol. Immunol. 268:185-208. [DOI] [PubMed] [Google Scholar]
- 6.Chiesa, A., E. Rapizzi, V. Tosello, P. Pinton, M. de Virgilio, K. E. Fogarty, and R. Rizzuto. 2001. Recombinant aequorin and green fluorescent protein as valuable tools in the study of cell signalling. Biochem. J. 355:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cline, S. W., W. L. Lam, R. L. Charlebois, L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35:148-152. [DOI] [PubMed] [Google Scholar]
- 8.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 9.Davis, S. J., and R. D. Vierstra. 1998. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36:521-528. [DOI] [PubMed] [Google Scholar]
- 10.Fenteany, G., and S. L. Schreiber. 1998. Lactacystin, proteasome function, and cell fate. J. Biol. Chem. 273:8545-8548. [DOI] [PubMed] [Google Scholar]
- 11.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 12.Flynn, J. M., I. Levchenko, M. Seidel, S. H. Wickner, R. T. Sauer, and T. A. Baker. 2001. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. USA 98:10584-10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda, H., M. Arai, and K. Kuwajima. 2000. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry 39:12025-12032. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann-Petersen, R., M. Seeger, and C. Gordon. 2003. Transferring substrates to the 26S proteasome. Trends Biochem. Sci. 28:26-31. [DOI] [PubMed] [Google Scholar]
- 16.Jolley, K. A., R. J. Russell, D. W. Hough, and M. J. Danson. 1997. Site-directed mutagenesis and halophilicity of dihydrolipoamide dehydrogenase from the halophilic archaeon, Haloferax volcanii. Eur. J. Biochem. 248:362-368. [DOI] [PubMed] [Google Scholar]
- 17.Kaczowka, S. J., and J. A. Maupin-Furlow. 2003. Subunit topology of two 20S proteasomes from Haloferax volcanii. J. Bacteriol. 185:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Maupin-Furlow, J. A., M. A. Gil, I. M. Karadzic, P. A. Kirkland, and C. J. Reuter. 2004. Proteasomes: perspecitves from the archaea [update 2004]. Front. Biosci. 9:1743-1758. [DOI] [PubMed] [Google Scholar]
- 20.Maupin-Furlow, J. A., S. J. Kaczowka, M. S. Ou, and H. L. Wilson. 2001. Archaeal proteasomes: proteolytic nanocompartments of the cell. Adv. Appl. Microbiol. 50:279-338. [DOI] [PubMed] [Google Scholar]
- 21.Mevarech, M., F. Frolow, and L. M. Gloss. 2000. Halophilic enzymes: proteins with a grain of salt. Biophys. Chem. 86:155-164. [DOI] [PubMed] [Google Scholar]
- 22.Navon, A., and A. L. Goldberg. 2001. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol. Cell 8:1339-1349. [DOI] [PubMed] [Google Scholar]
- 23.Nomura, S., and Y. Harada. 1998. Functional expression of green fluorescent protein derivatives in Halobacterium salinarum. FEMS Microbiol. Lett. 167:287-293. [DOI] [PubMed] [Google Scholar]
- 24.Ormo, M., A. B. Cubitt, K. Kallio, L. A. Gross, R. Y. Tsien, and S. J. Remington. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392-1395. [DOI] [PubMed] [Google Scholar]
- 25.Reuter, C. J., S. J. Kaczowka, and J. A. Maupin-Furlow. 2004. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J. Bacteriol. >186:7763-7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber-Ban, E. U., B. G. Reid, A. D. Miranker, and A. L. Horwich. 1999. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401:90-93. [DOI] [PubMed] [Google Scholar]
- 28.Wendoloski, D., C. Ferrer, and M. L. Dyall-Smith. 2001. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology 147:959-964. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, H. L., H. C. Aldrich, and J. A. Maupin-Furlow. 1999. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: purification, characterization, and gene sequence analysis. J. Bacteriol. 181:5814-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Withey, J. H., and D. I. Friedman. 2003. A salvage pathway for protein synthesis: tmRNA and trans-translation. Annu. Rev. Microbiol. 57:101-123. [DOI] [PubMed] [Google Scholar]
- 31.Woese, C. R., O. Kanlder, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]