Abstract
Fusobacterium nucleatum is an important oral anaerobic pathogen involved in periodontal and systemic infections. Studies of the molecular mechanisms involved in fusobacterial virulence and adhesion have been limited by lack of systems for efficient genetic manipulation. Plasmids were isolated from eight strains of F. nucleatum. The smallest plasmid, pKH9 (4,975 bp), was characterized and used to create new vectors for fusobacterial genetic manipulation. DNA sequence analysis of pKH9 revealed an open reading frame (ORF) encoding a putative autonomous rolling circle replication protein (Rep), an ORF predicted to encode a protein homologous to members of the FtsK/SpoIIIE cell division-DNA segregation protein family, and an operon encoding a putative toxin-antitoxin plasmid addiction system (txf-axf). Deletion analysis localized the pKH9 replication region in a 0.96-kbp fragment. The pKH9 rep gene is not present in this fragment, suggesting that pKH9 can replicate in fusobacteria independently of the Rep protein. A pKH9-based, compact Escherichia coli-F. nucleatum shuttle plasmid was constructed and found to be compatible with a previously described pFN1-based fusobacterial shuttle plasmid. Deletion of the pKH9 putative addiction system (txf-axf) reduced plasmid stability in fusobacteria, indicating its addiction properties and suggesting it to be the first plasmid addiction system described for fusobacteria. pKH9, its genetic elements, and its shuttle plasmid derivatives can serve as useful tools for investigating fusobacterial properties important in biofilm ecology and pathogenesis.
Fusobacterium nucleatum is the most numerous gram-negative bacterium isolated from dental plaque biofilms (26, 29); it is a central species in biofilm development and a pathogen in human infections, including periodontitis. Important properties of F. nucleatum in biofilm development and pathogenesis include the abilities to coaggregate to early and late colonizers of plaque biofilms (20, 21), adhere to and invade host tissue cells (13), induce proinflammatory cytokines (16), and produce proteases (2, 24). Of the bacterial species associated with periodontal disease, F. nucleatum is the oral organism most commonly found in systemic infections (26) and is strongly implicated in vaginal infections associated with preterm deliveries (15).
The study of fusobacteria has been hampered by a lack of systems for its genetic manipulation. As a result, very little is known at the molecular level about F. nucleatum properties. A homologous family of cryptic plasmids has been described to exist in F. nucleatum strains (12, 23), and sequence analyses have identified a putative plasmid origin of replication as well as replication and relaxase protein homologues encoded by one member of this family, pFN1. Construction of a shuttle plasmid, pHS17, by using pFN1 has enabled the transformation of F. nucleatum (12). A current limitation of the pHS17 shuttle plasmid is that it has been successfully established only in two strains of F. nucleatum. Because alternative vector systems will be important in molecular analysis of fusobacteria, we isolated additional native plasmids from F. nucleatum. One plasmid, pKH9, was characterized and found to possess features not previously identified in fusobacterial plasmids. We describe here the minimal replicon that was used to construct a pKH9-based shuttle plasmid and studies that strongly indicate a fusobacterial plasmid addiction system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
F. nucleatum oral clinical isolates used in this study were a kind gift from N. Ganeshkumar and A. Tanner of the Forsyth Institute. F. nucleatum ATCC 10953 and ATCC 23726 were from our laboratories' culture collections. The bacteria were grown in an anaerobic chamber (Coy Laboratory Products, Ann Arbor, Mich.) under an atmosphere of 85% N-10% H-5% CO2 at 37°C in brain heart infusion containing 0.25% glutamate or plated on Columbia agar (Oxoid, Basingstoke, Hampshire, United Kingdom). Antibiotics used for selection for F. nucleatum were clindamycin (0.4 μg/ml; Sigma-Aldrich, Steinhein, Germany) and thiamphenicol (chloramphenicol analogue, 5 μg/ml; Sigma Chemical Co., St. Louis, Mo.). Escherichia coli strain DH5α (Life Technologies, Carlsbad, Calif.) was used for plasmid construction and was grown in Luria-Bertani medium or on Luria-Bertani agar plates at 37°C under aerobic conditions. Antibiotics used to provide selection for E. coli strains were erythromycin (300 μg/ml; Sigma-Aldrich) and chloramphenicol (35 μg/ml; Sigma-Aldrich).
Computer analysis.
Public databases were searched for similar sequences with the BLASTN, BLASTP, and BLASTP/PSI algorithms using default parameters. The features of the predicted proteins were examined by the Pfam programs (http://www.sanger.ac.uk/Software/Pfam/search.shtml). Multiple alignment was performed using CLUSTAL W (14, 30).
Recombinant DNA techniques.
The plasmids used in this investigation and their relevant properties are listed in Table 1. Plasmid DNA was prepared using Qiaprep spin miniprep or maxiprep kits (QIAGEN, Hilden, Germany). PCR was performed using the Expand High Fidelity PCR system according to conditions specified by the manufacturer (Boehringer Mannheim, Indianapolis, Ind.). For all other DNA manipulations, standard protocols were followed (28). Plasmid electroporation into F. nucleatum was performed as described previously (12).
TABLE 1.
Plasmids used in this study
| Plasmid | Size (kb) | Relevant characteristic(s) | Resistancea | Reference or source |
|---|---|---|---|---|
| pHS17 | 10.0 | F. nucleatum-E. coli shuttle plasmid | Erm Cli | 12 |
| pHS30 | 7.3 | catP derivative of pHS17 | Cam | 19 |
| pKH9 | 5.0 | F. nucleatum KH9 native plasmid | This work | |
| pBluescript II KS+ | 3.0 | Blue-white screening cloning vector | Amp | Stratagene |
| pBK-erm | 5.0 | pBluescript II KS+ carrying ermF-ermAM from pHS17 | Erm Cli Amp | This work |
| pKH90 | 9.6 | pKH9 XbaI fragment (rep+axf+txf+oriI) inserted into the XbaI site of pBK-erm | Erm Cli Amp | This work |
| Derivatives of pKH90b | ||||
| pORI9 | 7.0 | rep+oriI | Erm Cli Amp | This work |
| pORI91 | 6.0 | pOR19ΔampR | Erm Cli | This work |
| pORI92 | 5.3 | oriI | Erm Cli Amp | This work |
| pORI93 | 5.0 | 5′ of oriI region | Erm Cli Amp | This work |
| pORI94 | 4.7 | 3′ of oriI region | Erm Cli Amp | This work |
Erm, erythromycin; Cli, clindamycin; Cam, thiamphenicol; Amp, ampicillin.
See also Fig. 3.
Plasmid construction.
For pBK-erm, the pHS17 (12) ermF-ermAM cassette (9) was cloned into pBluescript II KS+ (Stratagene Europe, Amsterdam, The Netherlands) by using KpnI-BamHI. For pKH90, a 4.8-kbp pKH9 fragment was amplified by inverse PCR with primers NH31 and NH32 (Table 2). The PCR amplification product was restricted with XbaI, and the 4.6-kbp restriction product was cloned into the XbaI site of pBK-erm. For pORI9, the pKH9 rep region was amplified by PCR from pKH90 with primers NH33 (containing a SacII site) and NH34. The 2.0-kb amplification product was restricted with BamHI and SacII and cloned into pBK-erm restricted with these enzymes. For pORI91, the ampicillin resistance gene was deleted from pORI9 by using RcaI digestion followed by self-ligation. For pORI92, pKH90 was used as a template for inverse PCR amplification with the EcoRI-containing primers RO1 and RO2. The 5.3-kb PCR product was restricted with EcoRI and self-ligated. For pORI93, pORI92 was digested with SacI and XbaI. The resulting 5.0-kb fragment was self-ligated after the ends were filled in with T4 DNA polymerase (New England BioLabs, Beverly, Mass.). For pORI94, pORI92 was digested with EcoRI and SacI and self-ligated as described above.
TABLE 2.
PCR primers used in this study
| Primer | Sequence | Used in plasmid construction |
|---|---|---|
| NH31 | 5′ CTTTTCCCAAATTGTCCAAGTTC | pKH90 |
| NH32 | 5′ GTAGGAGTTGAACCTTACTACAC | |
| NH33 | 5′ AATCCGCGGTACTGGTAACGGTGAGA | pORI9 |
| NH34 | 5′ CTACTGACAGCTTCGGGGGAT | |
| RO1 | 5′ GGAATTCCGCCAAATAACTCTATCTAAA | pORI92 |
| RO2 | 5′ GGAATTCGCGGAACCCCTATTTGTTTAT |
Plasmid stability assay.
The effect of the pKH9 operon comprising axf and txf on plasmid stability in F. nucleatum ATCC 23726 was measured as described previously for Enterococcus faecium (11). Four F. nucleatum transformant colonies were streaked onto Columbia agar containing clindamycin. One colony from each of the four streaks was restreaked onto a nonselective Columbia agar plate. The streaking onto nonselective plates was repeated for several successive subcultures. After 3, 6, 9, and 12 subcultures, colonies were tested for plasmid retention by stabbing 8 colonies from each streak (32 colonies in total) onto both nonselective and clindamycin-containing Columbia agar plates. The retention of the clindamycin-resistant phenotype was expressed as the percentage of colonies that were clindamycin resistant.
Plasmid compatibility assay.
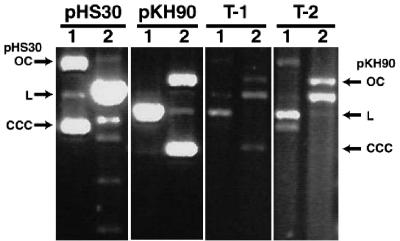
F. nucleatum ATCC 23726 carrying pHS30 (conferring chloramphenicol resistance) was electroporated with pORI91. The transformation outgrowth was plated on Columbia agar with clindamycin to select for the incoming pORI91 plasmid. All transformant colonies were subcultured on Columbia agar with clindamycin, followed by subculturing on Columbia agar with thiamphenicol and clindamycin to test for the resistance phenotype conferred by both plasmids. Plasmid minipreps were prepared from 20 transformants representing three separate transformations and analyzed by visualization of plasmid DNA on agarose gels, as described below (see Fig. 4).
FIG. 4.
Compatibility of pHS30 and pORI91. F. nucleatum ATCC 23726 harboring pHS30 was transformed with pORI91. Transformants were selected on media with clindamycin. Plasmid DNA isolated from representative transformants (T-1 and T-2) was examined by restriction enzyme digestion and compared with similarly digested pHS30 and pORI91 isolated from E. coli. DNA in lanes 1 was digested with KpnI to linearize pORI91. DNA in lanes 2 was digested with EcoRV to linearize pHS30. The open circular (OC), linear (L), and covalently closed circular (CCC) forms of pHS30 and pORI91 are indicated on the left and right, respectively.
Nucleotide sequence accession numbers.
pKH9 was submitted to GenBank under accession no. AF295336.
RESULTS
Isolation and characterization of F. nucleatum plasmid pKH9.
Thirty-four F. nucleatum clinical isolates from the Forsyth Dental Institute were screened for the presence of plasmids. Eight strains (UBC, IE10, KH9, IE3, IE7, IE5, IE24, and KI17) contained plasmids with estimated sizes of 5 to 15 kb, representing 24% of the strains examined. The smallest plasmid (pKH9, 4.975 kb) was isolated from F. nucleatum KH9 and was further analyzed.
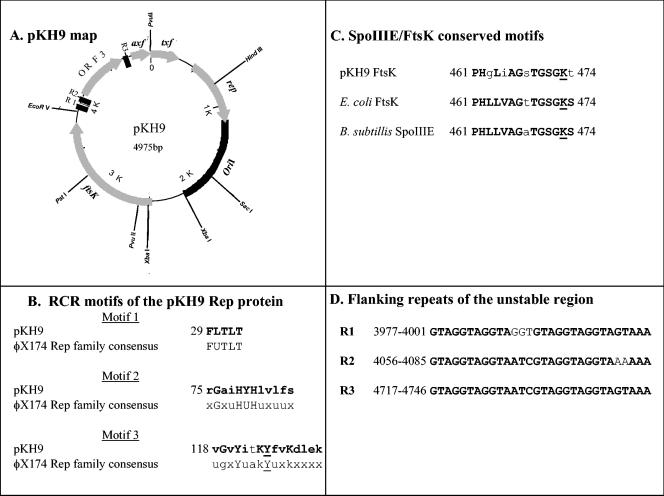
The KH9 plasmid, pKH9, was mapped by using restriction endonucleases (Fig. 1A), and both strands were sequenced. Computer analysis of the DNA sequence revealed a circular structure of 4,975 bp with a 29% G+C content, in agreement with the low G+C content of the fusobacterial genomic DNA (18) and with that of the previously described F. nucleatum plasmid pFN1 (12). A BLAST search (1) of the pKH9 translated sequences against sequences in the NCBI database revealed homology with five previously described proteins.
FIG. 1.
Genetic elements of the F. nucleatum pKH9 plasmid. (A) pKH9 map. ORFs are indicated by gray arrows. R1, R2, and R3 are the repeated sequences flanking the unstable region (D). The location of the pKH9 Rep-independent origin of replication (oriI) is indicated by a black rectangle. (B) Three RCR motifs found in the pKH9 Rep protein in comparison to the consensus sequences of the φX174 RCR family. U denotes hydrophobic residues (I, L, V, M, F, Y, and W). The conserved Tyr residue is underlined. (C) Conserved amino acid sequence of the pKH9 FtsK/SpoIIIE protein. The lysine essential for the ATP-dependent DNA pump function is underlined. (D) GTAG-rich repeat sequences flanking pKH9 ORF 3, which is unstable in E. coli. Bold and capital letters in panels B, C, and D denote conserved amino acids and nucleotides.
pKH9 replication protein.
The product of the first open reading frame (ORF) identified (rep, nucleotides 478 to 1179) is a predicted 266-amino-acid protein that is homologous to a family of rolling-circle replication (RCR) proteins. The 155 N-terminal amino acids of pKH9 Rep demonstrate 34% amino acid identity and 48% similarity to the characterized RCR protein of the Bacillus borstelensis pHT92 cryptic plasmid (7). Three conserved motifs typical of the rolling-circle initiator proteins (17) were identified in pKH9 Rep (Fig. 1B). These motifs include the FUTLT motif (U denotes bulky hydrophobic residues I, L, V, M, F, Y, and W), whose function is still unknown (17); the two-His motif (6), involved in DNA binding (motif 2); and the catalytic motif (motif 3), which contains the conserved Tyr residue (Fig. 1B) involved in generating the nick that initiates DNA replication (4).
pKH9 FtsK.
The product of the second ORF identified (ftsK, nucleotides 2483 to 3785) is a putative 434-amino-acid protein belonging to the FtsK/SpoIIIE protein family implicated in intercellular chromosomal DNA transfer (Pfam accession number PF01580). Alignment of the amino acid sequences of the predicted pKH9 FtsK protein against the FtsK-like DNA segregation ATPase YDCQ protein of Clostridium acetobutylicum (27) demonstrates 34% amino acid identity and 51% similarity in a stretch of 345 amino acids. Slightly less identity was observed with the characterized stage III sporulation protein E (SpoIIIE) of Bacillus subtilis (J. Wu and J. Errington, unpublished data). Moreover, pKH9 FtsK includes the FtsK/SpoIIIE C-terminal conserved domain containing the lysine residue (Fig. 1C) essential for the function of the ATP pump that actively transports DNA (3).
pKH9 FtsK is likely involved in maintaining the stability of the pKH9 plasmid by coordinating its segregation among dividing F. nucleatum KH9 cells. This hypothesis is based on well-characterized FtsK/SpoIIIE proteins. The E. coli FtsK cell division protein and homologues are important for proper DNA segregation during cell division. Likewise, SpoIIIE plays an important role in DNA segregation during sporulation in Bacillus subtilis (31).
pKH9 ORF 3.
The predicted product of the third ORF identified is a 184-amino-acid protein with 29% amino acid identity and 50% similarity to ORF 3 of the fusobacterial pFN1 plasmid (12). The function of ORF 3 remains to be determined.
GTAGGTAGGTA motifs.
Attempts to clone all of pKH9 in various plasmids of high and low copy numbers and by using different strains of E. coli (DH5α, ER1793, HB101, XL1-Blue, and SURE2) were unsuccessful, generally resulting in deletions. The A+T-rich content of pKH9 might be one of the factors causing instability in E. coli. Fusobacterium-specific methylation was ruled out as the cause of instability in E. coli because cloned PCR-amplified pKH9 DNA products, although unmethylated, showed no increase in stability. The ORF 3-containing DNA fragment between nucleotides 3977 and 4746 (Fig. 1A) was particularly unstable in E. coli. This fragment is flanked by, and contains within it, three 30-bp direct repeats (R1, R2, and R3), each containing two GTAGGTAGGTA motifs (Fig. 1D). A database search found no homologues to this repeated motif. DNA sequence analysis of an ORF 3 deletion construct isolated from E. coli revealed that a recombination of R1 and R3 (Fig. 1A) generated the deletion of the ORF 3-containing sequence.
pKH9 Axf-Txf addiction system.
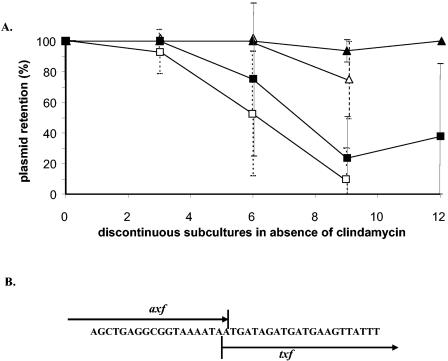
The product of the fourth ORF identified (txf, nucleotides 14 to 304) is a predicted 100-amino-acid protein. This ORF shares 44% amino acid identity and 57% similarity with the Txe protein. Txe (toxin from Enterococcus) is the toxin in the toxin-antitoxin system found in the pRUM plasmid of E. faecium U37 (11). Toxin-antitoxin (also referred to as addiction) systems are a common plasmid-encoded feature that promotes plasmid stability among segregating bacteria (8). The addiction loci contain an operon which encodes a stable toxin and a labile antitoxin. If a bacterial cell loses the plasmid, degradation of the antitoxin leads to the selective killing (or growth impairment) of plasmid-free cells by the stable toxin. In addiction systems, a short gene coding an antitoxin precedes and often overlaps the toxin gene. This feature was also found in pKH9. Upstream of and overlapping the txf (toxin from Fusobacterium) gene, a short ORF was detected (nucleotides 4768 to 15) (Fig. 1A). Based on its short length (73 amino acids) and on the fact that it overlaps txf, we hypothesized that this ORF encodes the Axf (antitoxin from Fusobacterium) antitoxin (Fig. 2B).
FIG. 2.
pKH9 axf-txf toxin-antitoxin system. (A) Stability of a pKH9 derivative containing axf-txf (pKH90) (triangles) versus that of a derivative lacking axf-txf (pORI9) (squares) in F. nucleatum ATCC 23726. Open symbols represent results of the first experiment, and filled symbols represent those of a second independent experiment. (B) Region of overlapping sequence between the axf and txf genes.
To test the function of the Axf-Txf system in F. nucleatum, the stability of pKH90, which possesses the axf-txf operon, was compared to that of pORI9, which lacks the putative addiction system. pKH90 was found to be more stable after subculturing in the absence of antibiotic selection than pORI9 under the same conditions (Fig. 2A). This finding strongly indicates that the pKH9 axf-txf operon functions as a toxin-antitoxin addiction system in fusobacteria.
Identification of the pKH9 replication region.
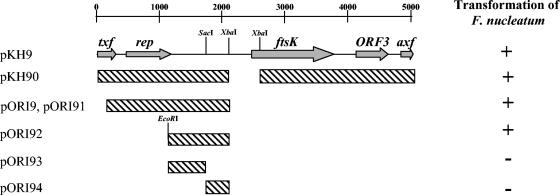
The 4,584-bp fragment released by digestion of pKH9 with XbaI was PCR amplified and cloned into the XbaI site of pBK-erm (Fig. 1A; Table 1) to create pKH90, which confers clindamycin resistance on F. nucleatum ATCC 23726 transformants isolated after electroporation (Fig. 3). A 2-kb fragment containing the pKH9 putative rep gene was amplified and used to construct the pORI9 plasmid (Fig. 3). For reasons yet unclear, pORI9 is more efficient than pKH90 in transforming F. nucleatum ATCC 23726, yielding 103 CFU of plasmid DNA per μg compared to 10 generated by pKH90.
FIG. 3.
Deletion analysis for the identification of the pKH9 minimal replication region. Hatched boxes represent pKH9 fragments present in each construct. pORI91 is a derivative of pORI9 lacking the ampicillin resistance gene. The EcoRI site in pORI92 was created during cloning.
Further deletion analysis suggested that pKH9 replication can occur independently of the Rep protein. The pORI92 plasmid is functional in F. nucleatum ATCC 23726 (Fig. 3), though its pKH9 fragment does not code for any ORF that can be annotated. This finding suggests that pKH9 replication can occur independently of plasmid-encoded proteins in F. nucleatum ATCC 23726, a strain in which no native plasmids have been detected. Additional attempts to minimize the pKH9 replication region (pORI93 and pORI94) (Fig. 3) failed to generate clindamycin-resistant F. nucleatum colonies.
Compatibility of pKH9 with pFN1.
F. nucleatum carrying the pFN1-based pHS30 plasmid conferring thiamphenicol resistance (19) was electroporated with pORI91 (expressing clindamycin resistance). Seventy-four transformants were recovered on media with clindamycin from three different transformations. All transformant colonies were subcultured on media with clindamycin and then tested for growth on media with both clindamycin and thiamphenicol. All transformant colonies grew on these selective media, demonstrating resistance to both antibiotics. Plasmid DNA was isolated from 20 representative transformants, and both plasmids were evident in these preparations, with no evidence of DNA deletion or rearrangement (Fig. 4).
DISCUSSION
In this study, pKH9, a novel 4,975-bp fusobacterial plasmid, has been described. Five ORFs were identified in the pKH9 sequence, and its replication region was determined (Fig. 1A). Previously, only two highly homologous F. nucleatum plasmids have been analyzed in depth. pPA52 (23) (GenBank accession no. AF022647) was isolated from F. nucleatum FDC 27-17, and pFN1 (12) (GenBank accession no. AF159249) was isolated from F. nucleatum 12230. Excluding ORF 3 (with the yet-unknown function), the pKH9 DNA sequence shows no identity to that of pFN1 or pPA52. In the latter two plasmids, replication is hypothesized to involve a theta mode (12). In contrast, the pKH9 Rep amino acid sequence suggests that it uses an RCR mechanism (Fig. 1B). pKH9 represents a new incompatibility group of F. nucleatum plasmids, as was confirmed by the incompatibility tests (Fig. 4).
The pORI92 construct carries a 0.96-kb pKH9 fragment that enables its replication in F. nucleatum ATCC 23726. No ORF can be annotated in this fragment, suggesting that pORI92 replication in this strain is independent of plasmid-encoded proteins. Whether this phenomenon is unique to strain ATCC 23726 (in which no other plasmid can be detected) is still under investigation. Since pORI92 does not carry pKH9 rep, the function of Rep seems redundant. A possible explanation is the fact that pKH9 has two modes of replication, as occurs in the phage-plasmid phasyl (10). The phasyl's first replication mode is independent of plasmid-encoded proteins and requires a specific origin of replication (named oriI). The second replication mode occurs by a rolling-circle mechanism and depends upon the phasyl's Arp replication protein. The homology of pKH9 Rep with the phasyl's Arp (26% amino acid identity and 41% similarity in the 134 N-terminal amino acids of Rep) supports this hypothesis of two replication modes. The location of the pKH9 Rep-dependent origin of replication might differ from that of pORI92, which utilizes the host protein-dependent replication mode, and remains to be identified.
Attempts to transform F. nucleatum ATCC 10953 with either pKH90 (rep+) or pORI92 (rep mutant) have been unsuccessful so far. This failure, together with the fact that only two F. nucleatum strains were successfully transformed with pHS17 (S. K. Haake, unpublished results), demonstrates the limited host range of F. nucleatum plasmids. The plasmid content of a pORI92 replication-proficient host might provide insights into the source of the replication initiator protein supplied by the host.
pKH9 Axf-Txf is the first fusobacterial addiction system tested. Axf-Txf increases plasmid segregational stability in fusobacteria (Fig. 2A). The toxic mechanism of Txf remains to be discovered. Sequence analysis reveals that Txf is most homologous to the enterococcal Txe toxin. Both proteins are associated with the DUF1224 protein family (Pfam accession no. PF06769). YoeB, a toxin of the E. coli YefM-YoeB toxin-antitoxin system and a member of the DUF1224 protein family, was recently shown to induce cleavage of translated mRNAs (5). The 73-amino-acid pKH9 antitoxin Axf is less conserved, as is typical among other antitoxins (25). However, a PSI-BLAST search with pKH9 Axf revealed a homology of 28% identity and 58% similarity over a stretch of 50 amino acids with a hypothetical F. nucleatum cytosolic protein (GenBank accession no. ZP_00145028). This hypothetical protein was found to belong to the Phd_YefM (22) antitoxin family (Pfam accession no. PF02604). Many members of the Phd_YefM antitoxin family are coupled with toxins of the DUF1224 family. This finding also supports the hypothesis that Axf-Txf is a toxin-antitoxin plasmid addiction system.
The pKH9- and pFN1-based E. coli-F. nucleatum shuttle plasmids are compatible in fusobacteria (Fig. 4), as expected from the sequence analysis. The chimeric shuttle plasmids are important for future fusobacterial research. Molecular approaches based on these vector systems will be critical for identification of fusobacterial genes involved in systemic infections and coaggregations with other human oral bacteria and for the elucidation of the role of fusobacterial bridging in the development of oral biofilms.
Acknowledgments
We thank N. Ganeshkumar and A. Tanner of the Forsyth Institute for supplying the strains of F. nucleatum and Tamar Kahan from the bioinformatic unit, Hebrew University Hadassah Medical School, for bioinformatic assistance.
This work was supported in part by United States-Israel Binational Science Foundation grant number 1999126 and by NIH/NIDCR PHS grant number DE12639 to S.K.H.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bachrach, G., G. Rosen, M. Bellalou, and M. Sela. 2004. Identification of a Fusobacterium nucleatum 65 kDa serine protease. Oral Microbiol. Immunol. 19:155-159. [DOI] [PubMed] [Google Scholar]
- 3.Bath, J., L. J. Wu, J. Errington, and J. C. Wang. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995-997. [DOI] [PubMed] [Google Scholar]
- 4.Campos-Olivas, R., J. M. Louis, D. Clerot, B. Gronenborn, and A. M. Gronenborn. 2002. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. USA 99:10310-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, S. K., G. Maenhaut-Michel, N. Mine, S. Gottesman, K. Gerdes, and L. Van Melderen. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 51:1705-1717. [DOI] [PubMed] [Google Scholar]
- 6.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarría, M. Espinosa, and R. Díaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebisu, S., Y. Murahashi, H. Takagi, K. Kadowaki, K. Yamaguchi, H. Yamagata, and S. Udaka. 1995. Nucleotide sequence and replication properties of the Bacillus borstelensis cryptic plasmid pHT926. Appl. Environ. Microbiol. 61:3154-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gielow, A., L. Diederich, and W. Messer. 1991. Characterization of a phage-plasmid hybrid (phasyl) with two independent origins of replication isolated from Escherichia coli. J. Bacteriol. 173:73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47:1419-1432. [DOI] [PubMed] [Google Scholar]
- 12.Haake, S. K., S. C. Yoder, G. Attarian, and K. Podkaminer. 2000. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J. Bacteriol. 182:1176-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, Y. W., W. Shi, G. T.-J. Huang, S. Kinder Haake, N.-H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins, D. G. 1994. CLUSTAL V: multiple alignments of DNA and protein sequences. Methods Mol. Biol. 25:307-318. [DOI] [PubMed] [Google Scholar]
- 15.Hill, G. B. 1998. Preterm birth: associations with genital and possibly oral microflora. Ann. Periodontol. 3:222-232. [DOI] [PubMed] [Google Scholar]
- 16.Huang, G. T.-J., D. Kim, J. K.-H. Lee, H. K. Kuramitsu, and S. K. Haake. 2001. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect. Immun. 69:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapatral, V., I. Anderson, N. Ivanova, G. Reznik, T. Los, A. Lykidis, A. Bhattacharyya, A. Bartman, W. Gardner, G. Grechkin, L. Zhu, O. Vasieva, L. Chu, Y. Kogan, O. Chaga, E. Goltsman, A. Bernal, N. Larsen, M. D'Souza, T. Walunas, G. Pusch, R. Haselkorn, M. Fonstein, N. Kyrpides, and R. Overbeek. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 184:2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinder Haake, S. 2003. Fusobacterium nucleatum-E. coli shuttle plasmid pHS30 confers resistance to chloramphenicol. J. Dent. Res. 82(Special issue B):B-154. [Google Scholar]
- 20.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E., and J. London. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175:3247-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehnherr, H., E. Maguin, S. Jafri, and M. B. Yarmolinsky. 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233:414-428. [DOI] [PubMed] [Google Scholar]
- 23.McKay, T. L., J. Ko, Y. Bilalis, and J. M. DiRienzo. 1995. Mobile genetic elements of Fusobacterium nucleatum. Plasmid 33:15-25. [DOI] [PubMed] [Google Scholar]
- 24.Mikamo, H., K. Kawazoe, Y. Sato, and T. Tamaya. 1999. Elastase activity of anaerobes isolated from amniotic fluid with preterm premature rupture of membranes. Am. J. Obstet. Gynecol. 180:378-380. [DOI] [PubMed] [Google Scholar]
- 25.Mittenhuber, G. 1999. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biotechnol. 1:295-302. [PubMed] [Google Scholar]
- 26.Moore, W. E. C., and L. V. H. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 27.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, GTC Sequencing Center Production, Finishing, and Bioinformatics Teams, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, L. J., and J. Errington. 1994. Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]