Abstract
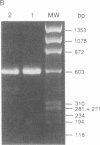
The RNA helicase gene family encodes a group of eight homologous proteins that share regions of sequence similarity. This group of evolutionarily conserved proteins presumably all utilize ATP (or some other nucleoside triphosphate) as an energy source for unwinding double-stranded RNA. Members of this family have been implicated in a variety of physiological functions in organisms ranging from Escherichia coli to human, such as translation initiation, mitochondrial mRNA splicing, ribosomal assembly, and germinal line cell differentiation. We have applied polymerase chain reaction technology to search for additional members of the RNA helicase family in the yeast Saccharomyces cerevisiae. Using degenerate oligonucleotide primers designed to amplify DNA fragments flanked by the highly conserved motifs V L D E A D and Y I H R I G, we have detected five putative RNA helicase genes. Northern and Southern blot analyses demonstrated that these genes are single copy and expressed in yeast. Several members of the RNA helicase family share sequence identity ranging from 49.2% to 67.2%, suggesting that they are functionally related. The discovery of such a multitude of putative RNA helicase genes in yeast suggests that RNA helicase activities are involved in a variety of fundamentally important biological processes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abramson R. D., Dever T. E., Lawson T. G., Ray B. K., Thach R. E., Merrick W. C. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J Biol Chem. 1987 Mar 15;262(8):3826–3832. [PubMed] [Google Scholar]
- Brow D. A., Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988 Jul 21;334(6179):213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- Cech T. R. A model for the RNA-catalyzed replication of RNA. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4360–4363. doi: 10.1073/pnas.83.12.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ford M. J., Anton I. A., Lane D. P. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature. 1988 Apr 21;332(6166):736–738. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- Geider K., Hoffmann-Berling H. Proteins controlling the helical structure of DNA. Annu Rev Biochem. 1981;50:233–260. doi: 10.1146/annurev.bi.50.070181.001313. [DOI] [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989 Jun 15;339(6225):562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- Hodgman T. C. A new superfamily of replicative proteins. Nature. 1988 May 5;333(6168):22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Konarska M. M., Grabowski P. J., Sharp P. A. Spliceosome assembly involves the binding and release of U4 small nuclear ribonucleoprotein. Proc Natl Acad Sci U S A. 1988 Jan;85(2):411–415. doi: 10.1073/pnas.85.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko P. F., Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988 Oct 13;335(6191):611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Leroy P., Alzari P., Sassoon D., Wolgemuth D., Fellous M. The protein encoded by a murine male germ cell-specific transcript is a putative ATP-dependent RNA helicase. Cell. 1989 May 19;57(4):549–559. doi: 10.1016/0092-8674(89)90125-6. [DOI] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. Birth of the D-E-A-D box. Nature. 1989 Jan 12;337(6203):121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Linder P., Slonimski P. P. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2286–2290. doi: 10.1073/pnas.86.7.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Slonimski P. P. Sequence of the genes TIF1 and TIF2 from Saccharomyces cerevisiae coding for a translation initiation factor. Nucleic Acids Res. 1988 Nov 11;16(21):10359–10359. doi: 10.1093/nar/16.21.10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K., Morel-Deville F., Hershey J. W., Leighton T., Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988 Dec 1;336(6198):496–498. doi: 10.1038/336496a0. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., Thach R. E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985 Jun 25;260(12):7651–7658. [PubMed] [Google Scholar]
- Scheffner M., Knippers R., Stahl H. RNA unwinding activity of SV40 large T antigen. Cell. 1989 Jun 16;57(6):955–963. doi: 10.1016/0092-8674(89)90334-6. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985 Sep;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Boulet A., Simon M., Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6810–6814. doi: 10.1073/pnas.84.19.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Boulet A., Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989 Jan 5;337(6202):84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]