Abstract
Non-small-cell lung cancer (NSCLC) may establish an immunosuppressive tumor microenvironment that is conducive to tumor growth. Natural killer (NK) cells play a pivotal role in immunological surveillance. Activation of NK cells partially depends on the interactions between killer-cell immunoglobulin-like receptors (KIRs) and human leukocyte antigen (HLA) class I ligands. We herein investigated the association of KIRs and HLA ligands with survival in metastatic NSCLC (mNSCLC) patients treated with chemotherapy in a Chinese Han population. Polymerase chain reaction with sequence-specific primers was used to type 15 KIRs at the DNA and mRNA level and 6 HLA ligands in 70 mNSCLC patients. Survival curves were estimated using the Kaplan-Meier method and compared with the log-rank test. Cox proportional hazard regression model was applied for multivariate survival analysis, with the stepwise selection, to determine independent predictors of survival. It was observed that patients with KIR2DS4del gene expression at the mRNA level or HLA-Bw4T80 exhibited poor overall survival (OS). The multivariate analysis revealed that HLA-Bw4T80 and KIR2DS4del expression were independent predictors of OS. This observation indicated that the KIR/HLA ligand is a promising predictor of survival in mNSCLC and may also provide a strategy for treatment stratification and patient management.
Keywords: killer-cell immunoglobulin-like receptor, human leukocyte antigen, non-small-cell lung cancer, prognosis
Introduction
Non-small-cell lung cancer (NSCLC) may establish an immunosuppressive tumor microenvironment that is conducive to tumor growth (1). Natural killer (NK) cells play a pivotal role in innate immunity and immunological surveillance (2). Activation of NK cells partly depends on the interactions between killer-cell immunoglobulin-like receptors (KIRs) and human leukocyte antigen (HLA) class I ligands (3). KIRs are a family of cell surface receptors expressed by NK cells and certain subpopulations of T lymphocytes. The KIR family consists of inhibitory as well as activating receptors characterized by both allelic (high numbers of variants) and haplotypic (different numbers of genes for inhibitory and activating receptors on individual chromosomes) polymorphisms (4). The ligands of KIRs are specific epitopes on HLA class I molecules (5). HLA-C allotypes fall into the C1 or C2 groups (asparagine and lysine, respectively, at position 80) that are recognized by KIR2DL2/KIR2DL3 and KIR2DL1/KIR2DS1, respectively. Two previous studies have implicated HLA-C1 and HLA-C2 as potential ligands for KIR2DS4 (6,7). KIR3DL1/KIR3DS1 recognizes HLA-Bw4 epitopes of HLA-A and HLA-B alleles, which are classified according to whether isoleucine or threonine is present at position 80 (4).
An increased understanding of the complexities of the biology of the KIR/HLA system has provided opportunities to leverage NK cell function as a novel avenue of immunotherapy for cancer (8). Epidemiological studies have associated particular KIR and HLA genotypes with the susceptibility to and clinical outcome of leukemia and certain types of solid tumors (9–11). There is a lack of clinical information on the role of KIR/HLA interactions in metastatic NSCLC (mNSCLC). Therefore, the present study was performed to investigate the effect of KIR/HLA ligand on the prognosis of Chinese Han mNSCLC patients treated with chemotherapy.
Patients and methods
Patients and follow-up
A total of 70 eligible Chinese Han patients who attended the Fudan University Shanghai Cancer Center (Shanghai, China) were enrolled in the present study. Eligible patients were aged ≥18 years and had pathologically confirmed metastatic or recurrent NSCLC. Patients were considered ineligible if they had received previous systemic anticancer therapy <1 year prior, had severe drug allergies, had an active infection or other serious diseases or conditions, or were unable to provide consent. Blood samples and laboratory blood test results were collected prior to chemotherapy. The performance status of the cancer patients was defined according to the Eastern Cooperative Oncology Group (ECOG). Eligible patients received platinum-based first-line chemotherapy. All the patients were followed up for 2 years after enrollment. Follow-up data for all patients were obtained from their most recent medical review, which consisted of a clinical examination and an assessment of chest X-rays or computed tomography scans. Progression-free survival (PFS) was defined as the time from the date of inclusion to the date of disease progression or death, whichever came first. Overall survival (OS) was defined as the time from the date of inclusion to the date of death. Data were censored if patients were non-progressing or alive at the time point of evaluation. If a patient missed a follow-up or had an unknown event date, the patient was censored at the last radiological assessment for PFS and the last contact date for OS. The Ethics Committee of Fudan University Shanghai Cancer Center approved this study and all the included patients provided written informed consent.
Sample preparation
Genomic DNA was extracted from peripheral blood mononuclear cells using the QIAamp DNA Mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. Total RNA was extracted from whole-blood samples with the PAXgeneÔ Blood RNA system (PreAnalytiX GmbH, Hombrechtikon, Switzerland). cDNA was then synthesized with the QuantiTect® reverse transcription kit (Qiagen).
KIR and HLA ligand typing
The presence or absence of 15 human KIR genes plus two pseudogenes at the level of genomic DNA and mRNA was analyzed by sequence-specific primer polymerase chain reaction (SSP-PCR) using the Miltenyi Biotec KIR typing kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) according to the manufacturer's protocol. Differentiation between KIR2DS4del (the 22 bp-deleted form of KIR2DS4) vs. KIR2DS4full (the full-length form of KIR2DS4) was also possible. The presence or absence of KIR-HLA ligands, including HLA-CAsn80 (HLA-C1), HLA-CLys80 (HLA-C2), HLA-BBw4+Thr80 (HLA-Bw4T80), HLA-BBw4+Ile80 (HLA-Bw4I80), HLA-ABw4+ and HLA-BBw4+Asp77,Thr80 was determined by SSP-PCR using the Olerup SSP KIR HLA Ligand kit (Olerup SSP, Stockholm, Sweden) according to the manufacturer's protocol.
Statistical analysis
Differences in categorical variables were analyzed with the Fisher's exact test. Frequencies of >20% and <80% for KIRs or HLA ligands were included in the univariate analysis. Survival curves were estimated using the Kaplan-Meier method and compared with the log-rank test. Only variables with P<0.1 in the univariate analysis (Tables II and III), apart from the epidermal growth factor receptor gene (EGFR) mutation status (17.1% of patients not tested), were entered in the multivariate model. The Cox proportional hazard regression model was applied for multivariate survival analysis, with the stepwise selection, to determine independent predictors of survival. The Benjamini-Hochberg method (12) was used to perform multitest correction. The contingency analysis was computed and the Fisher's exact test was used to investigate the independence between the significant KIR/HLA ligand and other variables. P<0.05 was considered to indicate statistically significant differences. All the analyses were conducted using SAS® 9.4 software (SAS Institute Inc., Cary, NC, USA).
Table II.
Association of KIR and HLA class I ligands with survival prognosis of patients with mNSCLC by univariate analysis.
| PFS (negative vs. positive) | OS (negative vs. positive) | |||
|---|---|---|---|---|
| KIR and HLA typing (n=70) | HR (95% CI) | P-value | HR (95% CI) | P-value |
| KIR genotype | ||||
| KIR2DL5all | 0.58 (0.32–1.05) | 0.072 | 1.05 (0.54–2.05) | 0.888 |
| KIR2DL5A | 0.59 (0.32–1.10) | 0.095 | 0.96 (0.49–1.90) | 0.912 |
| KIR2DL5B | 0.64 (0.35–1.17) | 0.148 | 1.06 (0.54–2.08) | 0.870 |
| KIR2DS4del | 1.07 (0.62–1.85) | 0.797 | 0.60 (0.31–1.16) | 0.126 |
| KIR2DS4full | 1.29 (0.69–2.41) | 0.430 | 1.39 (0.66–2.91) | 0.387 |
| KIR2DS4del+/KIR2DS4full− | 0.97 (0.50–1.91) | 0.935 | 0.60 (0.26–1.37) | 0.227 |
| KIR2DS4full+/KIR2DS4del− | 1.11 (0.64–1.92) | 0.716 | 1.57 (0.81–3.06) | 0.182 |
| KIR2DS4ful+/KIR2DS4del+ | 1.20 (0.67–2.16) | 0.540 | 0.85 (0.40–1.82) | 0.673 |
| KIR3DS1 | 0.70 (0.38–1.28) | 0.243 | 0.99 (0.50–1.95) | 0.979 |
| KIR2DS1 | 0.56 (0.30–1.05) | 0.071 | 0.93 (0.47–1.85) | 0.843 |
| KIR2DS2 | 0.80 (0.38–1.69) | 0.557 | 0.89 (0.38–2.11) | 0.795 |
| KIR2DS3 | 0.58 (0.34–2.14) | 0.736 | 1.48 (0.55–3.97) | 0.436 |
| KIR2DS5 | 0.60 (0.32–1.14) | 0.118 | 1.12 (0.56–2.26) | 0.744 |
| KIR cDNA type | ||||
| KIR2DL5all | 0.70 (0.39–1.27) | 0.244 | 0.90 (0.45–1.80) | 0.760 |
| KIR2DS1 | 0.72 (0.39–1.34) | 0.300 | 1.43 (0.62–3.27) | 0.399 |
| KIR2DS4del | 1.18 (0.64–2.18) | 0.588 | 0.51 (0.26–1.01) | 0.049 |
| KIR2DS4full | 1.22 (0.67–2.24) | 0.516 | 1.55 (0.77–3.11) | 0.219 |
| KIR3DL1 | 1.40 (0.80–2.46) | 0.231 | 1.16 (0.58–2.31) | 0.669 |
| KIR3DS1 | 0.59 (0.33–1.06) | 0.075 | 0.96 (0.48–1.91) | 0.908 |
| KIR2DS4full+/KIR2DS4del− | 1.19 (0.68–2.09) | 0.540 | 1.84 (0.93–3.66) | 0.082 |
| KIR HLA ligand | ||||
| HLA-C2 | 1.21 (0.67–2.18) | 0.523 | 1.23 (0.59–2.57) | 0.574 |
| HLA-Bw4T80 | 0.69 (0.39–1.22) | 0.199 | 0.54 (0.28–1.05) | 0.066 |
| HLA-Bw4I80 | 0.70 (0.39–1.25) | 0.225 | 0.93 (0.43–1.99) | 0.851 |
| HLA-ABw4 | 1.31 (0.73–2.35) | 0.364 | 1.72 (0.81–3.67) | 0.156 |
| HLA-C genotype | ||||
| C1C1 | 1.31 (0.69–2.47) | 0.614 | 1.33 (0.60–2.95) | 0.746 |
| C1C2 | 0.78 (0.24–2.57) | 0.86 (0.20–3.62) | ||
| C2C2 | 0.60 (0.17–2.12) | 0.64 (0.14–3.04) | ||
Bold print, P-values <0.1. mNSCLC, metastatic non-smallcell lung cancer; OS, overall survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval; KIR, killer-cell immunoglobulin-like receptor; HLA, human leukocyte antigen. Frequencies of >20% and <80% for KIRs or HLA ligands were included in the survival analysis.
Table III.
Patient characteristics and their value in predicting survival by univariate analysis.
| OS | PFS | |||||
|---|---|---|---|---|---|---|
| Variables | Subgroups | No. (%) | Median | P-value | Median | P-value |
| Age, years | ≤60 | 41 (58.6) | 19.3 | 0.583 | 10.5 | 0.178 |
| >60 | 29 (41.4) | 17.5 | 4.4 | |||
| Gender | Female | 29 (41.4) | NR | <0.001 | 12.1 | 0.006 |
| Male | 41 (58.6) | 11.9 | 4.5 | |||
| Smoking status | Never smoker | 32 (45.7) | NR | 0.001 | 10.5 | 0.152 |
| Ever smoker | 38 (54.3) | 11.9 | 4.4 | |||
| ECOG PS | 0 | 13 (18.8) | 23.2 | 0.073 | 6.5 | <0.001 |
| 1 | 49 (71.0) | 19.3 | 10.5 | |||
| 2/3 | 8 (11.4) | 4.5 | 2.2 | |||
| EGFRa | Mutation | 21 (36.2) | NR | <0.001 | 11.6 | 0.082 |
| Wildtype | 37 (63.8) | 15.6 | 6.3 | |||
| Histologyb | AD | 58 (86.6) | NR | <0.001 | 10.3 | 0.002 |
| SCC | 9 (13.4) | 10.0 | 2.8 | |||
| Metastatic site | Brain/liver | 13 (18.6) | 14.4 | 0.266 | 7.3 | 0.622 |
| Others | 57 (81.4) | 21.4 | 8.1 | |||
| CRP (mg/l)d | ≤10 | 45 (67.2) | NR | 0.001 | 10.3 | 0.140 |
| >10 | 22 (32.8) | 11.6 | 3.5 | |||
Data were unavailable for 12 patients.
Data were unavailable for 3 patients. PFS, progression-free survival; OS, overall survival; CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; EGFR, epidermal growth factor receptor gene; AD, adenocarcinoma; SCC, squamous cell carcinoma; S-1, fluorouracil derivative; CRP, C-reactive protein; NLR, neutrophil/lymphocyte ratio; PLR, platelet/lymphocyte ratio; NR, not reached.
Results
Concordance between KIR typing using DNA and mRNA in NSCLC patients
Raw frequency data of all tested KIRs at the DNA and mRNA level, as well as HLA ligands, were calculated (data not shown). The framework genes 3DL3, 3DP1, 2DL4 and 3DL2 were positive in all samples at the DNA level, while the framework genes 3DL3 (0%) and 3DP1 (4.3%) were absent in nearly all individuals at the mRNA level. The frequencies of 2DL1, 2DL2, 2DL3, 3DS1, 2DS4full and 2DP1 at the DNA level were similar to those at the mRNA level, whereas the frequencies of other genes at the DNA level were significantly higher compared with those at the mRNA level. Furthermore, the frequencies of the full-length and deleted versions of KIR2DS4 were analyzed and the difference between the DNA and mRNA levels was compared (Table I). The majority of the 70 samples [62 at the mRNA (88.6%) and 65 (92.9%) at the DNA level] from the Chinese Han population were positive for KIR2DS4. The KIR2DS4del variant was found in 47.1% of the 70 individuals at the DNA level, similar to what was previously reported (13). However, at the mRNA level, the presence of the KIR2DS4del variant decreased to 27.1% in this population. The ratio of deleted:full-length form of KIR2DS4 was 1:2.3 at the DNA level and 1:3.9 at the mRNA level (Table I).
Table I.
Frequency of KIR2DS4 alleles at the mRNA and DNA level.
| Type | 2DS4del | 2DS4full | 2DS4full-positive/2DS4del-positive | 2DS4full-positive/2DS4del-negative | 2DS4full-positive/2DS4del-positive | 2DS4negative | 2DS4dela/2DS4fullb |
|---|---|---|---|---|---|---|---|
| mRNA (n=70) | 27.1% (19) | 72.9% (51) | 15.7% (11) | 61.4% (43) | 11.4% (8) | 11.4% (8) | 1:3.9 |
| DNA (n=70) | 47.1% (33) | 72.9% (51) | 20.0% (14) | 45.7% (32) | 25.7% (18) | 7.1% (5) | 1:2.3 |
2DS4full-negative/2DS4del-positive.
2DS4full-positive/2DS4del-negative. KIR, killer-cell immunoglobulin-like receptor.
Effect of individual KIR and HLA class I ligands on the prognosis of mNSCLC
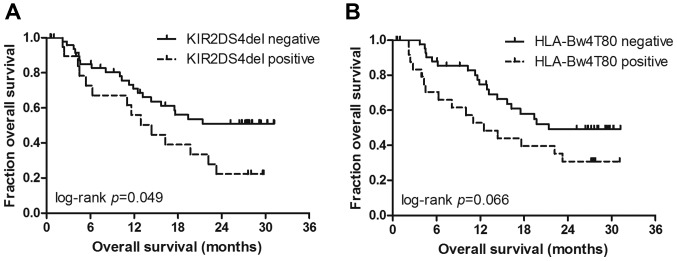
The vast majority of the patients (95.7%) were positive for HLA-C1, a frequency which is similar to that of the Japanese population (14). Among the 15 types of KIRs and the 6 types of HLA ligands, with the exception that positive HLA-Bw4I80 expression was associated with a poor performance status, there were no other significant associations between KIRs/HLA ligands and patient characteristics (data not shown). Patients without KIR2DS4del gene expression (Fig. 1A) exhibited a significantly better OS [hazard ratio (HR)=0.51, P=0.049]. The median OS was not reached in patients without KIR2DS4del gene expression vs. 14.5 months in patients with KIR2DS4del gene expression. As regards the HLA genotype, only HLA-Bw4T80 was found to be associated with decreased OS at the 10% level (HR=0.51, P=0.066) (Fig. 1B). The median OS was 12.5 months in the HLA-Bw4T80-positive group vs. 21.4 months in the HLA-Bw4T80-positive group. No other KIRs or HLA ligands were significantly associated with clinical outcome according to the univariate analysis (Table II).
Figure 1.
Kaplan-Meier overall survival curves for mNSCLC patients. (A) Kaplan-Meier curves of the association between KIR2DS4del and overall survival. (B) Kaplan-Meier curves of the association between HLA-Bw4T80 and overall survival. P-value was calculated by the log-rank test. mNSCLC, metastatic non-small-cell lung cancer. KIR, killer-cell immunoglobulin-like receptor; HLA, human leukocyte antigen.
Multivariate analysis considering patient characteristics
The effect of known clinicopathological confounding factors [age, gender, smoking status, performance status, EGFR mutation status, histology, metastatic site and C-reactive protein (CRP) level] on OS was evaluated by univariate analysis (Table III). The results revealed that male gender, adenocarcinoma and a performance status of 2 were significantly associated with disease progression. Male gender, adenocarcinoma, smoking, wild-type EGFR status and CRP level were factors that significantly affected OS. In the stepwise multivariate Cox model, which includes significant clinical factors and KIR/HLA ligands at the 10% level, positive KIR2DS4del gene expression (KIR2DS4del mRNA) (adjusted P=0.014, HR=3.33), positive HLA-Bw4T80 (adjusted P=0.012, HR=3.58), smoking (adjusted P=0.02, HR=2.87) and a CRP level >10 mg/l (adjusted P=0.004, HR=6.48) remained independent predictors of decreased OS; no KIR/HLA type or clinicopathological variables were selected as independent predictors of PFS following multitest correction (Table IV).
Table IV.
Multivariate Cox analysis of PFS and OS.
| Multivariate analysis | ||||
|---|---|---|---|---|
| Variables | Groups | HR (95% CI) | P-value | Adjusted P-value |
| OS (n=64) | ||||
| CRP, mg/l | >10 vs. ≤10 | 6.48 (2.11–19.91) | 0.001 | 0.004 |
| Smoking | Yes vs. no | 2.87 (1.18–6.96) | 0.020 | 0.020 |
| HLA-Bw4T8 | Positive vs. negative | 3.58 (1.51–8.47) | 0.004 | 0.012 |
| KIR2DS4del mRNA | Positive vs. negative | 3.33 (1.39–7.94) | 0.007 | 0.014 |
| PFS (n=64) | ||||
| CRP, mg/l | >10 vs. ≤10 | 2.94 (1.36–6.37) | 0.006 | 0.054 |
| Histology | SCC vs. AD | 2.38 (0.95–5.95) | 0.065 | 0.195 |
| Gender | Male vs. female | 1.71 (0.84–3.49) | 0.138 | 0.204 |
| ECOG PS | 2 vs. 0 | 4.20 (1.10–16.09) | 0.036 | 0.160 |
| HLA-Bw4T8 | Positive vs. negative | 2.41 (1.21–4.83) | 0.013 | 0.091 |
| KIR2DS1 DNA | Positive vs. negative | 17.08 (1.28–227.54) | 0.032 | 0.160 |
| KIR2DL5all DNA | Positive vs. negative | 34.14 (1.52–765.94) | 0.026 | 0.156 |
| KIR2DL5A DNA | Positive vs. negative | 0.047 (0.005–0.461) | 0.047 | 0.072 |
| KIR2DL5B DNA | Positive vs. negative | 0.063 (0.002–1.737) | 0.063 | 0.204 |
Bold print, P-values indicating statistically significant differences. OS, overall survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval-; CRP, C-reactive protein; SCC, squamous cell carcinoma; AD, adenocarcinoma; ECOG PS, Eastern Cooperative Oncology Group performance status; KIR, killer-cell immunoglobulin-like receptor; HLA, human leukocyte antigen.
Discussion
Several studies have investigated the association between KIR/HLA genotypes and autoimmune disease, infectious disease, transplantation, stem cell disease and malignancy (15–17). Regarding malignant diseases, certain KIR/HLA types have been associated with the susceptibility or worse clinical outcome in several types of cancer, such as leukemia, colorectal cancer and lung cancer (18–20).
In lung cancer, Wisniewski et al reported a possible protective effect of the HLA-C C1C2 genotype on susceptibility to NSCLC and its association with the KIR2DL2/KIR2DS2/C1C1 genotype, which was correlated with better response to therapy and longer survival in the Caucasian population (20). Due to the ethnic differences and small sample size, we did not observe these consistent findings in our samples. In the present study, both cDNA and genomic DNA were typed, in contrast to other reports where genomic DNA was used, considering that only expressed genes may play a biological role and non-expressed KIR gene(s) may exert an effect on a different, causative gene, due to linkage disequilibrium. We observed that the majority of KIR genes had a different typing profile at the DNA and mRNA levels. Furthermore, only KIR cDNA exerted a significant effect on the outcome of mNSCLC in this study population following multitest correction. We observed that KIR2DS4del expression was associated with a significantly decreased survival time, and KIR2DS4full-positive/KIR2DS4del-negative genotype was significantly associated with long-term survival according to the univariate analysis. However, following multivariate Cox regression analysis and multitest correction, only KIR2DS4del mRNA expression was considered as an independent factor associated with OS.
The activating KIR2DS4 receptor has been reported to bind to group 1 and 2 HLA-C, HLA-A11 and non-HLA class I ligands (21,22). KIR2DS4del is the mutant form of KIR2DS4 with a 22-bp deletion in exon 5, resulting in a frameshift mutation. This truncated KIR2DS4 protein would be secreted due to the loss of the transmembrane/cytoplasmic domains. Individuals who are homozygous for haplotype A and with a deletion in both alleles are likely to have a dysfunctional KIR receptor activator, considering that a proper allele of KIR2DL4 may function as an activating receptor. KIR2DS4del frequencies vary among different populations. In the Caucasian population, the ratio of full-length:deleted KIR2DS4 is ~1:2. However, in the Chinese, Japanese and Korean populations, the ratio of KIR2DS4full to KIR2DS4del is reversed (13), which is similar to what was observed in the present study.
The associations and functions of KIR2DS4 in infectious diseases have been extensively investigated. KIR2DS4full is associated with increased viral loads and transmission rates of HIV-1 (23,24). Additionally, KIR2DS4full promotes HIV-1 pathogenesis during chronic infection, probably through the maintenance of an excessive pro-inflammatory state (25). The combination of KIR2DS4 and KIR2DS4del was associated with disease progression from hepatitis to hepatocellular carcinoma (HCC) development via cirrhosis (26). In terms of malignant diseases, Giebel et al (27) reported that the absence of KIR2DS4full was associated with susceptibility to chronic myeloid leukemia, but the effect of KIR2DS4full appeared to be the opposite in endometrioid ovarian cancer (28). Although the associations of all known 15 human KIRs and their known HLA ligands with the prognosis of mNSCLC were evaluated in the present study, we only observed that the KIR2DS4full-positive/KIR2DS4del-negative type combined with its potential ligand HLA-C1 was significantly associated with improved OS in the univariate analysis (data not shown). Almost all patients expressed HLA-C1 (95.7% positive) in our samples; therefore, it was hypothesized that the improving effect of KIR2DS4full on OS may be associated with potentiating NK cell-mediated cytotoxicity and tumor surveillance through the interaction between HLA-C1 and activating KIR2DS4full.
KIR3DL1 will only inhibit NK cytotoxicity against target cells expressing a discrete subset of HLA-B and HLA-A allotypes. In consideration of the clinicopathological variables, we observed that HLA-Bw4T80 was the only HLA type significantly affecting OS in multivariate analyses following multitest correction. This observation is consistent with a previous report (29), which suggested an association of HLA-Bw4T80 with decreased OS in patients with HCC. HLA-Bw4T80 is an allotype of the HLA-Bw4 motif with a threonine at position 80, which exhibits lower affinity for KIR3DL1 compared with the HLA-Bw4 motif with an isoleucine at the same position (30). It is possible that HLA-Bw4T80 binds with KIR3DL1 to deliver inhibitory signals to NK cells, which would then enhance the ability of tumor cells to escape immune detection (31). However, a positive association of the combination of KIR3DL1/HLA-Bw4T80 with survival was not observed (data not shown).
Notably, the KIR/KLA system exhibits extensive genetic diversity. It would be interesting to perform a similar analysis of an association of KIR2DS4del mRNA and HLA-Bw4T80 with clinical outcome in other ethnic populations. To the best of our knowledge, this is the first study addressing the relevance of KIRs and HLA ligands regarding the clinical outcome of mNSCLC in a Chinese population. If confirmed in a larger cohort of patients in independent studies, KIR2DS4del mRNA and HLA-Bw4T80 may be prognostic indicators for treatment stratification and patient management. Furthermore, functional studies will be required to determine the role of KIR2DS4del mRNA and HLA-Bw4T80 in the immune response against mNSCLC.
Acknowledgements
The present study was supported by Transgene SA, France and by grants from the Wu Jieping Medical Foundation (no. 320675014278) and the Shanghai Municipal Commission of Health and Family Planning (no. 201440423).
Glossary
Abbreviations
- KIR
killer-cell immunoglobulin-like receptor
- HLA
human leukocyte antigen
- NK
natural killer
- NSCLC
non-small-cell lung cancer
- mNSCLC
metastatic non-small-cell lung cancer
- SSP-PCR
sequence-specific primer polymerase chain reaction
- OS
overall survival
- PFS
progression-free survival
- CRP
C-reactive protein
References
- 1.Carbone DP, Gandara DR, Antonia SJ, Zielinski C, Paz-Ares L. Non-small-cell lung cancer: Role of the immune system and potential for immunotherapy. J Thorac Oncol. 2015;10:974–984. doi: 10.1097/JTO.0000000000000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guillerey C, Smyth MJ. NK cells and cancer immunoediting. Curr Top Microbiol Immunol. 2016;395:115–145. doi: 10.1007/82_2015_446. [DOI] [PubMed] [Google Scholar]
- 3.Höglund P, Brodin P. Current perspectives of natural killer cell education by MHC class I molecules. Nat Rev Immunol. 2010;10:724–734. doi: 10.1038/nri2835. [DOI] [PubMed] [Google Scholar]
- 4.Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci. 2012;367:800–811. doi: 10.1098/rstb.2011.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 6.Campbell KS, Cella M, Carretero M, Lopez-Botet M, Colonna M. Signaling through human killer cell activating receptors triggers tyrosine phosphorylation of an associated protein complex. Eur J Immunol. 1998;28:599–609. doi: 10.1002/(SICI)1521-4141(199802)28:02<599::AID-IMMU599>3.3.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260–7267. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- 8.Benson DM, Jr, Caligiuri MA. Killer immunoglobulin-like receptors and tumor immunity. Cancer Immunol Res. 2014;2:99–104. doi: 10.1158/2326-6066.CIR-13-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al Omar S, Middleton D, Marshall E, Porter D, Xinarianos G, Raji O, Field JK, Christmas SE. Associations between genes for killer immunoglobulin-like receptors and their ligands in patients with solid tumors. Hum Immunol. 2010;71:976–981. doi: 10.1016/j.humimm.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Boyton RJ, Altmann DM. Natural killer cells, killer immunoglobulin-like receptors and human leucocyte antigen class I in disease. Clin Exp Immunol. 2007;149:1–8. doi: 10.1111/j.1365-2249.2007.03424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin MP, Borecki IB, Zhang Z, Nguyen L, Ma D, Gao X, Qi Y, Carrington M, Rader JS. HLA-Cw group 1 ligands for KIR increase susceptibility to invasive cervical cancer. Immunogenetics. 2010;62:761–765. doi: 10.1007/s00251-010-0477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreiraa JA, Nyangomab SO. A multivariate version of the Benjamini-Hochberg method. Journal of Multivariate Analysis. 2008;99:2108–2124. doi: 10.1016/j.jmva.2008.02.013. [DOI] [Google Scholar]
- 13.Bao X, Hou L, Sun A, Qiu Q, Yuan X, Chen M, Chen Z, He J. Distribution of killer cell immunoglobulin-like receptor genes and 2DS4 alleles in the Chinese Han population. Hum Immunol. 2010;71:289–292. doi: 10.1016/j.humimm.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz-Peña R, Vidal-Castiñeira JR, Mulero J, Sánchez A, Queiro R, López-Larrea C. Activating killer immunoglobulin-like receptors genes are associated with increased susceptibility to ankylosing spondylitis. Clin Exp Immunol. 2015;180:201–206. doi: 10.1111/cei.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Manna G, Corsini S, Iannelli S, Cappuccilli ML, Comai G, Iorio M, Todeschini P, Carretta E, Scolari MP, Bontadini A, Stefoni S. Influence of the immunogenetic KIR and HLA systems on long-term renal transplant outcome. Ann Transplant. 2013;18:611–621. doi: 10.12659/AOT.889157. [DOI] [PubMed] [Google Scholar]
- 17.Olvera A, Pérez-Álvarez S, Ibarrondo J, Ganoza C, Lama JR, Lucchetti A, Cate S, Hildebrand W, Bernard N, Gomez L, et al. The HLA-C*04: 01/KIR2DS4 gene combination and human leukocyte antigen alleles with high population frequency drive rate of HIV disease progression. AIDS. 2015;29:507–517. doi: 10.1097/QAD.0000000000000574. [DOI] [PubMed] [Google Scholar]
- 18.Babor F, Manser AR, Fischer JC, Scherenschlich N, Enczmann J, Chazara O, Moffett A, Borkhardt A, Meisel R, Uhrberg M. KIR ligand C2 is associated with increased susceptibility to childhood ALL and confers an elevated risk for late relapse. Blood. 2014;124:2248–2251. doi: 10.1182/blood-2014-05-572065. [DOI] [PubMed] [Google Scholar]
- 19.De Re V, Caggiari L, De Zorzi M, Talamini R, Racanelli V, Da M, Buonadonna A, Zagonel V, Cecchin E, Innocenti F, Toffoli G. Genetic diversity of the KIR/HLA system and outcome of patients with metastatic colorectal cancer treated with chemotherapy. PLoS One. 2014;9:e84940. doi: 10.1371/journal.pone.0084940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wisniewski A, Jankowska R, Passowicz-Muszyńska E, Wiśniewska E, Majorczyk E, Nowak I, Frydecka I, Kuśnierczyk P. KIR2DL2/S2 and HLA-C C1C1 genotype is associated with better response to treatment and prolonged survival of patients with non-small cell lung cancer in a Polish Caucasian population. Hum Immunol. 2012;73:927–931. doi: 10.1016/j.humimm.2012.07.323. [DOI] [PubMed] [Google Scholar]
- 21.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Aguilar AM Older, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz G, Gazit R, Arnon TI, Gonen-Gross T, Tarcic G, Markel G, Gruda R, Achdout H, Drize O, Merims S, Mandelboim O. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J Immunol. 2004;173:1819–1825. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- 23.Hong HA, Paximadis M, Gray GE, Kuhn L, Tiemessen CT. KIR2DS4 allelic variants: Differential effects on in utero and intrapartum HIV-1 mother-to-child transmission. Clin Immunol. 2013;149:498–508. doi: 10.1016/j.clim.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Merino A, Malhotra R, Morton M, Mulenga J, Allen S, Hunter E, Tang J, Kaslow RA. Impact of a functional KIR2DS4 allele on heterosexual HIV-1 transmission among discordant Zambian couples. J Infect Dis. 2011;203:487–495. doi: 10.1093/infdis/jiq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merino AM, Dugast AS, Wilson CM, Goepfert PA, Alter G, Kaslow RA, Tang J. KIR2DS4 promotes HIV-1 pathogenesis: New evidence from analyses of immunogenetic data and natural killer cell function. PLoS One. 2014;9:e99353. doi: 10.1371/journal.pone.0099353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan N, Jiang W, Sun H, Miao F, Qiu J, Jin H, Xu J, Shi Q, Xie W, Zhang J. KIR and HLA loci are associated with hepatocellular carcinoma development in patients with hepatitis B virus infection: A case-control study. PLoS One. 2011;6:e25682. doi: 10.1371/journal.pone.0025682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giebel S, Nowak I, Wojnar J, Krawczyk-Kulis M, Holowiecki J, Kyrcz-Krzemien S, Kusnierczyk P. Association of KIR2DS4 and its variant KIR1D with leukemia. Leukemia. 2008;22:2129–2131. doi: 10.1038/leu.2008.108. [DOI] [PubMed] [Google Scholar]
- 28.Giebel S, Boratyn-Nowicka A, Karabon L, Jedynak A, Pamula-Pilat J, Tecza K, Kula D, Kowal M, Frydecka I, Grzybowska E. Associations between genes for killer immunoglobulin-like receptors and their ligands in patients with epithelial ovarian cancer. Hum Immunol. 2014;75:508–513. doi: 10.1016/j.humimm.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Cariani E, Pilli M, Zerbini A, Rota C, Olivani A, Zanelli P, Zanetti A, Trenti T, Ferrari C, Missale G. HLA and killer immunoglobulin-like receptor genes as outcome predictors of hepatitis C virus-related hepatocellular carcinoma. Clin Cancer Res. 2013;19:5465–5473. doi: 10.1158/1078-0432.CCR-13-0986. [DOI] [PubMed] [Google Scholar]
- 30.Purdy AK, Campbell KS. Natural killer cells and cancer: Regulation by the killer cell Ig-like receptors (KIR) Cancer Biol Ther. 2009;8:2211–2220. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Smith AJ, Walsh KM, Ladner MB, Zhang S, Xiao C, Cohen F, Moore TB, Chokkalingam AP, Metayer C, Buffler PA, et al. The role of KIR genes and their cognate HLA class I ligands in childhood acute lymphoblastic leukemia. Blood. 2014;123:2497–2503. doi: 10.1182/blood-2013-11-540625. [DOI] [PMC free article] [PubMed] [Google Scholar]