ABSTRACT
Oligouridylate binding protein 1b (UBP1b), a marker protein of plant stress granules (SGs), plays a role in heat stress tolerance in plants. A previous microarray analysis revealed that the expression of several ABA signaling-related genes is higher in UBP1b-overexpressing Arabidopsis plants (UBP1b-ox) subjected to both non-stressed and heat stress conditions. Root elongation and seed germination assays demonstrated that UBP1b-ox exhibited hypersensitivity to ABA. RT-qPCR analysis confirmed that mitogen-activated protein kinase (MAPK) cascade genes, such as MPK3, MKK4, and MKK9 were upregulated in UBP1b-ox plants. ABA receptor genes, including PYL5 and PYL6, were also upregulated in UBP1b-ox plants. mRNA of WRKY33 – a downstream gene of MPK3 and an upstream gene of ethylene biosynthesis, exhibited high levels of accumulation, although the level of endogenous ABA was not significantly different between UBP1b-ox and control plants. In addition, RNA decay analysis revealed that WRKY33 was more stable in UBP1b-ox plants, indicating that the mRNA of WRKY33 was protected within UBP1b SGs. Collectively, these data demonstrate that UBP1b plays an important role in plant response to ABA.
KEYWORDS: ABA response, ABA sensitivity, RNA stability, UBP1b, UBP1b stress granule
Introduction
Plants have a variable ability to respond to and tolerate environmental stresses, such as drought, heat and high-salinity stress. Understanding the molecular mechanisms responsible for environmental stress tolerance in plants is essential for developing strategies to improve plant productivity under unfavorable environmental conditions. Recent studies have revealed that post-transcriptional regulatory mechanisms, such as the addition of 5′ cap structure,1 splicing,2 3′ poly A addition,3 and control of mRNA degradation4 and storage,5 are involved in stress response and adaptation.
Several papers have also reported that post-transcriptional events are linked to ABA responses. Mutants of genes involved in the post-transcriptional gene regulation exhibited an ABA hypersensitive phenotype. These included: 1) a Hyponastic leaves 1 (HYL1) gene encoding a nuclear localized double-stranded RNA (dsRNA) binding protein6; 2) genes encoding the mRNA cap binding proteins, ABH1 (ABA hypersensitive Arabidopsis 1), CBP80,1 and CBP207; 3) a ABA-hypersensitive germination2 (AHG2) gene encoding a poly (A)-specific ribonuclease (AtPARN)3; and 4) the Supersensitive to ABA and drought 1 (SAD1) gene encoding a polypeptide similar to multifunctional Sm-like snRNP protein.8 The function of the genes related to post-transcriptional gene regulation of ABA signaling and responses, however, is not well understood.
UBP1b, a component of and marker protein of stress granules, protects mRNAs from degradation under abiotic stress conditions by binding to their 3′-UTRs, U-rich introns, and poly(A) tails.9,10 Our previous research indicated that UBP1b plays an important role in plant heat stress tolerance.5 UBP1b–overexpressing plants exhibited increased heat tolerance, while ubp1b mutants exhibited heat sensitivity. Several potential targets of UBP1b, including mRNAs of a DNAJ binding protein and a zinc finger binding protein, were identified.5 Our microarray analysis of UBP1b-ox plants subjected to non-stressed or heat stress conditions revealed higher expression of several ABA signaling-related genes, suggesting that the UBP1b functions in the ABA signaling pathway. Root elongation and seed germination assays demonstrated that UBP1b-ox plants exhibit an ABA-hypersensitive phenotype. These data also indicate that UBP1b plays an important role in ABA response.
Results
Several ABA signaling-related genes are upregulated in UBP1b-ox plants
A previous microarray analysis revealed that the expression of 1,103 genes is higher in UBP1b-ox plants, relative to non-transformed controls, grown under non-stress or heat stress conditions.5 Genes that function in ABA signaling and heat stress response-related genes were among the upregulated genes (Fig. 1; Table 1). In particular, expression of the following ABA signaling-related genes was higher in UBP1b-ox than in control plants: 1) Pyrabactin-resistance 5 and 6 (PYL5, PYL6) genes encoding 2 members of PYR/PYL/RCAR family proteins, which function as ABA receptors11; 2) MAPK family genes such as Mitogen-activated protein kinase 3 (MPK3), and Mitogen-activated protein kinase kinase 4 and 9 (MKK4 and MKK9); all of which are important members of MAPK cascades involved in plant ABA signaling.12,13; and; 3) WRKY DNA-binding protein 33 (WRKY33) which is a member of the WRKY transcription factor family. WRKY33 has been reported to be phosphorylated by MPK3/614 and is involved in the regulation of ethylene biosynthesis15 (Table 1). Collectively, these data suggest that UBP1b may affect ABA signaling and the expression of related downstream genes.
Figure 1.
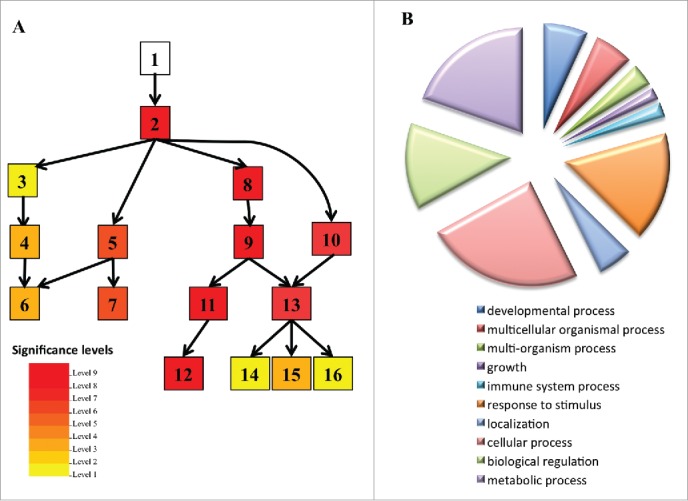
GO analysis of microarray data for biologic process categories. (A) Singular enrichment analysis of the genes with higher expression in UBP1b-ox plants under non-stressed and heat stress conditions. Analysis was performed using AgriGO (http://bioinfo.cau.edu.cn/agriGO/). The box colors indicate the level of statistical significance. 1, GO:0008150 biologic process; 2, GO:0050896 response to stimulus (p = 2.6e−28); 3, GO:0009628 response to abiotic stimulus (p = 5.4e−6); 4, GO:0009266 response to temperature stimulus (p = 7e−8); 5, GO:0006950 response to stress (p = 4.3e−14); 6, GO:0009409 response to cold (p = 2.4e−8); 7, GO:0006952 defense response (p = 5.1e−13); 8, GO:0042221 response to chemical stimulus (p = 8.1e−22); 9, GO:0010033 response to organic substance (p = 8.4e−26); 10, GO:0009719 response to endogenous stimulus (p = 2.6e−13); 11, GO:0009743 response to carbohydrate stimulus (p = 1.5e−20); 12, GO:0010200 response to chitin (p = 3.9e−21); 13, GO:0009725 response to hormone stimulus; 14, GO:009737 response to abscisic acid stimulus (p = 2.6e−13); 15, GO:009733 response to auxin stimulus (p = 5e−7); 16, GO:0032870 cellular response to hormone stimulus (p = 2.6e−13). (B) GO biologic process classification of 1,103 genes with higher expression in UBP1b-ox under non-stressed and heat stress conditions. GO-analysis was performed using the Gene Ontology tool (http://pantherdb.org).
Table 1.
List of ABA and ethylene-related genes whose expression was higher in UBP1b-ox plants than in empty vector, control (Venus) plants.
| Gene name/ | Non-stress |
Heat stress |
Ratio(ox/Venus) | Ratio(ox/Venus) | |||||
|---|---|---|---|---|---|---|---|---|---|
| AGI code | Encoded protein | Venus controla) | UBP1b-oxa) | Venus control a) | UBP1b-oxa) | under non-stressb) | under heat stressc) | Ratio(heat/non-stress) in Venusd) | Ratio(heat/non-stress) in oxe) |
| AT5G05440 | PYL5 | 9.2 | 10.3 | 8.6 | 10.5 | 1.2 | 1.9 | −0.6 | 0.2 |
| AT2G40330 | PYL6 | 7.7 | 8.1 | 5.7 | 7.3 | 0.4 | 1.6 | −2 | −0.8 |
| AT1G51660 | MKK4 | 9.3 | 9.9 | 8.9 | 9.8 | 0.6 | 0.9 | −0.4 | −0.1 |
| AT1G73500 | MKK9 | 10.7 | 12.4 | 9.8 | 10.4 | 1.7 | 0.6 | −0.9 | −2 |
| AT3G45640 | MPK3 | 11.3 | 12.4 | 10.7 | 11.7 | 1.1 | 1 | −0.6 | −0.7 |
| AT2G38470 | WRKY33 | 10.1 | 12.2 | 10.3 | 12.2 | 2.1 | 1.9 | 0.2 | 0 |
Average of signal intensity in 3 biologic replicates.
Average of the ratio of normalized signal value in UBP1b-ox versus Venus control under non-stress condition (p < 0.15; FDR < 0.0001).
Average of the ratio of normalized signal value in UBP1b-ox vs. Venus control under the heat stress condition (p < 0.15, FDR<0.0001).
Average of the ratio of normalized signal value in Venus control in the heat stress condition vs. non-stress condition vs. (p < 0.15, FDR < 0.0001).
Average of the ratio of normalized signal value in UBP1b-ox in the heat stress condition vs. non-stress condition (p < 0.15, FDR < 0.0001).
UBP1b-ox plants exhibit an ABA hypersensitive phenotype
The sensitivity of UBP1b-ox plants to exogenous ABA treatment was investigated using root elongation and seed germination assays to analyze the function of UBP1b in ABA signaling. Roots in UBP1b-ox plants subjected to a 10 μM ABA treatment were shorter than roots in control plants (Fig. 2). Seven days after being treated with ABA, roots in 2 lines of UBP1b-ox plants were approximately 2 cm in length, while roots in non-transformed control plants were about 3 cm. After 9 days, UBP1b-ox roots were less than 3 cm; while roots in control plants were approximately 4 cm (Fig. 2). Results of the seed germination assay indicated that germination of seeds from control plants was approximately 90% on MS medium, 90% on MS medium amended with 0.3 μM, and 50% on MS medium amended with 0.5 μM ABA (Fig. 3). In contrast, germination rates of UBP1b-ox seeds were 50–60% on normal MS medium, and 40% and 10% on MS medium amended with 0.3 μM or 0.5 μM ABA, respectively (Fig. 3). The data on UBP1b-ox seeds and roots indicate that UBP1b-ox plants are hypersensitive to ABA and that UBP1b may promote ABA response.
Figure 2.
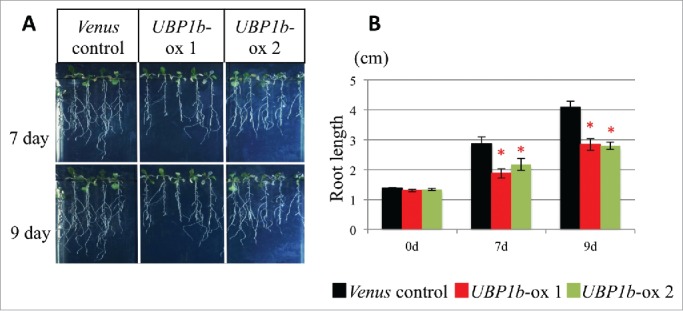
Root elongation assay. (A) Phenotype of UBP1b-ox and control plants in response to exogenous application of 10 μM ABA. Eight-day-old plants were transferred to vertical MS plates containing 10 μM ABA and grown in an environmental chamber. Phenotypes were observed after 7 and 9 d. (B) Root length of treated plants at 7 and 9 d. Data represent the mean ± sd. n = 15. An asterisk indicates a significant difference (p < 0.05) between UBP1b-ox and control plants as determined by a t-test.
Figure 3.
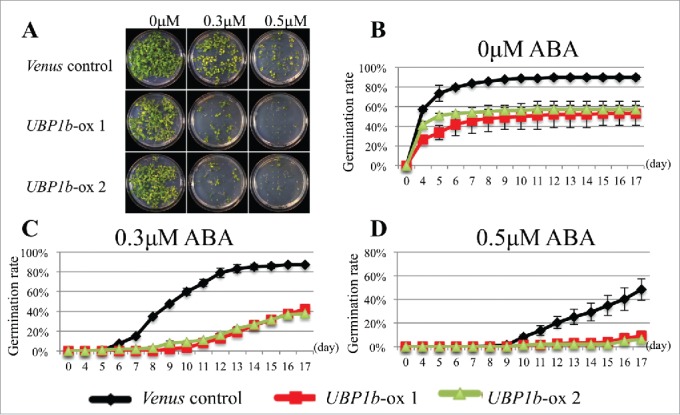
Seed germination assay. (A) Phenotype of 17 day-old seedlings of 2 lines of UBP1b-ox (1 and 2) and empty vector (Venus) control plants germinated on MS plates amended with ABA. Forty-nine seeds were placed on each MS plate amended with 0.3 μM ABA, or 0.5 μM ABA. (B) Seed germination rate of UBP1b-ox and control plants grown on non-amended MS plates. (C) Seed germination rate of UBP1b-ox and control plants on MS plates amended with 0.3 μM ABA. (D) Seed germination rate of UBP1b-ox and control plants on MS plates amended with 0.5 μM ABA. Data represent the mean ± sd. n = 3.
Endogenous ABA levels in UBP1b-ox plants is similar to control plants
The level of endogenous ABA was measured in UBP1b-ox and control plants to determine the role of UBP1b in ABA biosynthesis and the ABA signal transduction pathway. Results indicated that there was no significant difference in the transgenic and control plants under non-stress or heat stress (40oC for 1 h) conditions (Fig. 4B). Additionally, the expression level of Zeaxanthinepoxidase(ZEP), Nine-cis-epoxycarotenoiddioxygenase 3 (NCED3), and ABA-deficient 4 (ABA4), which are all ABA biosynthesis-related genes, was a little downregulated in UBP1b-ox compared with control (Fig. 4A). In contrast, an ABA catabolism-related gene, CYP707A3, was upregulated in non-stress UBP1b-ox, but was not different between UBP1b-ox and control plants subjected to heat stress (Fig. 4A). It is plausible that the differences of ABA biosynthesis and catabolism genes were not enough to cause the change of endogenous ABA level. These results suggest that UBP1b does not affect endogenous ABA levels but rather regulates ABA signaling-related genes.
Figure 4.
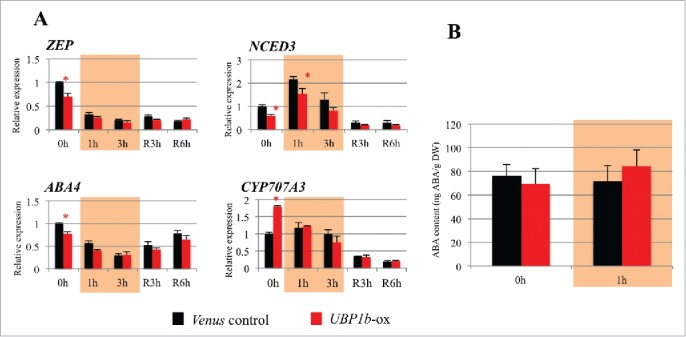
Expression level of ABA biosynthesis- and catabolism-related genes and endogenous ABA level in UBP1b-ox plants. (A) RT-qPCR analysis of the expression levels of ABA metabolism-related genes in UBP1b-ox and control plants. RT-qPCR analysis of ZEP, NCED3, ABA4, and CYP707A3 expression was performed using the RNA of plant samples collected under non-stress condition (0h), 1 hour and 3 hour treatment with 40°C (1h and 3h), and under recovery at 22°C for 3 hour and 6 hour (R3h and R6h). Data represent the mean ± sd. n = 3. An asterisk indicates a significant difference (p < 0.05) between UBP1b-ox and control plants as determined by a t-test. (B) Endogenous ABA level in UBP1b-ox and control plants under non-stressed and heat stress conditions. Data represent the mean ± sd. n = 6.
mRNA of WRKY33 is a potential target of UBP1b
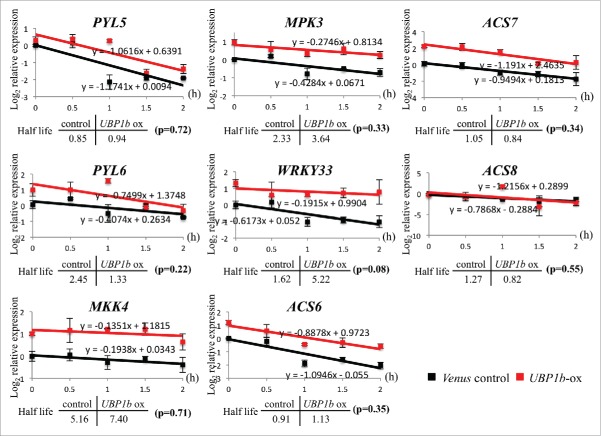
RT-qPCR analysis indicated that the expression of PYL5, PYL6, MPK3, MKK9 and WRKY33 was higher in UBP1b-ox plants grown under non-stress condition, while the expression of MKK4 was higher in UBP1b-ox plants subjected to heat stress (Fig. 5). The expression pattern of WRKY33 was significantly higher in non-stressed UBP1b-ox plants and in UBP1b-ox plants after 1 h of heat stress (Fig. 5). RNA decay analysis was performed to confirm whether or not those genes are target of UBP1b. If an mRNA interacts with a RNA-binding protein like UBP1b, the degradation of the interacting mRNA will be inhibited. It is possible to measure the rate of mRNA degradation in plants treated with a transcriptional inhibitor. In the present study, cordycepin was used as a transcriptional inhibitor. The degradation rate of mRNAs was measured in UBP1b-ox and control plants after they were treated with 0.6 mM cordycepin. Among the ABA-related genes investigated above, the degradation speed of WRKY33 mRNA was slower in UBP1b-ox plants than in control plants. The half-life of WRKY33 mRNA in UBP1b-ox plants was 5.22 h, compared with a degradation rate of 1.62 h in control plants (Fig. 6). The difference of regression coefficient in the linear regression model between overexpressed plants and control plants were calculated using F-test. Among all the target candidates, p-value of the F-test of WRKY33 was smallest and less than 0.1. Other candidates showed high p-values (Fig. 6). Therefore, it is possible that the mRNA of WRKY33 is protected from degradation by UBP1b, and that WRKY33 mRNA is a target of UBP1b. The mRNA of other ABA-related genes, however, did not show any differences in the degradation rate of UBP1b-ox vs. control plants (Fig. 6).
Figure 5.
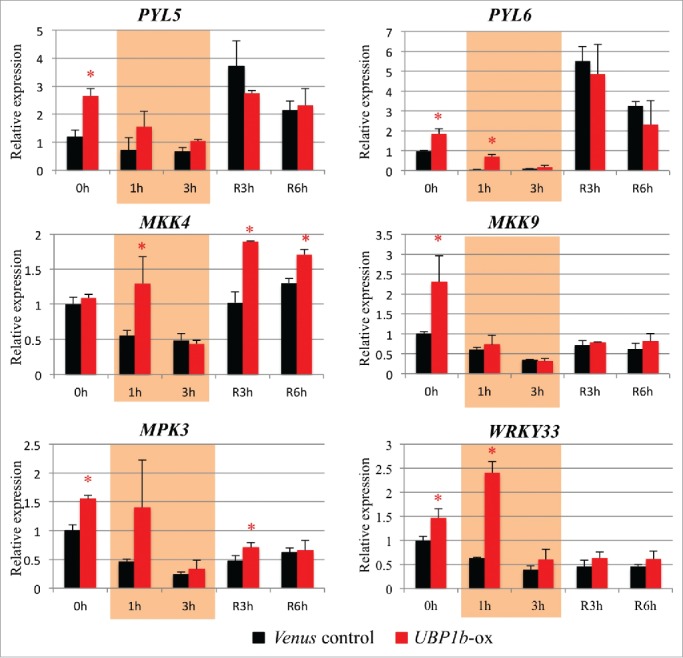
Expression level of UBP1b target candidates. (A) RT-qPCR analysis of target candidate genes was performed in UBP1b-ox and control plants under non-stressed and heat stress conditions; PYL5, PYL6, MKK4, MKK9, MPK3, and WRKY33. Data represent the mean ± sd. n = 3. An asterisk indicates a significant difference (p < 0.05) between UBP1b-ox and control plants as determined by a t-test.
Figure 6.
RNA decay assay of UBP1b target candidates. RNA decay rate of target candidate genes were checked in UBP1b-ox and control plants; PYL5, PYL6, MKK4, MPK3, WRKY33, ACS6, ACS7, and ACS8. Data represent the normalized log2 value of gene expression during cordycepin treatment. Data represent the mean ± sd. n = 3. F-test was performed to compare the RNA decay rate between parallel linear recognition model and individual linear recognition model and the p-value for each target candidate gene is shown within parenthesis.
Ethylene biosynthesis genes are upregulated in UBP1b-ox plants
WRKY33 is an important substrate of MPK3/6 in ABA signaling14 Under non-stressed conditions, WRKY33 affects plant root growth through the regulation of ethylene biosynthesis.15,16 WRKY33 binds directly to the W-box of 4 1-amino-cyclopropane-1-carboxylate synthases 2/6 (ACS2/ACS6) and thereby positively mediates the activation of these ethylene biosynthesis genes.15 High concentrations of ABA and/or ethylene inhibits plant root growth.16,17 Our microarray data indicated that 4 ACS genes (ACS6, ACS7, ACS8 and ACS11) which are key enzymes in ethylene biosynthesis, are also upregulated in UBP1b-ox plants (Table S1). The expression of ACS2, however, was not significantly changed in UBP1b-ox plants (data not shown). Therefore, the expression level of ACS genes was evaluated by RT-qPCR, to confirm the microarray data. Results indicated that the expression of ACS6, ACS7, and ACS8 was higher in UBP1b-ox plants. The expression of ACS11, however, was not significantly different in UBP1b-ox vs. control plants (Fig. 7).
Figure 7.
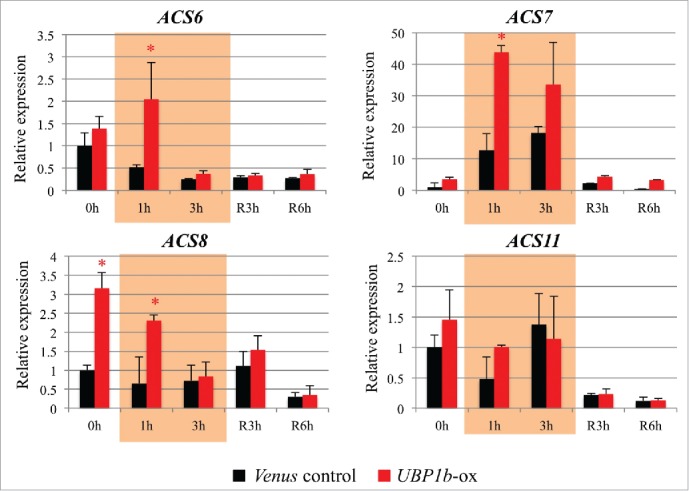
Expression level of ethylene biosynthesis-related genes. RT-qPCR analysis of ACS6, ACS7, ACS8, and ACS11 in UBP1b-ox and empty vector, control (Venus) plants under non-stressed and heat stress conditions. Data represent the mean ± sd. n = 3. An asterisk indicates a significant difference (p < 0.05) between UBP1b-ox and control plants as determined by a t-test.
Microarray data also showed that the mRNA expression of several ethylene treatment-upregulated genes, such as ethylene- responsive factor (ERF) 1, 2, 4 and 5, Auxin-Regulated Gene involved in Organ Size (ARGOS, AT3G59900),18 ARGOS-like (AT2G44080), and a peroxidase superfamily protein (PER10, AT1G49570) was higher in UBP1b-ox compared with control plants under normal condition (Fig. S1). On the other hand, the expression of several ethylene treatment-downregulated genes,18 such as glycosyl hydrolase 9C2 (GH9C2, AT1G64390), CYP71A16 (AT5G42590), and Expansin B3 (EXPB3, AT4G28250) was downregulated in UBP1b-ox under non-stress condition (Fig. S1). These results indicate the positive regulation of ethylene biosynthesis by UBP1b via the upregulation of ABA signaling. An RNA decay assay was conducted on all of the ACS genes, however, none of ACS genes exhibited a slower rate of mRNA degradation in UBP1b-ox plants compared with control plants. These data indicate that ACS genes are not the direct targets of UBP1b (Fig. 6).
Discussion
UBP1b functions as a component of SGs that play a role in protecting mRNAs from degradation. Previously generated microarray data indicated that UBP1b also functions under non-stressed conditions in up-regulating the expression level of various target genes5 (Fig. 1, Fig. 5, Fig. 7). Many ABA signaling genes, such as ABA receptors PYL5, PYL6, and MAPK cascades were upregulated in UBP1b-ox plants, suggesting that UBP1b plays a role in regulation in the ABA signaling pathway in plants (Fig. 1, Fig. 5, Fig. 7). Although UBP1b-ox exhibited hypersensitivity to exogenous application of ABA, endogenous ABA levels were not significantly different in UBP1b-ox plants than they were in vector transformed control plants (Fig. 4). Biosynthetic and metabolic genes expression was slightly changed in transgenic plants, however, the effect was not enough to regulate endogenous ABA (Fig. 4). These data indicate that the ABA hypersensitivity of UBP1b-ox plants was not due to high levels of ABA but rather due to the regulation of ABA signaling genes. It is possible that the upregulation of ABA signaling affected ABA biosynthesis and catabolism genes through a feedback regulation mechanism. ABA receptor genes have been previously reported to increase ABA sensitivity.19,20 Overexpression of MPK3 has also been reported to increase ABA sensitivity in plants,21 which is consistent with the data generated in the present study. Both germinating seeds and mature seedlings of UBP1b-ox exhibited an ABA hypersensitive phenotype in our study, indicating that UBP1b affects ABA signaling at various stages of plant development.
The expression level of WRKY33 was higher in UBP1b-ox plants and RNA decay analysis demonstrated that WRKY33 transcripts had a slower rate of degradation in UBP1b-ox plants than in control plants. These results suggest that WRKY33 mRNA is a target of UBP1b. A previous study reported that WRKY33-overexpressing plants were sensitive to ABA,22 which explains the phenotype of UBP1b-ox plants that were observed in response to exogenous application of ABA in the present study. Luo et al., 2014 reported that WRKY33 plays a key role in the inhibition of root growth by ABA through its effect on ethylene biosynthesis.16 Another study also suggested that WRKY33 regulates plant abiotic stress response through an ethylene-dependent process.23 In our data, ethylene biosynthesis genes, such as ACS6, ACS7, and ACS8, were upregulated in UBP1b-ox plants; and ABA treatment inhibited root growth in UBP1b-ox plants. Collectively, our data strongly suggest that UBP1b regulates plant ABA sensitivity by binding and stabilizing mRNAs of WRKY33.
Recent studies have reported crosstalk between ABA signaling and ethylene signaling in plants.15,16,24,25 The ethylene pathway is also important in plant abiotic stress response. Regulation of ethylene biosynthesis genes, as well as ethylene levels, results in changes in the timing of seed germination, stomata closure, and root growth, which in turn affect plant tolerance to salt, heat and osmotic stresses.16,26-29 ACC synthase (ACS) is a key enzyme in ethylene biosynthesis. There are 9 ACSs in Arabidopsis, including ACS1–2, ACS4–9, and ACS11; all of which are involved in the conversion of A-adenosylmethionine to 1-aminocyclopropane-1-carboxylic acid (ACC).25,30 ACS2 and ACS6 are phosphorylated by MPK3 and MPK6, which positively regulate ACS protein levels and activity, resulting in the enhancement of ethylene production under stress conditions. 15,24,31,32 ABA treatment activates calcium-dependent protein kinase 4 (CPK4) and 2 (CPK11), which phosphorylate ACS6.16 Overexpression of ACS6 resulted in the inhibition of root growth, an increase in the germination rate, and an increase in ethylene production when plants were treated with ABA.16 ACS7 is the only type 3 ACS protein in Arabidopsis, which is characterized by a very short C-terminus and the absence of a phosphorylation site.33,34 ACS7 expression is regulated by phytohormones, such as ethylene, ABA, and GA3; and abiotic stresses such as high light and salt.25,26 A loss-of-function mutant of ACS7 exhibited both earlier seed germination and a higher level of growth, as well as enhanced tolerance to salt, heat, and osmotic stresses.27 WRKY33, a substrate phosphorylated by MPK3/MPK6, regulates the expression of the ethylene biosynthesis genes, ACS2 and ACS615 via direct binding to the W-boxes of their promoters. Thus, WRKY33 plays a key role in the activation of ACS2 and ACS6 in response to abiotic stress conditions. 15 WRKY33 has also been reported to increase plant sensitivity to ABA under non-stressed conditions. 22
There is evidence for a link between ABA sensitivity and WRKY33, as well as between ABA and ethylene signaling. A splicing defect in WRKY33 was identified in a knockout mutant of sad1, which exhibited hypersensitivity to ABA8 and heat stress,35 and an activation of alternative splicing.36 The increased intron retention in WRKY33 transcripts in the sad1 mutant has been implicated as a factor that contributes to the stress sensitivity of this mutant.36 In contrast, WRKY33 expression levels in aba1–5 and abi4–137 plants is significantly reduced, indicating that WRKY33 expression is partially dependent on ABA biosynthesis and the ABA signaling pathway.22 During seed germination, ethylene acts as a negative regulator of plant ABA response.38 The impairment in ethylene signaling results in ABA hypersensitivity.39 After germination, ABA and ethylene signaling pathways exhibit a complex interaction.37 Both pathways inhibit root growth,38,39 and a disruption of ethylene signaling results in a decrease in the sensitivity of root growth to ABA.39 The present research provides new information pertaining to the function of UBP1b that helps to clarify the complex interaction between ABA signaling, ethylene biosynthesis, and WRKY33.
Materials and methods
Plant material and growth conditions
The present study used 35S::Venus-UBP1b-overexpressing (UBP1b-ox) A. thaliana plants (ecotype: Columbia), and empty vector 35S::Venus (Venus) A. thaliana plants (ecotype: Columbia) as a control.5 Plants were grown on Murashige and Skoog (MS) agar medium under long day conditions (16 h light/8 h dark) at 22°C in an environmental chamber (TOMY CF-405, Tokyo, Japan) and were used in all of the subsequent experiments.
Microarray analysis
Microarray data were obtained from a previous research study in which 14-day-old UBP1b-ox and control plants were sampled under non-stressed and heat stress conditions (40oC for 1h).5 The resulting microarray data were deposited in and are available on the GEO website (GEO ID: GSE78713). Microarray statistical analysis was performed using Pavlidis method.40 The genes whose expression level is significantly changed in the experimental conditions were selected as the candidate ones using one-way ANOVA analysis, with FDR <0.0001 as multiple test correction. And then, the gene expression data was subjected to the Student t-test as a post hoc test to find the conditions (UBP1b-ox non-stress, UBP1b-ox heat stress, control non-stress and control heat-stress) in which the gene expression was changed. The genes whose the expression data satisfies the following criteria were identified as ones with higher expression: log2 (expression ratio) > 0.7 and p-value of the Student's t-test < 0.15, relative to the control.
Root elongation assay
Eight-day-old UBP1b-ox and control plants were transferred to and grown on vertical MS plates containing 10μM ABA for 9 d under long-day conditions. Primary root length was measured on 3 biologic replicates in pictures of taken of the MS plates on 0, 3, and 7 d after plants were transferred to the ABA-containing media. Measurements in the photo were made using ImageJ (National Institutes of Health, http://rsb.info.nih.gov/ij/download.html) software.
Seed germination rate
Seeds from UBP1b-ox and control plants were imbibed in water for 48 hours before being placed on MS plates amended with 0 μM, 0.3 μM, or 0.5 μM ABA. A total of 49 seeds from each UBP1b-ox line and control plants were placed on 3 different plates. Seedlings with green cotyledons were counted as germinated seeds. Three biologic replicates were used in each test.
ABA measurements
All plant samples (leaf and stem) were frozen in liquid nitrogen and weighed after lyophilization. Samples were ground and homogenized in 80% (v/v) acetonitrile containing 1% (v/v) acetic acid with defined amounts of D6-ABA (Icon Isotopes, Summit, NJ, USA) as an internal standard. The solutions were incubated for 12 h at 4°C and then centrifuged at 3,000 g for 20 min at 4°C. The supernatants were dried under a vacuum and then dissolved in 1 mL of water containing 1% (v/v) acetic acid. Oasis WAX 1 mL solid-phase extraction cartridges (Waters) were conditioned with 1 mL acetonitrile followed by 1 mL methanol and 0.5 mL 0.1 M KOH, and then equilibrated with 1 mL of 1% acetic acid (v/v). Samples were loaded onto the cartridges and the cartridges were subsequently washed with 1 mL 1% (v/v) acetic acid and 1 mL acetonitrile. Fractions containing ABA were eluted from the cartridge with 1 mL 80% (v/v) acetonitrile containing 1% (v/v) acetic acid, dried under vacuum, and dissolved in 20 µL of water containing 1% (v/v) acetic acid. The LC–MS/MS system consisting of a quadrupole/time-of-flight tandem mass spectrometer (Triple TOF 5600, AB SCIEX, Toronto, Canada), and a Nexera HPLC system (SHIMADZU, Kyoto, Japan), were used to quantify the ABA concentration in each sample. LC-MS/MS was performed according to procedures as described previously.41 MultiQuant 2.0, (AB SCIEX, Toronto, Canada) software was used to calculate plant hormone concentrations from the LC–MS/MS data. Four seedlings were collected for each sample, and 6 biologic repeats were used in this experiment.
RT-qPCR
Fourteen-day-old plants were treated with heat (40°C) then recovered at 22°C. Samples were collected at 0 h, heat 1 h, heat 3 h, recovery 3 h and 6 h. Total RNA was extracted using the Plant RNA Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Each of biologic repeat consisted of a pool of 5 seedlings, and 3 biologic replicates were used. Following the manufacturer's protocol, cDNAs were synthesized with a Quantitech cDNA synthesis kit (Qiagen, Venlo, Netherlands) using RNA samples obtained for the microarray analysis and were used for the RT-qPCR analysis. RT-qPCR analyses were performed using Fast SYBR Green MasterMix (Thermo Fisher Scientific) and a StepOne Plus Real Time PCR system (Thermo Fisher Scientific). YLS8 was used as a reference gene to normalize expression data and RT-qPCR data were analyzed using StepOne Plus software (Thermo Fisher Scientific). Primers used in the RT-qPCR analyses are listed in Table S2.
RNA decay analysis
A total of 25 2-week-old plants were placed in a petri plate containing filter paper and 3 ml of water for 24 h. Subsequently, 2 ml of 1.2 mM cordycepin was added to each Petri dish, creating a final concentration of 0.6 mM cordycepin. The treatment was conducted at 22°C for 2 h and 5 seedlings were collected every 30 minutes (4 biologic replicates were used from a pool of 120 seedlings). Total RNA and cDNA were prepared from cordycepin-treated samples as described above. RT-qPCR was performed and decay rates were calculated based on the log2 value of the relative expression of the target genes. SUMO2 (AT5G55160), a transcript with a long half-life,42 was used as a reference gene for the normalization of the expression data. For the test of the difference of 2 regression coefficients, the F-statistic is computed using parallel linear model (Y1 = β0 +β1×1, Y2 = β2 +β1×2, where X1 and Y1 indicate time of RNA decay and log2 copy number in UBP1b-ox plants, respectively, and X2 and Y2 indicate those in control plants) and individual linear model (Y1 = β0 +β1×1, Y2 = β3 +β4×2). The formula for the F statistic is F = {SSrb – (SSr1 + SSr2) / (dF2- dF1)}/ {(SSr1 + SSr2) / dF2)}, where SSrb is the residual sum of squares for the parallel linear model, SSr1 and SSr2 are the residual sum of squares for the individual linear model of control plants and UBP1b-ox plants, respectively. And dF is N − V degrees of freedom, where N is the number of data points and V is the number of parameters being estimated. When the p-value is less than 0.1, we decided that the 2 lines of RNA decay were not parallel.
Supplementary Material
Disclosure of potential conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be considered as a potential conflict of interest.
Funding
This work was supported by a grant from the RIKEN Center for Sustainable Resource Science (to M.Seki), the Japan Science and Technology Agency (JST) [Core Research for Evolutionary Science and Technology (CREST)] (to M.Seki), and Grants-in-Aid for Scientific Research in Innovative Areas (no. 16H01476) by the Ministry of Education, Culture, Sports, and Technology of Japan (to M.Seki), and the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 25850247 to K.N.. ABA measurement were supported by the Japan Advanced Plant Science Research Network.
References
- 1.Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 2001; 106:477-87; PMID:11525733; http://dx.doi.org/ 10.1016/S0092-8674(01)00460-3 [DOI] [PubMed] [Google Scholar]
- 2.Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res 2004; 32:5096-103; PMID:15452276; http://dx.doi.org/ 10.1093/nar/gkh845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T. Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 2005; 44:972-84; PMID:16359390; http://dx.doi.org/ 10.1111/j.1365-313X.2005.02589.x [DOI] [PubMed] [Google Scholar]
- 4.Nguyen AH, Matsui A, Tanaka M, Mizunashi K, Nakaminami K, Hayashi M, Iida K, Toyoda T, Nguyen DV, Seki M. Loss of Arabidopsis 5′-3′ Exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol 2015; 56:1762-72; PMID:26136597; http://dx.doi.org/ 10.1093/pcp/pcv096 [DOI] [PubMed] [Google Scholar]
- 5.Nguyen CC, Nakaminami K, Matsui A, Kobayashi S, Kurihara Y, Toyooka K, Tanaka M, Seki M. Oligouridylate binding protein 1b plays an integral role in plant heat stress tolerance. Front Plant Sci 2016; 7:853; PMID:27379136; http://dx.doi.org/ 10.3389/fpls.2016.00853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 2000; 12:2351-66; PMID:11148283; http://dx.doi.org/ 10.1105/tpc.12.12.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol Biol 2004; 55:679-86; PMID:15604709; http://dx.doi.org/ 10.1007/s11103-004-1680-2 [DOI] [PubMed] [Google Scholar]
- 8.Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y,Xu W, Galbraith D, Zhu JK. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 2001;1:771-81;PMID:11740939;http://dx.doi.org/ 10.1016/S1534-5807(01)00087-9 [DOI] [PubMed] [Google Scholar]
- 9.Lambermon MH, Simpson GG, Wieczorek Kirk DA, Hemmings-Mieszczak M, Klahre U, Filipowicz W. UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J 2000; 19:1638-49; PMID:10747031; http://dx.doi.org/ 10.1093/emboj/19.7.1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber C, Nover L, Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 2008; 56:517-30;PMID:18643965;http://dx.doi.org/ 10.1111/j.1365-313X.2008.03623.x [DOI] [PubMed] [Google Scholar]
- 11.Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 2009; 60:575-88; PMID:19624469; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03981.x [DOI] [PubMed] [Google Scholar]
- 12.Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci U S A 1996; 93:765-9; PMID:8570631; http://dx.doi.org/ 10.1073/pnas.93.2.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tena G, Asai T, Chiu WL, Sheen J. Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 2001; 4:392-400; PMID:11597496; http://dx.doi.org/ 10.1016/S1369-5266(00)00191-6 [DOI] [PubMed] [Google Scholar]
- 14.Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011; 23:1639-53;PMID:21498677; http://dx.doi.org/ 10.1105/tpc.111.084996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 2012; 8:e1002767; PMID:22761583; http://dx.doi.org/ 10.1371/journal.pgen.1002767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo X, Chen Z, Gao J, Gong Z. Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J 2014; 79:44-55; PMID:24738778; http://dx.doi.org/ 10.1111/tpj.12534 [DOI] [PubMed] [Google Scholar]
- 17.Joshi-Saha A, Valon C, Leung J. Abscisic acid signal off the STARting block. Mol Plant 2011; 4:562-80; PMID:21746700; http://dx.doi.org/ 10.1093/mp/ssr055 [DOI] [PubMed] [Google Scholar]
- 18.Markakis MN, De Cnodder T, Lewandowski M, Simon D, Boron A, Balcerowicz D, Doubbo T, Taconnat L, Renou JP, Höfte H, et al.. Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biol 2012; 12:208; PMID:23134674; http://dx.doi.org/ 10.1186/1471-2229-12-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al.. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 2010; 61:290-9; PMID:19874541; http://dx.doi.org/ 10.1111/j.1365-313X.2009.04054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al.. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science (New York, NY) 2009; 324:1068-71; PMID:19407142; http://dx.doi.org/12434021 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu C, Han MH, Guevara-Garcia A, Fedoroff NV. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci U S A 2002; 99:15812-7;PMID:12434021;http://dx.doi.org/ 10.1073/pnas.242607499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Deyholos MK. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 2009; 69:91-105; PMID:18839316; http://dx.doi.org/ 10.1007/s11103-008-9408-3 [DOI] [PubMed] [Google Scholar]
- 23.Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011; 233:1237-52; PMID:21336597; http://dx.doi.org/ 10.1007/s00425-011-1375-2 [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 2004; 16:3386-99; PMID:15539472; http://dx.doi.org/ 10.1105/tpc.104.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang NN, Shih MC, Li N. The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J Exp Botany 2005; 56:909-20; PMID:15699063; http://dx.doi.org/ 10.1093/jxb/eri083 [DOI] [PubMed] [Google Scholar]
- 26.Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science (New York, NY) 2006; 311:91-4; PMID:16400150; http://dx.doi.org/21765163 10.1126/science.1118642 [DOI] [PubMed] [Google Scholar]
- 27.Dong H, Zhen Z, Peng J, Chang L, Gong Q, Wang NN. Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J Exp Botany 2011; 62:4875-87; PMID:21765163; http://dx.doi.org/ 10.1093/jxb/err143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkindale J, Hall JD, Knight MR, Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 2005; 138:882-97; PMID:15923322; http://dx.doi.org/ 10.1104/pp.105.062257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 2009; 183:979-1003; PMID:19752216; http://dx.doi.org/ 10.1534/genetics.109.107102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annual Rev Plant Physiol 1984; 35:155-89; http://dx.doi.org/ 10.1146/annurev.pp.35.060184.001103 [DOI] [Google Scholar]
- 31.Joo S, Liu Y, Lueth A, Zhang S. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J 2008; 54:129-40; PMID:18182027; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03404.x [DOI] [PubMed] [Google Scholar]
- 32.Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 2010; 64:114-27; PMID:20659280; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04318.x [DOI] [PubMed] [Google Scholar]
- 33.Chae HS, Kieber JJ. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci 2005; 10:291-6; PMID:15949763; http://dx.doi.org/ 10.1016/j.tplants.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 34.Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol 2005; 5:14; PMID:16091151; http://dx.doi.org/ 10.1186/1471-2229-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto M, Matsui A, Tanaka M, Morosawa T, Ishida J, Iida K, Mochizuki Y, Toyoda T, Seki M. Sm-like protein-mediated RNA metabolism is required for heat stress tolerance in arabidopsis. Front Plant Sci 2016; 7:1079; PMID:27493656; http://dx.doi.org/ 10.3389/fpls.2016.01079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui P, Zhang S, Ding F, Ali S, Xiong L. Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol 2014; 15:R1; PMID:24393432; http://dx.doi.org/ 10.1186/gb-2014-15-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002; 14:S15-45; PMID:12045268; http://doi.org/ 10.1105/tpc.010441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 2000; 12:1117-26; PMID:10899978; http://dx.doi.org/ 10.1105/tpc.12.7.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beaudoin N, Serizet C, Gosti F, Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 2000; 12:1103-15; PMID:10899977; http://dx.doi.org/ 10.1105/tpc.12.7.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavlidis P. Using ANOVA for gene selection from microarray studies of the nervous system. Methods (San Diego, Calif) 2003; 31:282-9; PMID:14597312; http://dx.doi.org/ 10.1016/S1046-2023(03)00157-9 [DOI] [PubMed] [Google Scholar]
- 41.Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, Sano N, Koshiba T, Kamiya Y, Ueda M, Seo M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Communs 2016; 7:13245; PMID:27782132; http://dx.doi.org/ 10.1038/ncomms13245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 2007; 19:3418-36; PMID:18024567; http://dx.doi.org/ 10.1105/tpc.107.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.