ABSTRACT
Cell cycle regulation is fundamental to growth and development, and Cyclin-Dependent Kinase Inhibitors (CKIs) are major negative regulators of the cell cycle. Plant genomes encode substantially more CKIs than metazoan or fungal genomes. Plant CKIs fall into 2 distinct families, KIP-RELATED PROTEINS (KRPs) and SIAMESE-RELATED proteins (SMRs). SMRs can inhibit both S-phase and M-phase CDK complexes in vitro and are transcribed throughout the cell cycle, yet SMRs do not inhibit DNA replication in vivo. This suggests that SMRs must be activated post transcriptionally after the start of S-phase, but the mechanism of this hypothesized activation is unknown. Recent work indicates that even distantly related SMRs have the same biochemical function, and that differential transcriptional regulation likely maintains their distinct roles in integrating various environmental and developmental signals with the cell cycle.
KEYWORDS: Arabidopsis thaliana, CDK inhibitors, cell cycle, endoreduplication, endoreplication
Introduction
Coordination between cell division and growth is required for proper development of multicellular organisms. In response to this need, complex systems of cell cycle regulation have evolved in plants and animals. Cyclin-dependent kinases (CDKs), a conserved class of serine/threonine kinases, along with their regulatory subunit cyclins (CYCs) drive unidirectional and irreversible progression from one cell cycle phase to the next by phosphorylating target proteins. CDK inhibitors (CKIs) negatively control cell cycle progression and are involved in instituting cell cycle checkpoints. In some circumstances, CKIs also promote a modified cell cycle known as endoreplication or endoreduplication, in which mitosis and cytokinesis are bypassed, but DNA replication continues, resulting in cells with increased ploidy.1,2
The number of genome-encoded CKIs varies among different species and kingdoms, and some lineages have more than one distinct type of CKI. For example, mammals have 2 CKI families, CDK interacting protein/kinase inhibitory protein (CIP/KIP) and INHIBITOR of CDK4 (INK4) and these 2 families share no sequence similarity. The CIP/KIP (Kinase inhibitor protein) family has 3 members (p21Cip1, p27Kip1, p57Kip2) that act as broad-spectrum CDK inhibitors, while the INK4 family has 4 members, p15INK4a, P16INK4b, P18INK4c, P19INK4d, that inhibit only CDK4 and CDK6.3 Drosophila melanogaster has only 2 CDK inhibitors, Dacapo (Dap), which negatively regulate the G1/S transition, and roughex (rux), which is a negative regulator of CYCA/CDK activity during G1.4,5 Both Drosophila CKIs are distantly related to mammalian Kip proteins. In contrast, fungal CKIs appear to be unrelated to CKIs from other organisms. Budding yeast, Saccharomyces cerevisiae, has 2 cell cycle-related CKIs, p40Sic1 and Far1.6 Fission yeast, Saccharomyces pombe, has only a single CKI, p25RUM17.
Land plants have 2 well-established CKI families, the INTERACTOR/INHIBITOR OF CDK/KIP-RELATED PROTEINs (ICK/KRPs) and the SIAMESE-RELATED PROTEINs (SMRs), that play a variety of roles in cell cycle regulation.7-10 ICK/KRPs have limited sequence similarity with mammalian Kip proteins, while SMRs have no recognizable homologs outside of the plant kingdom.10,11 Furthermore, ICK/KRPs and SMRs share only a single 6 amino acid motif, which is thought to be a cyclin-binding motif.8,12 The ICK/KRP gene family was initially discovered based on similarity to metazoan KIP genes. The SMR gene family was identified based on the mutant phenotype of the sim gene, which results in trichomes (shoot epidermal hairs) that divide instead of endoreplicating (Fig. 1). Most land plant genomes contain multiple genes in both CKI families; for example the Arabidopsis genome contains 7 KRP genes and 17 SMR genes, numbers that are typical of angiosperm genomes.10,13 The large number of ICK/KRPs and SMRs encoded by plant genomes raises the question of why plants need so many CKI genes.
Figure 1.
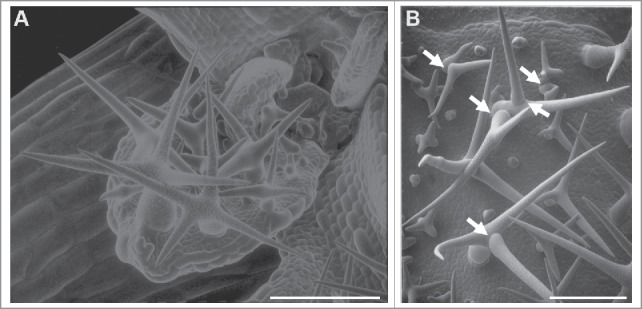
Scanning electron micrographs of (A) wild-type and (B) sim developing trichomes on Arabidopsis leaves. Scale bars = 100 μm. Note cell junctions in sim multicellular trichomes (arrows).
SMRs and KRPs play overlapping but distinct roles in the cell cycle
Although proteins of both families are CKIs, KRPs and SMRs appear to play distinct roles in the cell cycle. The clearest evidence that KRPs and SMRs have distinct cell cycle roles comes from ectopic overexpression studies.8,10,14 While overexpression of either type of CKI results in a similar overall reduced growth phenotype, they have differential effects on the specific phases of the cell cycle. KRPs function as a dose-dependent cell cycle inhibitors, with transgenic plants showing low levels of ectopic expression suppressing mitosis and promoting endoreplication and higher expression levels blocking both mitosis and DNA replication, sometimes resulting in cell death.15-17 In contrast, while SIM overexpression can induce endoreplication, resulting in DNA contents as high as 128C, overexpression of SIM or other SMRs has never been observed to inhibit DNA replication or cause cell death.8,13 Thus, available evidence indicates that SMRs only inhibits M-phase, while KRPs can block entry into both M- and S-phases.
Both KRPs and SMRs inhibit CDK activity in vitro.10,12,13,17 Plants contain 2 types of CDKs, CDKA and CDKBs. In Arabidopsis, the sole CDKA kinase, CDKA;1, primarily regulates the G1/S transition, while CDKBs are required for mitosis and are only expressed during G2 and M.9 KRPs are thought to primarily inhibit CDKA;1.10,17 A complex feedback loop in which KRPs inhibit G1/S CDK activity until degraded by an SCF E3 ubiquitin ligase complex containing the F-box protein FBL17 is a key regulator of the G1/S transition, as illustrated in Fig. 2.19 Thus KRPs play a key role in establishing the G1 checkpoint.
Figure 2.
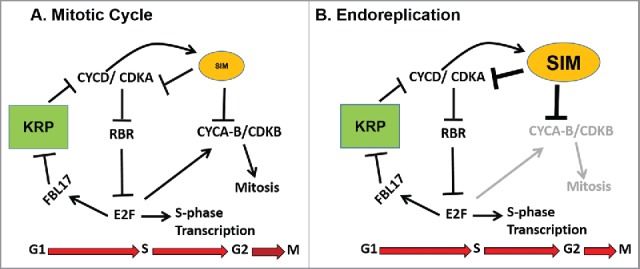
Roles of KRPs and SMRs in the mitotic and endoreplication cell cycles. (A) During mitosis, a feedback loop including KRPs that establishes the 2 alternative states of no CDK activity during G1, and increased CDK activity triggering S phase. SMRs are proposed to restrict S phase CDK activity after the initiation of S phase to prevent premature entry in mitosis, which depends on CDK phosphorylation of G2/M transcription factors. (B) During endoreplication, the cyclical inhibition of CDK activity by KRPs continues unchanged, resulting in cycles of DNA replication. Increased expression of SIM or other SMRs prevents activation of G2/M transcription factors by S phase CDKs, and inhibits any M phase CDKs that are synthesized, thus preventing mitosis. Proposed requirement for SIM or SMRs to be activated via phosphorylation by CYCD/CDKA complexes introduces a time delay that allows initiation of S-phase, but results in inhibition of these complexes before they can contribute to activation of M-phase.
Unlike KRPs, SMRs appear to be exclusively involved in establishing a G2 checkpoint. Recent work firmly establishes that SMRs interact with and inhibit both CDKA:1 and CDKB1;1 both in vitro and in vivo.13 Because CDKA;1 is the main Arabidopsis G1/S CDK, yet SMR overexpression does not inhibit S-phase entry in vivo, this recent work suggests that SMRs are inactive throughout G1 and the G1/S transition, allowing S phase to proceed, but that in mitotically arrested or endoreplicating cells, SMRs are activated to block entry into mitosis. Transcription of SIM or other SMRs does not appear to be regulated relative to cell cycle phase, and thus it seems likely that SMRs are activated post-transcriptionally after the initiation of S-phase. One reasonable possibility is activation by direct phosphorylation of SMRs by G1/S CDK activity. A simplified model of the cell cycle that emphasizes the roles of KRPs and SMRs in the mitotic and endoreplication versions of the cell cycle is shown in Fig. 2.
KRPs and SMRs play diverse roles in plant growth and development
SMRs and KRPs play central roles in balancing cell proliferation and differentiation in response to development and environmental signals. Perhaps the extra complexity of the plant cell cycle, including the large number of CKIs, derives from the sessile nature of plants, requiring them to respond a changing environment by altering growth. There is substantial functional redundancy in both CKI gene families, suggesting that divergence in biochemical function plays a minor role in maintaining the number of genes.18,20,21 Of particular note, Kumar and coworkers found that 10 different SMRs, including an SMR from the bryophyte Physcomitrella patens, can complement the trichome cell division phenotype of Arabidopsis sim mutants.13 Additionally, individual KRP and the SMR genes show distinct patterns of tissue-specific and stress-regulated expression, consistent with the individual genes playing roles in integrating the cell cycle with a wide variety of signals.12,22,23 This level of transcriptional control is presumably secondary to the post-transcriptional control of CKI protein levels acting within the cell cycle itself that was described in the previous section.
While individual krp knockouts have minimal affects on plant morphology, knocking out multiple KRP genes results in larger organ size due to increased cell proliferation via increased expression of E2F target genes,20 consistent with the proposed G1 checkpoint role of KRPs. However, unique roles for some individual KRPs have emerged. KRP5 is expressed in the root apical meristem and acts as a rate-limiting factor in the primary root growth.24 ICK2/KRP2 overexpression inhibits lateral root initiation by preventing cell division in Arabidopsis xylem pericycle.25 In rice, KRP overexpression (osiICK6) affects pollen viability, seed-setting rate, and the dorsal-ventral plane of leaf blades.26 Ectopic expression of ICK1/KRP1 and ICK2/KRP2 reduces gall size and nematode offspring number by impeding cell cycle progression at Arabidopsis root-knot nematode infection sites.27 Unexpectedly, KRP6 appears to induce rather than restrict division, and may play a role in the formation of multinucleate giant cells in nematode-induced root knots.28
SMRs are involved in a particularly rich array of developmental and environmental cell cycle responses. As noted earlier, SIM blocks mitosis and induces endoreplication during Arabidopsis trichome development (Fig. 1), and is a direct target of the trichome development transcription factors. Similarly, SMR1, also known as LGO, is involved in initiating endoreplication during development of giant cells in the Arabidopsis sepal epidermis.15 SIM, SMR1, and SMR2, along with KRP2, have been implicated in gibberellin signaling to regulate root meristem size, perhaps as direct positive targets of DELLA transcription factors.29 SMR2 also plays a role in restricting cell proliferation early in leaf development, negatively regulating leaf size.13 Expression of several SMRs is regulated by biotic or abiotic stress.12,22 SMR5 and SMR7 are direct targets of the DNA damage-responsive transcription factor SOG1, and inhibit cell proliferation and promote endoreplication in response to DNA damage.22
An unexpected link between CKIs and plant pathogen responses
Early hints of a connection between the cell cycle and pathogen responses came from the observation that either genetic manipulation of plant defense pathways30 or pathogen infection31 can trigger endoreplication. More recent work shows that smr1 mutants have increased pathogen susceptibility21 and that both SMR1 and KRP2 play roles in Arabidopsis effector-triggered immunity to bacterial and fungal pathogens through a physical interaction with a nuclear envelope protein CONSTITUTIVE EXPRESSOR OF PATHOGENESIS-RELATED GENES5 (CPR5), apparently by contributing to the hyperphosphorylation of the key cell cycle regulator RETINOBLASTOMA-RELATED1 (RBR1).32 The observation that CKIs contribute to hyperphosphorylation of RBR1 is somewhat paradoxical in light of the evidence that SMRs and KRPs are well-characterized inhibitors of CDK kinase activity, as discussed above. Consistent with such a role of CKIs in plant immune responses, overexpression of SMR1/LGO in the sepal epidermis results in overexpression of a suite of defense-response genes that overlap substantially with the set of genes upregulated in cpr5 mutants.33 CPR5 appears to be an integral component of the plant nuclear pore complex, and a model for the role of KRPs and SMRs in effector triggered immunity has been proposed in which the CKIs are associated with CPR5 in the nuclear pore complex until released for effector-triggered immune signaling by a conformational change in CPR534 It remains to be resolved are how the role of KRPs and SMRs in the mitotic and endoreplication cell cycles, which presumably require free CKIs in the nucleoplasm, are related to this model.
Conclusions
Both families of plant CKIs coordinate cell division, cell expansion and organ growth with developmental and environmental cues. This crucial role of the CKIs in integrating various environmental and developmental signals with the cell cycle may be the primary reason that plants maintain such large CKI gene families, particularly in light of their sessile lifestyle. SMR gene families in plants seem particularly large, perhaps because, unlike KRPs, they play a less essential role in the core mitotic cycle and are instead adapted to act as modifiers that fine-tune cell cycle responses. Much exciting work remains to be done, including determining the mechanisms of KRP and SMR function in the G1 and G2 checkpoints, determining the roles of the individual CKIs in plant growth and development, and understanding the role of CKIs in plant immune responses.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the NSF under the grant MCB1615782 to J.C.L.
References
- 1.de Veylder L, Larkin JC, Schnittger A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 2011; 16:624-34; PMID:21889902; http://dx.doi.org/ 10.1016/j.tplants.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 2.Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development 2013; 140:3-12; PMID:23222436; http://dx.doi.org/ 10.1242/dev.080531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999; 13:1501-12; PMID:10385618; http://dx.doi.org/ 10.1101/gad.13.12.1501 [DOI] [PubMed] [Google Scholar]
- 4.Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell 1996; 87:1225-35; PMID:8980229; http://dx.doi.org/ 10.1016/S0092-8674(00)81818-8 [DOI] [PubMed] [Google Scholar]
- 5.Thomas BJ, Zavitz KH, Dong X, Lane ME, Weigmann K, Finley RL Jr, Brent R, Lehner CF, Zipursky SL. roughex Down-regulates G2 cyclins in G1. Genes Dev 1997; 11:1289-98; PMID:9171373; http://dx.doi.org/ 10.1101/gad.11.10.1289 [DOI] [PubMed] [Google Scholar]
- 6.Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 1998; 62:1191-243; PMID:9841670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correa-Bordes J, Nurse P. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell 1995; 83:1001-9; PMID:8521500; http://dx.doi.org/ 10.1016/0092-8674(95)90215-5 [DOI] [PubMed] [Google Scholar]
- 8.Churchman ML, Brown ML, Kato N, Kirik V, Hülskamp M, Inzé D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, et al.. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 2006; 18:3145-57; PMID:17098811; http://dx.doi.org/ 10.1105/tpc.106.044834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polyn S, Willems A, De Veylder L. Cell cycle entry, maintenance, and exit during plant development. Curr Opin Plant Biol 2015; 23:1-7; PMID:25449720; http://dx.doi.org/ 10.1016/j.pbi.2014.09.012 [DOI] [PubMed] [Google Scholar]
- 10.De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 2001; 13:1653-68; PMID:11449057; http://dx.doi.org/ 10.1105/tpc.13.7.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres Acosta JA, Fowke LC, Wang H. Analyses of phylogeny, evolution, conserved sequences and genome-wide expression of the ICK/KRP family of plant CDK inhibitors. Ann Bot 2011; 107:1141-57; PMID:21385782; http://dx.doi.org/ 10.1093/aob/mcr034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peres A, Churchman ML, Hariharan S, Himanen K, Verkest A, Vandepoele K, Magyar Z, Hatzfeld Y, Van Der Schueren E, Beemster GT, et al.. Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J Biol Chem 2007; 282:25588-96; PMID:17599908; http://dx.doi.org/ 10.1074/jbc.M703326200 [DOI] [PubMed] [Google Scholar]
- 13.Kumar N, Harashima H, Kalve S, Bramsiepe J, Wang K, Sizani BL, Bertrand LL, Johnson MC, Faulk C, Dale R, et al.. Functional conservation in the SIAMESE-RELATED family of cyclin-dependent kinase inhibitors in land plants. Plant Cell 2015; 27:3065-80, in press; PMID:26546445; http://dx.doi.org/ 10.1105/tpc.15.00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC. Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 2000; 24:613-23; PMID:11123800; http://dx.doi.org/ 10.1046/j.1365-313x.2000.00899.x [DOI] [PubMed] [Google Scholar]
- 15.Roeder AH, Chickarmane V, Cunha A, Obara B, Manjunath BS, Meyerowitz EM. Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol 2010; 8:e1000367; PMID:20485493; http://dx.doi.org/ 10.1371/journal.pbio.1000367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnittger A, Weinl C, Bouyer D, Schobinger U, Hulskamp M. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 2003; 15:303-15; PMID:12566574; http://dx.doi.org/25944099 10.1105/tpc.008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 2005; 17:1723-36, PMID:15863515; http://dx.doi.org/ 10.1105/tpc.105.032383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Harashima H, Dissmeyer N, Pusch S, Weimer AK, Bramsiepe J, Bouyer D, Rademacher S, Nowack MK, Novak B, Sprunck S, Schnittger A A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet 2012; 8:e1002847; PMID:22879821; http://dx.doi.org/ 10.1371/journal.pgen.1002847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noir S, Marrocco K, Masoud K, Thomann A, Gusti A, Bitrian M, Schnittger A, Genschik P. The control of arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-Box protein FBL17. Plant Cell 2015; 27:1461-76; PMID:25944099; http://dx.doi.org/ 10.1105/tpc.114.135301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, Cao L, Wang S, Li Y, Shi X, Liu H, Li L, Zhang Z, Fowke LC, Wang H, et al.. Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J 2013; 75:642-55; PMID:23647236; http://dx.doi.org/ 10.1111/tpj.12228 [DOI] [PubMed] [Google Scholar]
- 21.Hamdoun S, Zhang C, Gill M, Kumar N, Churchman M, Larkin JC, Kwon A, Lu H. Differential roles of two homologous cyclin-dependent kinase inhibitor genes in regulating cell cycle and innate immunity in arabidopsis. Plant Physiol 2016; 170:515-27; PMID:26561564; http://dx.doi.org/ 10.1104/pp.15.01466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi D, Alvim Kamei CL, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama KO, Larkin J, Van den Daele H, et al.. The arabidopsis SIAMESE-RELATED Cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 2014; 26:296-309; PMID:24399300; http://dx.doi.org/ 10.1105/tpc.113.118943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC. ICK1, a cyclin-dependent protein kinase inhibitor from .Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 1998; 15:501-10; PMID:9753775; http://dx.doi.org/ 10.1046/j.1365-313X.1998.00231.x [DOI] [PubMed] [Google Scholar]
- 24.Wen B, Nieuwland J, Murray JA. The Arabidopsis CDK inhibitor ICK3/KRP5 is rate limiting for primary root growth and promotes growth through cell elongation and endoreduplication. J Exp Bot 2013; 64:1135-44; PMID:23440171; http://dx.doi.org/ 10.1093/jxb/ert009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inz e D, Beeckman T. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 2013; 14:2339-51; PMID:12368490; http://dx.doi.org/10.1105/ tpc.004960 21558459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang R, Tang Q, Wang H, Zhang X, Pan G, Wang H, Tu J. Analyses of two rice (Oryza sativa) cyclin-dependent kinase inhibitors and effects of transgenic expression of OsiICK6 on plant growth and development. Ann Bot 2011; 107:1087-101; PMID:21558459; http://dx.doi.org/ 10.1093/aob/mcr057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira P, Engler G, de Almeida Engler J. Enhanced levels of plant cell cycle inhibitors hamper root-knot nematode-induced feeding site development. Plant Signal Behav 2013; 8:e26409; PMID:24056043; http://dx.doi.org/ 10.4161/psb.26409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vieira P, De Clercq A, Stals H, Van Leene J, Van De Slijke E, Van Isterdael G, Eeckhout D, Persiau G, Van Damme D, Verkest A, et al.. The cyclin-dependent kinase inhibitor KRP6 induces mitosis and impairs cytokinesis in giant cells induced by plant-parasitic nematodes in arabidopsis. Plant Cell 2014; 26:2633-47; PMID:24963053; http://dx.doi.org/ 10.1105/tpc.114.126425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. Gibberellin signaling controls cell proliferation rate in arabidopsis. Curr Biol 2009; 19:1188-93; PMID:19576768; http://dx.doi.org/ 10.1016/j.cub.2009.05.059 [DOI] [PubMed] [Google Scholar]
- 30.Vanacker H, Lu H, Rate DN, Greenberg JT. A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J 2001; 28:209-16; PMID:11722764; http://dx.doi.org/ 10.1046/j.1365-313X.2001.01158.x [DOI] [PubMed] [Google Scholar]
- 31.Chandran D, Inada N, Hather G, Kleindt CK, Wildermuth MC. Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site-specific processes and regulators. Proc Natl Acad Sci U S A 2010; 107:460-5; PMID:20018666; http://dx.doi.org/ 10.1073/pnas.0912492107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X. A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe 2014; 16:787-94; PMID:25455564; http://dx.doi.org/ 10.1016/j.chom.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz EM, Roeder AH. Transcriptomic effects of the cell cycle regulator LGO in arabidopsis sepals. Front Plant Sci 2016; 7:1-22; PMID:26858731; http://dx.doi.org/ 10.3389/fpls.2016.01744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu Y, Zebell SG, Liang Z, Wang S, Kang BH, Dong X. Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell 2016; 166:1526-38; PMID:27569911; http://dx.doi.org/ 10.1016/j.cell.2016.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]


