Abstract
Despite its key position in central metabolism, l-serine does not support the growth of Corynebacterium glutamicum. Nevertheless, during growth on glucose, l-serine is consumed at rates up to 19.4 ± 4.0 nmol min−1 (mg [dry weight])−1, resulting in the complete consumption of 100 mM l-serine in the presence of 100 mM glucose and an increased growth yield of about 20%. Use of 13C-labeled l-serine and analysis of cellularly derived metabolites by nuclear magnetic resonance spectroscopy revealed that the carbon skeleton of l-serine is mainly converted to pyruvate-derived metabolites such as l-alanine. The sdaA gene was identified in the genome of C. glutamicum, and overexpression of sdaA resulted in (i) functional l-serine dehydratase (l-SerDH) activity, and therefore conversion of l-serine to pyruvate, and (ii) growth of the recombinant strain on l-serine as the single substrate. In contrast, deletion of sdaA decreased the l-serine cometabolism rate with glucose by 47% but still resulted in degradation of l-serine to pyruvate. Cystathionine β-lyase was additionally found to convert l-serine to pyruvate, and the respective metC gene was induced 2.4-fold under high internal l-serine concentrations. Upon sdaA overexpression, the growth rate on glucose is reduced 36% from that of the wild type, illustrating that even with glucose as a single substrate, intracellular l-serine conversion to pyruvate might occur, although probably the weak affinity of l-SerDH (apparent Km, 11 mM) prevents substantial l-serine degradation.
Cometabolism is often observed for xenobiotic compounds which do not enable growth as a single carbon- and energy source (21). This is due, for example, to long degradation pathways and unnatural structures. On the other hand, microorganisms with a restricted metabolism, such as Lactococcus lactis, are dependent on cometabolism of essential natural compounds, e.g., amino acids (35). The amino acid l-serine is characterized by the fact that several organisms have the ability to introduce it into the central metabolism via pyruvate (35, 2, 16). Another distinguishing feature is its high cellular demand, exceeding the simple provision of l-serine for protein synthesis, since l-serine is additionally required for glycine, cysteine, tryptophan, and phospholipid synthesis as well as for 1-carbon-unit generation (52). Glycine, in turn, is a precursor for purines and heme. In Corynebacterium glutamicum about 7.5% of the total carbon flux toward l-serine is utilized for these purposes (28). Prior estimates for Escherichia coli determined that as much as 15% of the carbon assimilated from glucose involves l-serine (42). Due to the high demand and its key position in the precursor supply, l-serine has to be regarded as an intermediate of the central metabolism (52).
In spite of the presence of l-serine dehydratase (l-SerDH) (47) and the key position of l-serine, growth of E. coli on l-serine as a carbon source is very poor, allowing doubling times of about 60 h only (63). When the organism is additionally exposed to low concentrations of glycine, isoleucine, and threonine, growth is enhanced (36). Interestingly, during growth on tryptone broth, where a number of amino acids are present, l-serine is utilized immediately and earlier than any other amino acid (43). Taken together, l-serine is clearly a poor growth substrate for E. coli and is preferably cometabolized. Klebsiella aerogenes and Salmonella enterica serovar Typhimurium can also grow only slowly on l-serine as a carbon source (30), and modest growth of K. aerogenes is supported with l-serine as a nitrogen source (58). Utilization of l-serine has also been reported for Helicobacter pylori (31). This bacterium exhibits a strict respiratory form of metabolism and is unable to utilize glucose but prefers amino acids such as l-serine and l- or d-alanine, which are oxidized, thereby serving as an important energy source (32).
C. glutamicum is able to grow on a variety of mixed carbon sources (3, 44, 61), which, with the exception of the sequential consumption of glucose and glutamate (25), are metabolized in parallel. It has been shown that C. glutamicum is also able to degrade a xenobiotic compound in cometabolism with readily metabolizable carbon sources (10).
Our interest was to study l-serine utilization by C. glutamicum. This bacterium is used for industrial production of l-glutamate and l-lysine (5); the latter amino acid accumulates to as much as 170 g liter−1 in the medium with mutant strains (41). One reason for the exceptional l-lysine-synthesizing property of this bacterium is its inability to degrade this amino acid. In exploring the ability to produce l-serine with C. glutamicum (38), it is therefore obviously necessary to assay for utilization of this amino acid also. Here we report studies which revealed that l-serine is a cometabolized substrate with very high utilization rates, enabling its channeling into the central metabolism.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
The wild-type C. glutamicum strain ATCC 13032 was used for construction of recombinant strains. For plasmid construction, E. coli DH5αMCR (13) was used. Plasmids used were pGEM-T (Promega, Madison, Wisc.) for subcloning of PCR fragments in E. coli, pK19mobsacB (48) for construction of the l-SerDH-negative mutant, and pXMJ19 (19) for construction of the sdaA-overexpressing strain of C. glutamicum. For overexpression of metC (aecD), plasmid pSL173 (24) was used, and for metC deletion, plasmid pCR007d (46) was used.
Luria-Bertani medium was used as the standard medium for cultivation of E. coli, while all C. glutamicum strains were precultured on brain heart infusion medium (Difco). The CgXII minimal medium used for growth of C. glutamicum has been described previously (23); it contained 30 μg of protocatechuic acid ml−1 and was supplemented with the respective carbon source. When appropriate, E. coli strains received carbenicillin or kanamycin (each at 50 μg ml−1) or chloramphenicol (30 μg ml−1). C. glutamicum strains received kanamycin (50 μg ml−1) or chloramphenicol (10 μg ml−1). After transformation, C. glutamicum received reduced kanamycin or chloramphenicol concentrations of 15 or 4 μg ml−1, respectively. In sdaA overexpression experiments using pXMJ19 as a vector, 1 mM isopropyl-β-d-thiogalactopyranoside was added for induction of Ptac. All C. glutamicum cultures (60 ml in 500-ml baffled Erlenmeyer flasks) were inoculated to give an optical density at 600 nm (OD600) of about 1 and then incubated aerobically at 30°C on a rotary shaker at 120 rpm. For determination of l-serine utilization, C. glutamicum was precultured in CgXII medium containing 200 mM glucose as a carbon source. The cells were harvested in the early-stationary phase and used to inoculate fresh CgXII medium with glucose or glucose plus l-serine at the concentrations described in Results. The substrate consumption rate for growing cells is given as growth rate (μ) × (substrate concentration at t1 − substrate concentration at t0)/(dry weight at t1 − dry weight at t0), where t1 is the time of complete glucose depletion. For nongrowing cells in phase 2, l-serine consumption rates were determined by the following formula: (serine concentration at t2 − serine concentration at t1)/(mean dry weight at phase 2) × (t2 − t1), where t2 is the time of complete serine depletion. The relation established previously (an OD600 of 1 is equivalent to 0.3 mg [dry weight] ml−1) was used to calculate the dry weight of the cultures.
NMR spectroscopy.
For the 13C-labeling experiments, l-[U-13C]serine (98% enrichment) from Cambridge Isotope Laboratories, Andover, Mass., was used. Wild-type C. glutamicum and the ΔsdaA mutant were harvested after growth for 20 h, cells lyophilized, and hydrolyzed, and the hydrolysate was used to record a heteronuclear single-quantum correlated two-dimensional nuclear magnetic resonance (NMR) spectrum as described previously (37). Isotopomer distributions of proteinogenic amino acids and glycerol were determined from this two-dimensional spectrum. NMR measurements were performed on an AMX-400 WB spectrometer system (Bruker, Karlsruhe, Germany).
Gene overexpression.
Plasmids were constructed in E. coli DH5αMCR from PCR-generated fragments (Expand High Fidelity PCR kit; Roche Diagnostics) by using C. glutamicum DNA, prepared as previously described (6), as a template. E. coli was transformed by the RbCl2 method (14). C. glutamicum was transformed via electroporation (56). All transformants were analyzed by plasmid analysis and/or PCR with appropriate primers.
In order to construct pXMJ19sdaA, the sdaA gene from C. glutamicum was amplified by PCR using the upstream primer 5′-GCTCTAGAAAGGAGATATAGAT[r]ATGGCTATCAGTGTTGTTGAT-3′ (nucleotide 1744884 of NC003450 is underlined) and the reverse primer 5′-GCGAATTCGCCAAGCAAGACAAAATCCAAGCC-3′ (nucleotide 1746274 of NC003450 is underlined). Boldfaced nucleotides correspond to an XbaI and an EcoRI restriction site, respectively, and italicized nucleotides correspond to the ribosome binding site of T7 gene 10. The PCR product was subcloned into the pGEM-T vector by using the pGEM-T vector system (Promega). The resultant plasmid, pGEM-TsdaA, was digested with EcoRI and XbaI, and the sdaA-containing insert obtained was ligated with EcoRI- and XbaI-treated pXMJ19, resulting in plasmid pXMJ19sdaA.
Gene inactivation.
The sdaA gene was inactivated by modified gene replacement methods as described previously (27, 48). According to the sequence of the sdaA gene, four primers (primers ΔsdaA_1 [5′-TCGTGCAACTTCAGACTC-3′], ΔsdaA_2 [5′-CCCATCCACTAAACTTAAACACGTCATAATGAACCCACC-3′], ΔsdaA_3 [5′-TGTTTAAGTTTAGTGGATGGGCCGACTAATGGTGCTGCG-3′], and ΔsdaA_4 [5′-CGGGAAGCCCAAGGTGGT-3′]) were designed, with primers ΔsdaA_2 and ΔsdaA_3 containing homologous extensions of 21 bp (underlined) at the 5′ end used as a linker sequence in order to allow crossover PCR. The primer pair ΔsdaA_1 and ΔsdaA_2 was used to amplify a 504-bp fragment of the 5′ end, and primer pair ΔsdaA_3 and ΔsdaA_4 was used to amplify a 509-bp fragment of the 3′ end, of the sdaA gene by PCR from denatured cells from C. glutamicum. The resulting PCR fragments were used as templates for PCR with primer pair ΔsdaA_1 and ΔsdaA_4 to amplify the sdaA gene harboring a 415-bp deletion plus the 21-bp linker sequence. The resulting 1,034-bp fragment was ligated into the SmaI restriction site of the mobilizable E. coli vector pK19mobsacB, which is nonreplicative in C. glutamicum, leading to pK19mobsacBΔsdaA. By use of a method described previously (39), the resulting vector pK19mobsacBΔsdaA was used to replace the intact chromosomal sdaA gene in C. glutamicum ATCC 13032 with the truncated sdaA gene. PCR with primers located upstream and downstream of the truncated gene was performed to verify the replacement at the chromosomal sdaA locus (data not shown). The sdaA mutant was designated 13032ΔsdaA. Site-specific deletion of the metC gene of strain 13032ΔsdaA was performed by using plasmid pCR007d (46). The desired deletion was also verified by PCR (data not shown), and the resulting mutant was designated 13032ΔsdaAΔmetC.
Enzyme assays.
l-Serine dehydratase activity was measured by the formation of pyruvate from l-serine via high-performance liquid chromatography (HPLC) after derivatization with 4,5-dimethoxy-1,2-diaminobenzene (DDB) (22). Crude extracts were acquired by ultrasonication in 50 mM HEPES (pH 8.0)-10% glycerol-3 mM FeSO4-10 mM dithiothreitol. Assays were performed in mixtures (1.5 ml) containing 50 mM HEPES (pH 8.0), 1% glycerol, 10 mM dithiotreitol, 0 to 500 mM l-serine, and different amounts of crude extract (10 to 100 μg of protein ml−1). d-Serine, l-threonine, d-threonine, l-allo-threonine, or d-allo-threonine (50 mM each) was included in the mixture instead of l-serine, respectively, for determination of the substrate specificity. The reaction was stopped after 10 min by adding 150 μl of DDB solution (80 μg of DDB ml−1, 0.5 M HCl, 0.21 M β-mercaptoethanol) to a 150-μl reaction mixture. Derivatization was performed by 2 h of incubation at 102°C.
Cystathionine β-lyase activity was determined by a method analogous to that described for l-serine dehydratase by measuring pyruvate formation via HPLC. Crude extracts were prepared by resuspension of the cell pellets in 1 ml of 100 mM Tris-HCl, pH 8.5 (24), and disruption by ultrasonic treatment. Enzyme activity was determined immediately in the cell-free supernatant. The assay mixture contained 100 mM Tris-HCl (pH 8.5), 200 μM pyridoxal-5′-phosphate, 5 mM cystathionine, 5 mM l-cystine, 50 mM cysteine or 50 mM l-serine, respectively, and an appropriate amount of crude extract. Pyruvate formation was analyzed as described above.
Gene expression analysis.
For the DNA microarray analysis (60), wild-type C. glutamicum was grown after precultivation in CgXII medium in two parallel cultures with 100 mM glucose as the carbon source. After 4 h of growth, from an OD600 of 0.8 to an OD600 of 3.5, 1 mM seryl-tripeptide was added to one of the cultures and the cells were further incubated to reach an OD600 of 7 before harvest. Intracellular quantification of the amino acid pools (51) confirmed that addition of 1 mM seryl-tripeptide to the exponentially growing culture resulted in an intracellular l-serine concentration of 95 mM, whereas the concentration in the control culture without peptide was 1 mM. The harvesting of 25-ml aliquots of the cultures and the entire procedure of RNA preparation, cDNA synthesis, DNA microarray hybridization, washing, data normalization, and gene expression analysis have been described previously (18, 26).
Analytical methods.
Amino acid concentrations in the culture supernatant were determined by reversed-phase liquid chromatography (HPLC) after derivatization with ortho-phthaldialdehyde as described previously (49). Glucose concentrations were determined with the d-glucose determination test kit (R-Biopharm, Darmstadt, Germany).
RESULTS
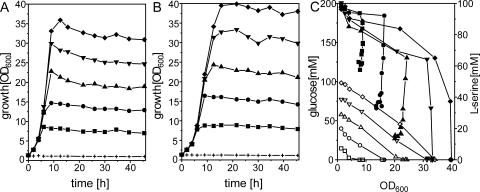
Utilization of l-serine by C. glutamicum. Initial growth experiments showed that wild-type C. glutamicum was not able to utilize l-serine either as a sole carbon source or as a sole nitrogen source (data not shown), although l-serine was utilized in the presence of glucose. We therefore analyzed coutilization of l-serine in detail. For this purpose, C. glutamicum was grown on minimal medium with different glucose concentrations in the absence (Fig. 1A) or presence (Fig. 1B) of 100 mM l-serine. As can be seen, with all glucose concentrations, an increase in the final biomass was attained due to addition of l-serine. After 30 h of cultivation, the increase in the OD600 was 18 to 22%. Addition of l-serine reduced the growth rate slightly, i.e., from 0.37 h−1 at 60 mM glucose to 0.30 h−1. Cometabolism of l-serine with glucose is therefore different from that for E. coli, where 1 mM l-serine abolishes growth on glucose or lactate (36).
FIG. 1.
Growth (A, B) and substrate consumption (C) of wild-type C. glutamicum in minimal medium containing different glucose concentrations (+, 0 mM; ▪, 20 mM; •, 40 mM; ▴, 60 mM; ▾, 80 mM; ♦, 100 mM) in the absence (A) or presence (B) of l-serine. (C) Consumption of glucose (open symbols) and serine (closed symbols) as a function of OD600.
For all cultures grown on the glucose-l-serine mixtures (Fig. 1B), consumption of both substrates was monitored and the respective concentrations determined were plotted against the OD600 reached at the time when the sample was taken (Fig. 1C). Interestingly, we observed different phases of l-serine utilization. For instance, on 60 mM glucose plus 100 mM l-serine, glucose was utilized completely by the cells, whereas at the same time only about 40% of the l-serine was consumed. However, after exhaustion of glucose (at an OD600 of 24), the culture still continued to consume l-serine without additional growth to a concentration of approximately 15 mM, where utilization stopped. This result shows that distinct phases of cometabolism exist: in phase 1, growth occurs during coutilization of glucose with l-serine; in phase 2, after growth has stopped, l-serine is still utilized; in phase 3, l-serine is not utilized at all. As can be seen from Fig. 1C, the largest part of l-serine is consumed during phase 2, and this depends on the amount of biomass present. The average consumption rate of glucose for all cultures was 43.8 ± 4.9 nmol min−1 (mg [dry weight])−1. For l-serine it was 19.4 ± 4.0 nmol min−1 (mg [dry weight])−1 in phase 1 and 8.2 ± 2.6 nmol min−1 (mg [dry weight])−1 in phase 2.
Intracellular fate of l-serine.
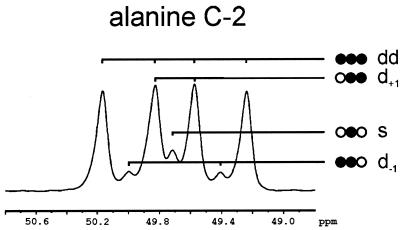
In order to obtain direct information on the fate of l-serine, a tracer analysis was performed. For this purpose C. glutamicum was grown on 100 mM glucose with 90 mM l-[U-13C]serine. Cells were harvested after 20 h, when all glucose and 80% of the l-serine had been consumed. The cell pellet was hydrolyzed, and a sample was analyzed by two-dimensional (1H, 13C) HSQC NMR (54) to quantify 13C fractional enrichments and isotopomer distributions of glycine, alanine, aspartate, phenylalanine, glycerol, and histidine in the biomass hydrolysate (37). Table 1 shows that 92.6% of the l-serine was labeled uniformly, giving rise to a doublet of doublet (dd) signals in the NMR 13C spectrum. The fact that almost all labeled cellular l-serine was labeled uniformly corresponded to the situation expected when the l-[U-13C]serine is taken up and integrated into the cellular protein. More interesting was the analysis of the proteinogenic l-alanine, because this gives direct information about the labeling in pyruvate (29). The fine structure of the 13C NMR signal from C-2 of l-alanine is shown in Fig. 2. It is apparent that the dominant signal was again a doublet of doublets resulting from the isotopomer in which all three carbons were labeled. Also the singlet signal(s) from the isotopomer in which only C-2 of l-alanine is labeled was visible, as well as signals due to the two isotopomers in which two carbon atoms were labeled. The results of the quantitative analysis of selected NMR signals are given in Table 1. It shows that 85.5% ± 2.0% of the labeled l-alanine was uniformly labeled, which was further confirmed by the result for C-3 of l-alanine, showing 86.8% ± 0.5% coupling with C-2. This result verified that the carbon skeleton of l-serine was converted as an entity to pyruvate and used for different purposes such as transamination to l-alanine. Glycine was almost uniformly labeled in both C atoms (96.3%), indicating that it was also derived directly from l-serine. Moreover, significant 13C label was detected in further cellular compounds (Table 1) such as l-phenylalanine or l-aspartate, from which it can be concluded that label derived from l-serine was further transferred to cellular metabolites via pyruvate.
TABLE 1.
Isotopomer labeling patterns of central metabolites from wild-type C. glutamicum
| Central metabolite and carbon atom position | Measured from biomass compound | Fine structure of 13C multiplet signals (%)
|
||||
|---|---|---|---|---|---|---|
| s | d−1 | d−1 + d+1a | d+1 | dd | ||
| Serine C-2 | Serine C-2 | 2.0 ± 1.0 | 3.4 ± 0.5 | 2.0 ± 1.0 | 92.6 ± 2.0 | |
| 3-Phosphoglycerate C-2 | Glycine C-2 | 3.7 ± 0.5 | 96.3 ± 0.5 | |||
| Pyruvate C-2 | Alanine C-2 | 5.0 ± 2.0 | 5.9 ± 1.0 | 3.6 ± 2.0 | 85.5 ± 2.0 | |
| Pyruvate C-3 | Alanine C-3 | 13.2 ± 0.5 | 86.8 ± 0.5 | |||
| Phosphoenolpyruvate C-2 | Phenylalanine C-2 | 21.0 ± 5.0 | 15.0 ± 1.0 | 5.0 ± 2.0 | 59.0 ± 4.0 | |
| Triose phosphates C-1/C-3 | Glycerol C-1/C-3 | 40.0 ± 3.0 | 60.0 ± 2.0 | |||
| Triose phosphates C-2 | Glycerol C-2 | 38.0 ± 3.0 | 19.0 ± 2.0 | 43.0 ± 4.0 | ||
| Pentose phosphates C-3 | Histidine C-3 | 39.0 ± 2.0 | 61.0 ± 2.0 | |||
| Oxaloacetate C-2 | Aspartate C-2 | 14.7 ± 0.5 | 25.4 ± 0.5 | 12.9 ± 1.0 | 47.0 ± 1.0 | |
Since glycerol is a symmetrical molecule, only the sum of d−1 and d+1 is determined.
FIG. 2.
13C NMR spectra of the C-2 of alanine and illustration of the signal fine structure composition. s, singlet peak of [2-13C]alanine (no neighboring labels); d−1, 13C in the preceding position ([1,2-13C2]alanine) produces a doublet peak, split by scalar coupling; d+1, 13C in the following position ([2,3-13C2]alanine) yields another doublet split with a different coupling constant; dd, “doublet of doublet” signal of [13C3]alanine.
Identification, deletion, and overexpression of sdaA.
Based on the tracer experiment, we searched the genome of C. glutamicum for an l-SerDH which could be a candidate enzyme for generating pyruvate from l-serine. By using the E. coli SdaA, SdaB, or TdcG sequence invariably, one single protein sequence was identified in C. glutamicum (encoded by NCgl1583) exhibiting high identities (about 40%) to all three E. coli polypeptides. The protein encoded by C. glutamicum NCgl1583 exhibits three conserved cysteine residues which could serve for the coordination of a [4Fe-4S] cluster as postulated for the l-SerDH of the anaerobic bacterium Peptoniphilus asaccharolyticus (formerly known as Peptostreptococcus asaccharolyticus) (17). In order to investigate whether the gene identified encodes a functional l-SerDH, the corresponding gene, termed sdaA, was amplified and cloned into the E. coli-C. glutamicum shuttle vector pXMJ19 (19). The resulting plasmid was used to transform wild-type C. glutamicum to yield the recombinant strain 13032(pXMJ19sdaA). In addition, a deletion plasmid was constructed which was used in two rounds of positive selection (48) to enable deletion of sdaA in the genome of C. glutamicum. The deletion in the resulting 13032ΔsdaA strain was confirmed by PCR (data not shown).
Since known l-SerDHs often exhibit only weak activities (7), we used a highly sensitive assay based on the direct quantification of pyruvate via HPLC (see Materials and Methods). Cells were harvested from a glucose-plus-l-serine culture, and crude extracts prepared were immediately used in the l-SerDH assay. From the linear increase in pyruvate formation, the specific activities were calculated. With plasmid-encoded sdaA a high specific activity of 210.7 ± 8.7 nmol min−1 (mg of protein)−1 was obtained, and the activity was dependent on reducing conditions and iron (data not shown), as has been described for E. coli (53). This result clearly showed that sdaA encodes a functional l-SerDH. As expected, the mutant 13032ΔsdaA(pXMJ19) strain, with sdaA deleted, did not give any activity (specific activity, 0.7 ± 0.9 nmol min−1 [mg of protein]−1). Surprisingly, with the wild type carrying the empty plasmid also, no significant activity was observed (specific activity, 1.0 ± 0.6 nmol min−1 [mg of protein]−1), suggesting that the enzyme is not active under the chosen conditions.
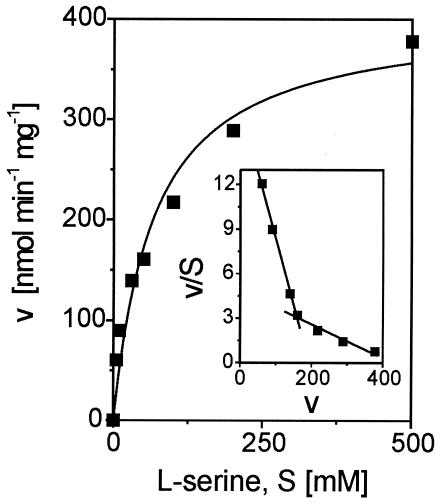
In order to determine the substrate constant for l-serine, the activity of the l-SerDH was quantified with an extract of the 13032(pXMJ19sdaA) strain at different substrate concentrations. As shown in Fig. 3, substrate conversion does not follow classical Michaelis-Menten type kinetics. Instead, the Eadie-Hofstee plot (Fig. 3 inset) revealed a clear biphasic dependence of the activity on the l-serine concentration. Whereas at the lower substrate concentrations (up to 50 mM) an apparent Km of 11 mM was determined, at the higher concentrations (up to 500 mM) the apparent Km determined was 90 mM. The possibility that l-serine itself influences the assay at exceptionally high concentrations cannot be excluded. Nevertheless, the l-SerDH of C. glutamicum exhibited a relatively low affinity for its substrate l-serine, as was shown for the E. coli SdaA enzyme with its estimated Km of 42 mM (34). When d-serine, l-threonine, d-threonine, l-allo-threonine, and d-allo-threonine were assayed as possible substrates, in no case did the activity exceed 1.6% of the activity obtained with l-serine.
FIG. 3.
Specific activity (v) of l-SerDH in crude extracts of C. glutamicum 13032(pXMJ19sdaA) as a function of the l-serine concentration (S). Inset represents the respective Eadie-Hofstee plot.
Physiological consequences of altered sdaA expression.
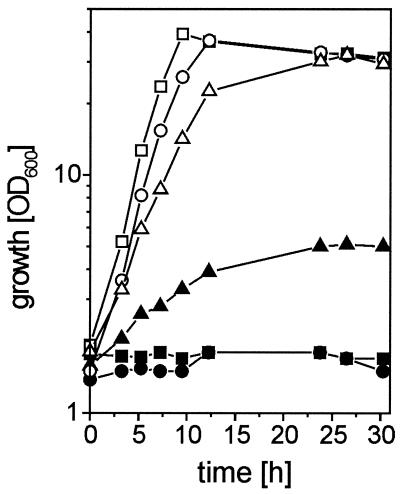
We performed growth experiments on minimal medium containing 100 mM glucose and/or 100 mM l-serine with the recombinant strains constructed. Most interestingly, the 13032(pXMJ19sdaA) strain was able to grow on l-serine as a sole carbon source, albeit at a low growth rate of 0.07 h−1 (Fig. 4) and not entirely consuming the substrate (data not shown). This indicates that sdaA expression enables utilization of l-serine, but that at the same time an unknown regulatory mechanism might prevent its entire consumption. Furthermore, growth of the 13032(pXMJ19sdaA) strain on glucose was significantly affected, since its growth rate was reduced from 0.36 h−1 for the 13032(pXMJ19) strain as a control to 0.23 h−1 (Fig. 4). These effects were confirmed with separate clones and in separate growth experiments, and they illustrate the in vivo activity of l-SerDH in the overexpressing strain.
FIG. 4.
Growth of different C. glutamicum strains in minimal medium containing 100 mM glucose (open symbols) or 100 mM l-serine (closed symbols). Triangles, 13032(pXMJ19sdaA); circles, 13032(pXMJ19); squares, 13032ΔsdaA(pXMJ19).
The consequences of altered sdaA expression on l-serine coutilization were derived from the cultures grown on glucose plus l-serine. Upon overexpression of sdaA, the l-serine consumption rate was increased about twofold to 30.0 nmol min−1 (mg [dry weight])−1, compared to 16.4 nmol min−1 (mg [dry weight])−1 obtained with the control strain 13032(pXMJ19). Moreover, l-serine utilization by the sdaA deletion mutant was reduced to 8.7 nmol min−1 (mg [dry weight])−1. This shows that l-SerDH contributes significantly to l-serine utilization. However, due to the clearly observable residual l-serine consumption by the sdaA deletion mutant, we analyzed the fate of l-[U-13C]serine in the same manner as that described for the wild type. Indeed, the 13C enrichments in cellular metabolites determined by two-dimensional (1H, 13C) HSQC NMR were comparable to that for the wild type. For instance, for the multiplet signals obtained for C-2 of alanine, s = 6% ± 2%, d−1 = 6% ± 2%, d+1 = 7% ± 3%, and dd = 81% ± 4%. This demonstrates that l-serine is still converted via pyruvate to l-alanine in the deletion mutant. Furthermore, other multiplet signals also were almost identical. The dd obtained for C-2 of aspartate was 43% ± 1% in the mutant compared to 47% ± 1% in the wild type. This indicates that the pyruvate resulting from l-serine is used to synthesize oxaloacetate by the anaplerotic pyruvate carboxylase (39, 40), which is further converted to l-aspartate. These data confirm that besides l-serine dehydratase, further l-serine-converting reactions yielding pyruvate are operating.
Global gene expression analysis.
To get access to such reactions, we used DNA microarrays to probe for altered mRNA levels as a further approach to identify l-serine-degrading reactions (26). To enable a high intracellular l-serine concentration, we added the tripeptide Ser-Ser-Ser, which is hydrolyzed upon uptake and thus ensures a high intracellular l-serine concentration (51). Total RNA was isolated 70 min after peptide feeding, and relative RNA levels were determined by hybridization to DNA microarrays representing 95.5% of the open reading frames of C. glutamicum (NCBI NC003450). In total only a relatively small number of genes (69 genes) exhibited an mRNA level exceeding a twofold alteration, with 62 of them showing increased expression due to peptide addition. Table 2 lists expression changes of eight open reading frames (ORFs) related to amino acid metabolism. NCgl2241 encodes the putative ATP-binding protein of an oligopeptide transporter. Its increased mRNA level could thus be a direct consequence of the peptide present to enable its efficient uptake. With the others, a correlation to l-serine is probably less apparent (see Discussion). Most interestingly, the cystathionine β-lyase mRNA level (metC) is increased 2.4-fold, and the respective enzyme of l-methionine synthesis (24), catalyzing a β-elimination reaction, has been reported to have a broad substrate specificity in C. glutamicum and in E. coli, also reacting with l-cysteine, which is structurally related to l-serine (1, 45, 59).
TABLE 2.
ORFs showing altered relative mRNA levels in response to l-serine in wild-type C. glutamicum
| ORF | NCBI no.a | Function or annotationb | Genec | Increase in mRNA level in response to l-serine (fold) |
|---|---|---|---|---|
| 2793 | NCgl1222 | Acetolactate synthase large chain | ilvB | 4.6 |
| 3236 | NCgl2133 | Glutamine synthetase I | glnA | 2.4 |
| 3118 | NCgl2227 | Cystathionine β-lyase | metC | 2.4 |
| 844 | NCgl0371 | Strong similarity to purU (Shigella flexneri), encoding formyltetrahydrofolate deformylase | 2.3 | |
| 1500 | NCgl0895 | Similarity to livM (E. coli), encoding a leucine transport protein | 2.2 | |
| 3696 | NCgl2139 | Threonine synthase | thrC | 2.1 |
| 1396 | NCgl0811 | Inositol monophosphatase family protein; similarity to cysQ (E. coli) | 2.0 | |
| 3102 | NCgl2241 | Strong similarity to oppD (L. lactis), encoding an oligopeptide transport ATP-binding protein | 1.9 |
Number for the corresponding ORF in the revised C. glutamicum genome in the National Center for Biotechnology Information (NCBI) database.
Most similar gene in public databases.
Only characterized C. glutamicum genes are given.
Activity of cystathionine β-lyase.
Based on these observations, the possibility that cystathionine β-lyase also uses l-serine as a substrate was tested. We therefore used plasmid pSL173, encoding metC (24), to overexpress metC in C. glutamicum 13032ΔsdaA. The resulting 13032ΔsdaA(pSL173) strain and the control 13032ΔsdaA(pZ1) strain were grown in minimal medium with 200 mM glucose as the carbon source. Enzyme activities were determined with crude extracts of both strains with the natural substrate cystathionine (5 mM) as well as with l-cystine (5 mM), l-cysteine (50 mM), and l-serine (50 mM), and pyruvate formation was quantified. With cystathionine and l-cystine, the control exhibited comparable specific activities of 0.13 and 0.11 μmol min−1 mg−1, respectively. With metC overexpressed, the specific activities were increased to 1.05 and 1.34 μmol min−1 mg−1 for the respective substrate. With l-cysteine as a substrate and an extract of the overexpressing strain, the specific activity was 0.35 ± 0.07 μmol min−1 mg−1, corroborating the finding that cystathionine β-lyase has l-cysteine desulfhydration activity (59). Also with l-serine a significant activity of 0.04 ± 0.01 μmol min−1 mg−1 was determined. This result showed that cystathionine β-lyase of C. glutamicum is capable of deaminating l-serine to pyruvate in vitro, as was shown for E. coli (2) and Neurospora crassa (8).
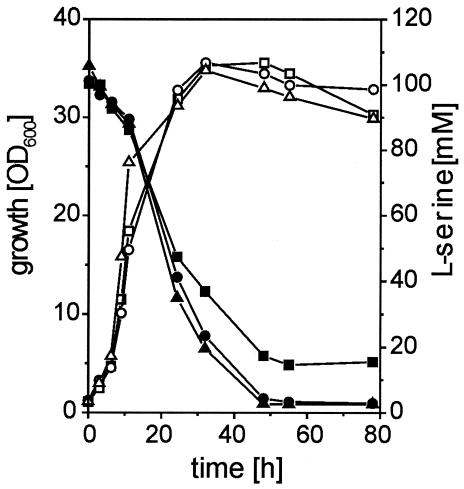
To test the influence of cystathionine β-lyase on l-serine degradation, a ΔsdaA ΔmetC double mutant was constructed. The 13032ΔsdaAΔmetC strain was cultivated on minimal medium containing 100 mM glucose plus 100 mM l-serine, and growth and l-serine utilization were monitored (Fig. 5). The metC-overexpressing 13032ΔsdaApSL173 strain and the 13032ΔsdaApZ1 strain were analyzed as controls. After 55 h of cultivation, the remaining concentration of l-serine in the medium was 15 mM for the double-mutant 13032ΔsdaAΔmetC strain, whereas for the 13032ΔsdaA(pSL137) and 13032ΔsdaA(pZ1) strains, the remaining concentration for both cultures was below 3 mM. This result shows that chromosomally encoded metC has in fact a significant influence on l-serine degradation.
FIG. 5.
Growth (open symbols) and l-serine consumption (closed symbols) of different C. glutamicum strains in minimal medium containing 100 mM glucose plus 100 mM l-serine. Triangles, 13032ΔsdaA(pSL173); circles, 13032ΔsdaA(pZ1); squares, 13032ΔsdaAΔmetC.
DISCUSSION
Cometabolism of l-serine by C. glutamicum can be divided into distinct phases and therefore clearly differs from the strict parallel metabolism of glucose and acetate (61) as well as from the sequential metabolism of glucose and glutamate (25). In phase 1 at a growth rate of 0.30 h−1, the demand for l-serine for cell material formation, including the many metabolites derived from this amino acid, is 7.5 nmol min−1 (mg [dry weight])−1 (28). Thus, about half of the l-serine consumed could be directly incorporated into cell material in this phase, resulting in an increased growth yield. This is consistent with the finding that a large amount of l-serine is metabolized via pyruvate. Integration of l-serine into biomass occurs even though C. glutamicum is not able to grow on l-serine as a sole carbon source. A similar situation is observed with Campylobacter jejeuni, which utilizes this amino acid in the presence of pyruvate, resulting in an increased growth yield of 46% (57). Also, it has been reported for E. coli that l-serine as an auxiliary carbon source results in a greatly increased growth yield (33). In phase 2, l-serine utilization was decoupled from growth, and thus, other reasons for l-serine utilization must exist, as, for instance, its degradation to derive energy for maintenance. Indeed, an E. coli mutant is known which has a growth advantage in the stationary phase and which at the same time has an increased capability to grow with l-serine, enabling generation times of 6.7 h instead of 61.2 h (63). Another discernible consequence with C. glutamicum is that after consumption of glucose, the cultures which received l-serine appear to maintain a higher OD600 for a longer time than cultures without l-serine (compare Fig. 1A and B). Considering that after depletion of glucose, l-serine is still consumed, its metabolism might serve to increase the maintenance of the culture under starvation conditions. Although wild-type E. coli is not capable of effective l-serine utilization (47), mutants with increased utilization are known, where mutations increase the fitness in starved cultures (58).
As shown by incorporation of 13C-labeled l-serine in protein, the advantage of coutilization consists in part in a reduced requirement for the synthesis of l-serine from glucose. However, as revealed by the high uptake rate, together with the labeling information, the ΔsdaA ΔmetC mutant studies, and the enzyme analysis, the largest part of externally added l-serine is converted to pyruvate. This was also shown for L. lactis, where carbon flux analysis with 14C-labeled l-serine revealed that 72% of the initial l-serine flux is found in metabolites originating from pyruvate (35). While L. lactis has a restricted metabolism and is therefore necessarily dependent on the cometabolism of l-serine with glucose, an advantage for C. glutamicum in cometabolism could be utilization of the generated pyruvate for biosynthetic purposes as well as its oxidation to produce energy for maintenance. Although the current label information alone cannot give an answer to whether substantial quantities of l-serine are oxidized via pyruvate up to carbon dioxide, thus contributing to energy generation, it is clear that a large part of pyruvate contributes to synthesis of cell material, since the 13C label from l-serine-derived pyruvate is present in phosphoglycerate, triose phosphates, and even pentose phosphates (Table 1). Therefore, it can be concluded that the pyruvate skeleton is converted to glyceraldehyde-3-phosphate in vivo in sufficiently high concentrations to enable reverse metabolic flux from pyruvate up to fructose-1,6-bisphosphate.
As found for other l-SerDHs (12, 47, 57), the enzyme from C. glutamicum has high substrate specificity and does not accept substrates other than l-serine. Unlike the pyridoxal-5-phosphate-containing d-serine and l-threonine dehydratases, it is most likely that all highly specific bacterial l-SerDHs contain an [4Fe-4S]-cluster as the prosthetic group (11). The C. glutamicum l-SerDH polypeptide also exhibits three of the four possible conserved cysteine residues (9), and the activity is dependent on reducing conditions. Accordingly, the l-SerDH is oxygen sensitive, as demonstrated for the enzymes of the strictly anaerobic organisms P. asaccharolyticus (12) and Clostridium propionicum (16) as well as for those of the microaerophilic bacterium C. jejeuni (57). Although C. glutamicum possesses a functional l-serine dehydratase, no activity was observed in crude extracts of the wild type under the chosen conditions, but since deletion of the enzyme resulted in 30% decreased l-serine degradation, it must be concluded that the enzyme is active in vivo in the wild type. This is similar to the situation for E. coli, where activity in crude extracts was detectable only after treatment with iron and reducing agents (53, 34). Even though this dependence was confirmed for the overexpressed C. glutamicum l-SerDH, the possibility that other, yet unknown factors are necessary to stabilize the activity cannot be ruled out.
The respective l-SerDHs of P. asaccharolyticus (12), C. propionicum (16), and C. jejeuni (57) were isolated from cells grown on complex media containing l-serine, implying that coutilization of this amino acid might occur. Whereas these organisms have a restricted metabolism, it is astonishing that E. coli is not able to grow on l-serine, although it has three l-SerDHs, encoded by sdaA (53), sdaB (50), and tdcG (15), which are subject to complex regulation by a number of different effectors (for a review, see reference 47). With the sdaA-overexpressing strain of C. glutamicum, we found growth on l-serine, albeit at a lower growth rate and final biomass yield than on pyruvate (data not shown). This indicates that although the specific activity of l-SerDH in this strain would be sufficient for growth on l-serine comparable to growth on pyruvate, there are either regulatory phenomena or l-SerDH-specific biochemical features that prevent the entire conversion of l-serine. Since the conversion of l-serine to pyruvate liberates ammonium, a high internal ammonium concentration might be toxic for the cell as well.
This assumption is further substantiated by the result of the DNA-Chip experiment, where the glnA transcript of glutamine synthetase I, the only ammonia-fixing enzyme in C. glutamicum (20), is increased 2.4-fold at an elevated intracellular l-serine concentration. The notably increased level of ilvB (acetolactate synthase) could be an indirect consequence of an increased pyruvate pool, since the synthase utilizes two pyruvate molecules. Interestingly, the metC transcript level is also increased, presumably as an indirect consequence of increased l-serine concentrations. This gene encodes the cystathionine β-lyase of l-methionine synthesis (24), but we could clearly demonstrate that it is also able to deaminate l-serine to pyruvate in vitro. This result shows that the enzyme not only degrades sulfur-containing amino acids (59), but is also able to degrade l-serine in a β-elimination reaction, as has been found for cystathionine β-lyases from N. crassa (8) and E. coli (2). Deletion of the metC gene in the ΔsdaA background resulted in a strain that degraded significantly less l-serine than the ΔsdaA single mutant, demonstrating that l-serine is also converted by this enzyme in vivo. Nevertheless, l-serine degradation still occurs in that strain, indicating that at least one additional activity is present. For instance, threonine dehydratase (55) or the β-subunit of l-tryptophan synthase from E. coli and Salmonella serovar Typhimurium (4, 62) is known to deaminate l-serine to pyruvate in vitro. Whether these enzymes are involved in l-serine degradation in C. glutamicum remains to be elucidated.
Acknowledgments
We thank H. Etterich for exellent technical assistance, A. A. de Graaf for performing the NMR experiments, C. Rückert of the University of Bielefeld, Bielefeld, Germany, for providing plasmid pCR007d, and H.-S. Lee of the Korea University, Seoul, Korea, for providing plasmid pSL173. V. F. Wendisch is thanked for fruitful discussions and critical reading of the manuscript.
This work was financed in part by the Deutsche Bundesstiftung Umwelt (DBU AZ 13037).
REFERENCES
- 1.Awano, N., M. Wada, A. Kohdoh, T. Oikawa, H. Takagi, and S. Nakamori. 2003. Effect of cysteine desulfhydrase gene disruption on l-cysteine overproduction in Escherichia coli. Appl. Microbiol. Biotechnol. 62:239-243. [DOI] [PubMed] [Google Scholar]
- 2.Brown, E. A., R. D'Ari, and E. B. Newman. 1990. A relationship between l-serine degradation and methionine biosynthesis in Escherichia coli K12. J. Gen. Microbiol. 136:1017-1023. [DOI] [PubMed] [Google Scholar]
- 3.Cocaign, M., C. Monnet, and N. D. Lindley. 1993. Batch kinetics of Corynebacterium glutamicum during growth on various substrates: use of substrate mixtures to localize metabolic bottlenecks. Appl. Microbiol. Biotechnol. 40:526-530. [Google Scholar]
- 4.Crawford, I. P., and J. Ito. 1964. Serine deamination by the B protein of Escherichia coli tryptophan synthetase. Proc. Natl. Acad. Sci. USA 51:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eggeling, L., and H. Sahm. 1999. l-Glutamate and l-lysine: traditional products with impetuous developments. Appl. Microbiol. Biotechnol. 52:146-153. [Google Scholar]
- 6.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K. Lüdtke, and H. Sahm. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 7.Fariás, M. E., A. M. Strasser de Saad, A. A. Pesce de Ruiz Holgado, and G. Oliver. 1991. Purification and properties of l-serine dehydratase from Lactobacillus fermentum ATCC 14931. Curr. Microbiol. 22:205-211. [Google Scholar]
- 8.Flavin, M., and C. Slaughter. 1964. Cystathionine cleavage enzymes of Neurospora. J. Biol. Chem. 239:2212-2219. [PubMed] [Google Scholar]
- 9.Flint, D. H., and R. M. Allen. 1996. Iron-sulfur proteins with nonredox functions. Chem. Rev. 96:2315-2334. [DOI] [PubMed] [Google Scholar]
- 10.Girbal, L., D. Hilaire, S. Leduc, L. Delery, J. L. Rols, and N. D. Lindley. 2000. Reductive cleavage of demeton-S-methyl by Corynebacterium glutamicum in cometabolism on more readily metabolizable substrates. Appl. Environ. Microbiol. 66:1202-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabowski, R., A. E. Hofmeister, and W. Buckel. 1993. Bacterial l-serine dehydratases: a new family of enzymes containing iron-sulfur clusters. Trends Biochem. Sci. 18:297-300. [DOI] [PubMed] [Google Scholar]
- 12.Grabowski, R., and W. Buckel. 1991. Purification and properties of an iron-sulfur-containing and pyridoxal-phosphate-independent l-serine dehydratase from Peptostreptococcus asaccharolyticus. Eur. J. Biochem. 199:89-94. [DOI] [PubMed] [Google Scholar]
- 13.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1985. Techniques for transformation of Escherichia coli, p. 109-136. In D. M. Glover (ed.), DNA cloning, vol. 1. IRL Press, Oxford, United Kingdom. [Google Scholar]
- 15.Heßlinger, C., S. A. Fairhurst, and G. Sawers. 1998. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades l-threonine to propionate. Mol. Microbiol. 27:477-492. [DOI] [PubMed] [Google Scholar]
- 16.Hofmeister, A. E., R. Grabowski, D. Linder, and W. Buckel. 1993. l-Serine and l-threonine dehydratase from Clostridium propionicum. Two enzymes with different prosthetic groups. Eur. J. Biochem. 215:341-349. [DOI] [PubMed] [Google Scholar]
- 17.Hofmeister, A. E., S. Textor, and W. Buckel. 1997. Cloning and expression of the two genes coding for l-serine dehydratase from Peptostreptococcus asaccharolyticus: relationship of the iron-sulfur protein to both l-serine dehydratases from Escherichia coli. J. Bacteriol. 179:4937-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakoby, M., C. E. Ngouoto-Nkili, and A. Burkovski. 1999. Construction and application of new Corynebacterium glutamicum vectors. Biotechnol. Techniques 13:437-441. [Google Scholar]
- 20.Jakoby, M., M. Tesch, H. Sahm, R. Krämer, and A. Burkovski. 1997. Isolation of the Corynebacterium glutamicum glnA gene encoding glutamine synthetase I. FEMS Microbiol. Lett. 154:81-88. [DOI] [PubMed] [Google Scholar]
- 21.Janke, D., and W. Fritsche. 1985. Nature and significance of microbial cometabolism of xenobiotics. J. Basic Microbiol. 25:603-619. [DOI] [PubMed] [Google Scholar]
- 22.Keen, R. E., C. H. Nissenson, and J. R. Barrio. 1993. Analysis of femtomole concentrations of α-ketoisocaproic acid in brain tissue by precolumn fluorescence derivatization with 4,5-dimethoxy-1,2-diaminobenzene. Anal. Biochem. 213:23-28. [DOI] [PubMed] [Google Scholar]
- 23.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. W., H. J. Kim, Y. Kim, M. S. Lee, and H. S. Lee. 2001. Properties of the Corynebacterium glutamicum metC gene encoding cystathionine β-lyase. Mol. Cells 11:220-225. [PubMed] [Google Scholar]
- 25.Krämer, R., C. Lambert, C. Hoischen, and H. Ebbighausen. 1990. Uptake of glutamate in Corynebacterium glutamicum. 1. Kinetic properties and regulation by internal pH and potassium. Eur. J. Biochem. 194:929-935. [DOI] [PubMed] [Google Scholar]
- 26.Lange, C., D. Rittmann, V. F. Wendisch, M. Bott, and H. Sahm. 2003. Global expression profiling and physiological characterization of Corynebacterium glutamicum grown in the presence of l-valine. Appl. Environ. Microbiol. 69:2521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marx, A., A. A. deGraaf, W. Wiechert, L. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 49:111-129. [DOI] [PubMed] [Google Scholar]
- 29.Marx, A., K. Striegel, A. A. deGraaf, H. Sahm, and L. Eggeling. 1997. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol. Bioeng. 56:168-180. [DOI] [PubMed] [Google Scholar]
- 30.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 31.Mendz, G. L., and S. L. Hazell. 1995. Amino acid utilization by Helicobacter pylori. Int. J. Biochem. Cell. Biol. 27:1085-1093. [DOI] [PubMed] [Google Scholar]
- 32.Nagata, K., Y. Nagata, T. Sato, M. A. Fujino, K. Nakajima, and T. Tamura. 2003. l-Serine, d- and l-proline and alanine as respiratory substrates of Helicobacter pylori: correlation between in vitro and in vivo amino acid levels. Microbiology 149:2023-2030. [DOI] [PubMed] [Google Scholar]
- 33.Newman, E. B., and C. Walker. 1982. l-Serine degradation in Escherichia coli K-12: a combination of l-serine, glycine, and leucine used as a source of carbon. J. Bacteriol. 151:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman, E. B., and V. Kapoor. 1980. In vitro studies on l-serine deaminase activity of Escherichia coli K12. Can. J. Biochem. 58:1292-1297. [DOI] [PubMed] [Google Scholar]
- 35.Novak, L., and P. Loubiere. 2000. The metabolic network of Lactococcus lactis: distribution of 14C-labeled substrates between catabolic and anabolic pathways. J. Bacteriol. 182:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa, W., T. Kayahara, M. Tsuda, T. Mizushima, and T. Tsuchiya. 1997. Isolation and characterization of an Escherichia coli mutant lacking the major serine transporter, and cloning of a serine transporter gene. J. Biochem. 122:1241-1245. [DOI] [PubMed] [Google Scholar]
- 37.Petersen, S., A. A. de Graaf, L. Eggeling, M. Mollney, W. Wiechert, and H. Sahm. 2000. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 275:35932-35941. [DOI] [PubMed] [Google Scholar]
- 38.Peters-Wendisch, P., R. Netzer, L. Eggeling, and H. Sahm. 2002. 3-Phosphoglycerate dehydrogenase from Corynebacterium glutamicum: the C-terminal domain is not essential for activity but is required for inhibition by l-serine. Appl. Microbiol. Biotechnol. 60:437-441. [DOI] [PubMed] [Google Scholar]
- 39.Peters-Wendisch, P. G., V. F. Wendisch, A. A. de Graaf, B. J. Eikmanns, and H. Sahm. 1996. C3-carboxylation as an anaplerotic reaction in phosphoenolpyruvate carboxylase-deficient Corynebacterium glutamicum. Arch. Microbiol. 165:387-396. [DOI] [PubMed] [Google Scholar]
- 40.Peters-Wendisch, P. G., V. F. Wendisch, S. Paul, B. J. Eikmanns, and H. Sahm. 1997. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology 143:1095-1103. [DOI] [PubMed] [Google Scholar]
- 41.Pfefferle, W., B. Möckel, B. Bathe, and A. Marx. 2003. Biotechnological manufacture of lysine. Adv. Biochem. Eng. Biotechnol. 79:59-112. [DOI] [PubMed] [Google Scholar]
- 42.Pizer, L. I., and M. L. Potochny. 1964. Nutritional and regulatory aspects of serine metabolism in Escherichia coli. J. Bacteriol. 88:611-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prüß, B. M., J. M. Nelms, C. Park, and A. J. Wolfe. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 176:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riedel, C., D. Rittmann, P. Dangel, B. Möckel, S. Petersen, H. Sahm, and B. J. Eikmanns. 2001. Characterization of the phosphoenolpyruvate carboxykinase gene from Corynebacterium glutamicum and significance of the enzyme for growth and amino acid production. J. Mol. Microbiol. Biotechnol. 3:573-583. [PubMed] [Google Scholar]
- 45.Rossol, I., and A. Pühler. 1992. The Corynebacterium glutamicum aecD gene encodes a C-S lyase with α,β-elimination activity that degrades aminoethylcysteine. J. Bacteriol. 174:2968-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rückert, C., A. Pühler, and J. Kalinowski. 2003. Genome-wide analysis of the l-methionine biosynthetic pathway in Corynebacterium glutamicum by targeted gene deletion and homologous complementation. J. Biotechnol. 104:213-228. [DOI] [PubMed] [Google Scholar]
- 47.Sawers, G. 1998. The anaerobic degradation of l-serine and l-threonine in enterobacteria: networks of pathways and regulatory signals. Arch. Microbiol. 171:1-5. [DOI] [PubMed] [Google Scholar]
- 48.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 49.Schrumpf, B., A. Schwarzer, J. Kalinowski, A. Pühler, L. Eggeling, and H. Sahm. 1991. A functionally split pathway for lysine synthesis in Corynebacterium glutamicum. J. Bacteriol. 173:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao, Z., R. T. Lin, and E. B. Newman. 1994. Sequencing and characterization of the sdaC gene and identification of the sdaCB operon in Escherichia coli K12. Eur. J. Biochem. 222:901-907. [DOI] [PubMed] [Google Scholar]
- 51.Simic, P., H. Sahm, and L. Eggeling. 2001. l-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J. Bacteriol. 183:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stauffer, G. V. 1996. Biosynthesis of serine, glycine, and one-carbon units, p. 506-513. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 53.Su, H., J. Moniakis, and E. B. Newman. 1993. Use of gene fusions of the structural gene sdaA to purify l-serine deaminase 1 from Escherichia coli K-12. Eur. J. Biochem. 211:521-527. [DOI] [PubMed] [Google Scholar]
- 54.Szyperski, T. 1995. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids. An efficient analytical tool to investigate intermediary metabolism. Eur. J. Biochem. 232:433-448. [DOI] [PubMed] [Google Scholar]
- 55.Umbarger, H. E. 1996. Biosynthesis of the branched-chain amino acids, p. 442-457. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 56.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 57.Velayudhan, J., M. A. Jones, P. A. Barrow, and D. J. Kelly. 2004. l-Serine catabolism via an oxygen-labile l-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 72:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vining, L. C., and B. Magasanik. 1981. Serine utilization by Klebsiella aerogenes. J. Bacteriol. 146:647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wada, M., N. Awano, K. Haisa, H. Takagi, and S. Nakamori. 2002. Purification, characterization and identification of cysteine desulfhydrase of Corynebacterium glutamicum, and its relationship to cysteine production. FEMS Microbiol. Lett. 217:103-107. [DOI] [PubMed] [Google Scholar]
- 60.Wendisch, V. F. 2003. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J. Biotechnol. 104:273-285. [DOI] [PubMed] [Google Scholar]
- 61.Wendisch, V. F., A. A. de Graaf, H. Sahm, and B. J. Eikmanns. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, Y., and R. H. Abeles. 1993. Inhibition of tryptophan synthase by (1-fluorovinyl)glycine. Biochemistry 32:806-811. [DOI] [PubMed] [Google Scholar]
- 63.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]