Abstract
Novel, vacuolate sulfur bacteria occur at shallow hydrothermal vents near White Point, Calif. There, these filaments are attached densely to diverse biotic and abiotic substrates and extend one to several centimeters into the surrounding environment, where they are alternately exposed to sulfidic and oxygenated seawater. Characterizations of native filaments collected from this location indicate that these filaments possess novel morphological and physiological properties compared to all other vacuolate bacteria characterized to date. Attached filaments, ranging in diameter from 4 to 100 μm or more, were composed of cylindrical cells, each containing a thin annulus of sulfur globule-filled cytoplasm surrounding a large central vacuole. A near-complete 16S rRNA gene sequence was obtained and confirmed by fluorescent in situ hybridization to be associated only with filaments having a diameter of 10 μm or more. Phylogenetic analysis indicates that these wider, attached filaments form within the gamma proteobacteria a monophyletic group that includes all previously described vacuolate sulfur bacteria (the genera Beggiatoa, Thioploca, and Thiomargarita) and no nonvacuolate genera. However, unlike for all previously described vacuolate bacteria, repeated measurements of cell lysates from samples collected over 2 years indicate that the attached White Point filaments do not store internal nitrate. It is possible that these vacuoles are involved in transient storage of oxygen or contribute to the relative buoyancy of these filaments.
Introduction
Representatives of only a few bacterial genera have been found to possess large internal vacuoles, which can comprise 40 to 98% or more of a cell's biovolume (10, 12, 18, 20, 21, 26, 33). These representatives, which include all wider marine Beggiatoa spp., marine and some freshwater Thioploca spp., and Thiomargarita namibiensis, all belong to the colorless sulfur bacteria and form a tight cluster within the gamma proteobacteria, based on 16S rRNA phylogenetic analyses (1a, 12, 13, 21, 37, 38). They have been found in high density in both shallow marine and deep-sea surficial sediments and on vent chimney structures where biogenic or thermogenic sulfide concentrations are high. These sites include deep-sea hydrothermal vents, cold seeps, and areas of coastal upwelling (9, 16, 20, 21, 26, 27, 32, 33).
Prior to this study, all the vacuolate sulfur bacteria tested were found to contain high internal levels of nitrate (15 to 800 mM), representing a 400- to 20,000-fold increase over ambient levels (3, 12, 20, 21, 28, 32, 33). Although never specifically proven, it is logically assumed that much or all of this nitrate is stored in the vacuole. There, it serves as an electron acceptor, permitting oxidation of sulfide under anoxic conditions. The majority of evidence suggests that stored nitrate is reduced to ammonia (4, 28, 32, 42); however, the initial suggestion of denitrification (3) is still supported by some data (10, 28), and no pure cultures of vacuolate bacteria exist to provide unequivocal answers.
Members of the genus Thiothrix are also colorless sulfur oxidizers in the gamma proteobacteria, and they form a monophyletic group at some evolutionary distance from the vacuolate bacteria. Recognized pure-culture strains are narrow filaments (1 to 2 μm in diameter) that live attached to substrates and form rosettes and gliding gonidia (14). Wide, filamentous sulfur bacteria (4 to 200 μm in diameter) that show superficial resemblance to narrower cultured Thiothrix spp. have been observed in abundance attached to surfaces at shallow and deep-sea hydrothermal vents (5, 8, 35; D. C. Nelson, unpublished data). Their prominence at certain deep-sea hydrothermal vents, where organic carbon input is low, suggests that they may be an important source of food for the local macrofauna. The morphology of these wide filaments seems to lie between that of known vacuolate, filamentous sulfur bacteria (nonattached) which form extensive mats on sediment surfaces and that of nonvacuolate, narrow filamentous sulfur bacteria of the genus Thiothrix, which grow attached to surfaces via “holdfast” cells.
Initial observations of the attached bacteria from White Point, Calif., suggest that the bacteria with larger-diameter filaments have extensive internal vacuoles and are morphologically similar to those of the attached sulfur bacteria found at deep-sea vents (K. M. Kalanetra, unpublished data). Therefore, an investigation was initiated to determine the extent of vacuolization in these vacuolate, attached filaments (VAF), the possible correlation between the presence of vacuoles and the accumulation of nitrate, and the phylogenetic position of these bacteria relative to nonvacuolate Thiothrix spp. and vacuolate, nitrate-accumulating sulfur bacteria.
MATERIALS AND METHODS
Sample collection and preservation.
Samples were obtained from White Point by scuba divers in 2001 and 2002. Rocks with attached filaments were collected from the immediate vicinity of venting sulfidic flows (15 to 30°C) at depths of 5 to 7 m. The rocks were kept submerged in seawater and transported to the laboratory on ice. The filamentous bacteria attached to the rocks were harvested within 48 h of collection by cutting them off with a scalpel. The detached filaments were resuspended in sterile, filtered, natural seawater, and debris was separated from the more slowly settling filaments. Repeating this process several times for each collection yielded a single suspension of filaments in a known volume of filtered seawater. Aliquots (1 to 2 ml) were removed, placed in microcentrifuge tubes, and pelleted loosely by centrifugation at 6,000 × g for 5 to 10 min, and the supernatant was reserved. Filaments were relatively buoyant, and neither higher-speed nor longer centrifugation yielded a very compact pellet. Cell pellets and supernatants were stored at −80°C until use. From each suspension, aliquots of known volumes were preserved in glutaraldehyde (final concentration, 2.5%) for phase-contrast and epifluorescence microscopies and stored at 4°C. Other aliquots were prepared for confocal microscopy by the method of Amann (2). Briefly, freshly prepared 4% paraformaldehyde was added (final concentration, 3%), and the samples were chilled (4°C) for a minimum of 4 h and then rinsed three times in a phosphate-buffered saline (PBS) solution. Between rinses, filaments were allowed to settle to the bottom of the vial, and the old PBS was decanted. After the last rinse, ethanol was added to a final concentration of 50%, and the samples were stored at −20°C.
Sediment cores covered by Beggiatoa mats were collected from a depth of about 900 m at Carmel Canyon, Calif., in September 2002 by using the remotely operated vehicle Ventana. Samples were transported on ice and held at 4°C for 3 days to allow migration of filaments to the core surface. Beggiatoa filaments were gently collected with a Pasteur pipette; suspended in sterile, filtered, natural seawater; and processed as described above.
Control strains.
Thiothrix fructosivorans strain Q (strain 49748) was obtained from the American Type Culture Collection (Manassas, Va.) and grown in LTH medium (7, 41). Cultures were preserved for fluorescence microscopy as described above.
Escherichia coli XL1-Blue (Stratagene) was grown in MOPS (morpholinepropanesulfonic acid) minimal medium (23). Cells were harvested by centrifugation (6,000 × g for 2 min) and processed for analysis by ion chromatography as described below.
Measurement of internal anion concentrations.
Frozen cell pellets, each representing a known volume of a filament suspension, were resuspended in a known volume of ultrapurified water (Barnstead NANOpure ultrapure water system; 17.9 MΩ cm−1). Cells were lysed by sonication (several brief pulses at the minimum setting) with a model 1000L sonicator (Ultrasonic Power Corporation). Lysis was confirmed by microscopic examination, and two 50-μl aliquots were removed for protein determination (see below). The remaining cell extract was heated (90 to 100°C) for 10 min to precipitate protein and then clarified by centrifugation (16,100 × g; 15 min). Supernatants were diluted with ultrapurified water, passed through a Dionex OnGuard II Ag cartridge to remove Cl− ions, and then passed through an OnGuard II H cartridge to remove residual Ag+ counterions. Processed lysates were then analyzed by ion chromatography using an Omnipac Pax-500 analytical column (Dionex Corporation) and an anion micromembrane suppressor. The isocratic elution solvent was 2% methanol-40.6 mM sodium hydroxide. Nitrate and phosphate concentrations were calculated based on peak areas compared to standard curves and were converted to intracellular concentrations by using protein/biovolume ratios (20).
Biovolume, width, and protein determinations.
For each suspension of VAF (White Point) or Beggiatoa sp. (Carmel Canyon), biovolume measurements and frequencies were calculated for filaments of different diameters as described previously (24, 26) with glutaraldehyde-preserved samples.
Corresponding protein values were determined following hot trichloroacetic acid hydrolysis and protein precipitation by using the Coomassie brilliant blue dye-binding technique of Bradford (2b) with previously published modifications (20, 26).
FITC staining.
A 1-ml suspension of paraformaldehyde-preserved filaments was stained in the dark (30 to 60 min) with 10 μl of a 10-mg/ml concentration of fluorescein isothiocyanate (FITC; Sigma) (31). The filaments were then rinsed three times with fresh PBS. The sample was viewed and images were recorded with a Leica TCS-SP laser-scanning confocal microscope fitted with an argon ion laser (488-nm excitation wavelength).
16S rRNA gene sequence.
Chromosomal DNA was extracted from White Point and Carmel Canyon frozen cell pellets by a guanidine thiocyanate method (29). White Point pellet DNA was amplified in three parts by using PCR primer pairs 341f-534r, 8fpl-464r, and 445f-1492rpl (Table 1) to amplify almost all of the rest of the White Point VAF 16S rRNA gene (40). Amplification was carried out with AmpliTaq Gold (Applied Biosystems) as follows: initial denaturation at 95°C for 4 min; 30 cycles of 95°C for 45 s, 45°C for 45 s, and 72°C for 45 s; and a final extension at 72°C for 20 min. Carmel Canyon pellet DNA was amplified by using the following pairs of PCR primers: 8fpl-VSO673r and VSO656f-1492rpl (Table 1). This amplification was carried out as described above, except annealing cycles (45 s) were carried out at 59°C.
TABLE 1.
rRNA-targeted oligonucleotide primers and fluorescent probes used in this study
| Probe or primer | Positionsa | Oligonucleotide sequence | Probe or primer specificity |
| 16S rRNA primer | |||
| 341fb | 341-357 | 5′-CCTACGGGAGGCAGCAG-3′ | Eubacteria |
| 534rb | 534-518 | 5′-ATTACCGCGGCTGCTGG-3′ | Eubacteria |
| 8fplc | 8-27 | 5′-AGAGTTTGATCCTGGCTCAG-3′ | Eubacteria |
| 1492rplc | 1510-1492 | 5′-GGTTACCTTGTTACGACTT-3′ | Eubacteria |
| WPF445fd | 445-464 | 5′-GGGAAGAAAAACTTAAAGCT-3′ | White Point VAF (≥10 μm) |
| WPF464rd | 464-445 | 5′-AGCTTTAAGTTTTTCTTCCC-3′ | White Point VAF (≥10 μm) |
| VSO656fd | 656-673 | 5′-GTACAGTAGAGGGAAGCG-3′ | Unattached, vacuolate, sulfur-oxidizing spp.e |
| VSO673rd | 673-656 | 5′-CGCTTCCCTCTACTGTAC-3′ | Unattached, vacuolate, sulfur-oxidizing spp.e |
| 16s RNA probe | |||
| Eub338f | 338-355 | 5′-GCTGCCTCCCGTAGGAGT-3′ | Eubacteria |
| WPF464d | 445-464 | 5′-AGCTTTAAGTTTTTCTTCCC-3′ | White Point wide, attached filaments |
| VSO673d | 656-673 | 5′-CGCTTCCCTCTACTGTAC-3′ | Unattached, vacuolate, sulfur-oxidizing spp.e |
| G123Tg | 697-714 | 5′-CCTTCCGATCTCTATGCA-3′ | Thiothrix spp. |
| G123T-Cg | 697-714 | 5′-CCTTCCGATCTCTACGCA-3′ | Nonsense competitor to probe G123T |
| 23S rRNA probe | |||
| GAM42ah | 1027-1043 | 5′-GCCTTCCCACATCGTTT-3′ | Gamma proteobacteria |
| BET42ah | 1027-1043 | 5′-GCCTTCCCACTTCGTTT-3′ | Beta proteobacteria |
a Based on E. coli 16S rRNA base pair numbering.
b Primer from reference 22.
c Primer from reference 40.
d Primer or probe designed in this study.
e Designed to target Monterey Canyon, Bay of Concepcion, and Carmel Canyon Beggiatoa spp., vacuolate Thioploca spp., and Thiomargarita namibiensis (see Materials and Methods).
f Probe from reference 39.
g Probe from reference 11.
h Probe from reference 19.
Amplified DNA was cloned by using a TOPO TA cloning kit with a pCR 2.1-TOPO vector and TOP10 cells (Invitrogen Corp.) per the manufacturer's instructions or by blunt-end cloning into the EcoRV site of pBluescript. Plasmid DNA from positive transformants was prepared for sequencing with a Qiaprep spin miniprep kit (QIAGEN). All sequencing was carried out at the Division of Biological Sciences Sequencing Facility at the University of California, Davis, by using an ABI 3730 capillary electrophoresis genetic analyzer and ABI BigDye Terminator version 3.1 cycle sequencing chemistry. Sequence data were edited and analyzed with BioEdit Sequence Alignment Editor version 5.0.9 software (6).
FISH.
Fluorescent in situ hybridization (FISH) was performed with VAF (White Point), Beggiatoa sp. (Carmel Canyon), and cultured Thiothrix fructosivorans bacteria by use of published methods (2) modified as follows. Filaments were spotted on silylated glass slides (PGC Scientifics) and air dried. After dehydration of filaments in a series of ethanol baths, hybridization wells (Hybriwell sealing system; PGC Scientifics) were fixed to the slides. For each spot to be hybridized, a hybridization mixture containing a volume ratio of 1:1:7 (probe 1-probe 2-hybridization buffer) was added to fill the well. Hybridizations were carried out at 40 or 46°C in hybridization buffers with formamide concentrations ranging from 0 to 60%. Because some vacuolate sulfur bacteria are known to be autofluorescent (1a), controls were run by subjecting preserved filaments to the hybridization conditions without a probe. After hybridization, slides were mounted in Citifluor antifadent solution AF3. Fluorescence was detected and images were recorded with a Leica TCS-SP laser-scanning confocal microscope fitted with argon and krypton ion lasers (488- and 568-nm excitation wavelengths, respectively). The fluorescence intensities of the filaments were determined by using Leica Confocal software (version 2.0). Competitive hybridizations were carried out for probe pairs GAM42a-BET42a and G123T-G123T-C (each bearing a different fluorochrome), in which each pair targets the same region but there is a 1-bp difference between the two competing sequences. Fluorescent probes and their specificities are listed in Table 1.
Phylogenetic analysis.
The 16S rRNA gene sequences from DNA derived from White Point VAF and the Carmel Canyon Beggiatoa sp. were obtained by combining two or three bidirectional sequences. These were manually aligned with sequences of other gamma and epsilon proteobacterial outgroups (National Center for Biotechnology Information website [http://www.ncbi.nlm.nih.gov/]) by using MacClade version 4.05 (15). Base pair positions 126 to 1376 (E. coli numbering) were used for the main phylogenetic analysis. Large-mask analyses using base pair positions 358 to 828 were also carried out to allow inclusion of partial sequences from marine Thioploca spp., Thiomargarita namibiensis, and Beggiatoa spp. from Tokyo Bay (sites A and C). Parsimony and minimum evolutionary phylogenetic trees were inferred with the program PAUP* 4.0 (36). The minimum evolutionary trees were obtained by using the Kimura two-parameter model, and all trees were checked with 1,000 bootstrap replicates.
Statistics.
Unless otherwise stated, mean values ± 1 standard error (standard deviation of the mean) are presented.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the White Point VAF bacterium and the Carmel Canyon Beggiatoa sp. have been deposited in the GenBank database under accession numbers AY496953 and AY580013, respectively.
RESULTS
Sample collection and microscopic observations.
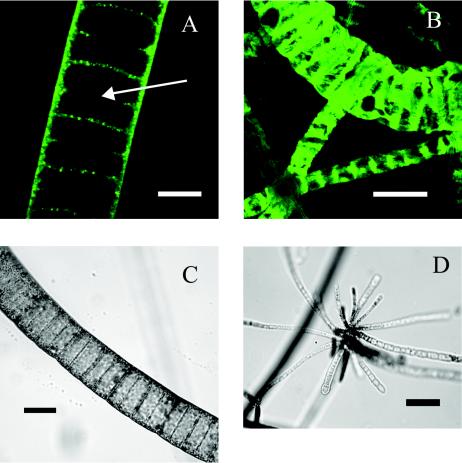
The target bacteria were collected in 2001 and 2002 from shallow (5- to 7-m-deep) hydrothermal vents in White Point, Calif. There, they formed dense, macroscopically visible mats, which covered biotic and abiotic substrates immediately adjacent to areas of venting. Microscopic examination of collected samples showed nonmotile, attached filaments which were fairly uniform in diameter along their length but whose diameter increased by 1 to 2 μm towards the base. Most filaments had sulfur inclusions, and in some, the inclusions graduated from nonexistent at the base to quite dense at the tip. In suspension, the filaments appeared stiff and buoyant and would not pellet easily via centrifugation. The narrowest filaments collected measured less than 2 μm in diameter, were nonvacuolate as judged by FITC staining (see below), and contained sulfur inclusions. Filaments that appeared, under phase-contrast light microscopy, to contain vacuoles ranged from 4 to 112 μm in diameter (Fig. 1 and 2). Rosettes and gonidia were observed in filaments measuring from 2 to 10 μm in diameter (Fig. 1) but not in the larger ones.
FIG 1.
(A) White Point filament, stained with FITC, showing large internal vacuoles. Bar, 20 μm. (B) White Point filaments hybridized with White Point filament-specific probe WPF464 in 20% formamide. Bar, 50 μm. (C) Light micrograph of large White Point filament. Bar, 50 μm. (D) Light micrograph of White Point filaments forming a rosette. Bar, 40 μm.
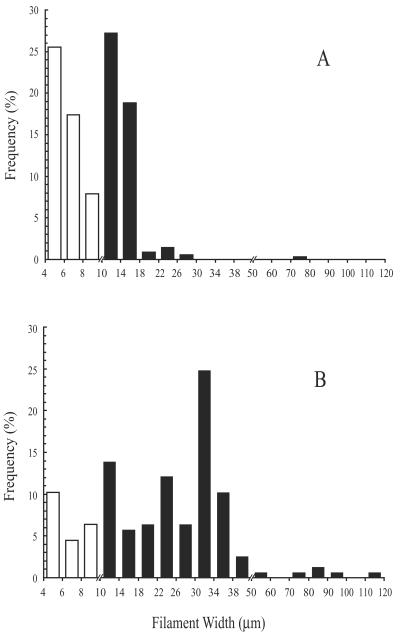
FIG 2.
Relationship of frequency to diameter for VAF collected from rocks at White Point vents (water depth, ca. 6 m). Filaments less than 10 μm in diameter (white bars) did not hybridize with any fluorescent probe, including WPF464, while all wider, attached filaments (black bars) positively hybridized with WPF464. Suspensions were collected in July 2001 (A) and February 2002 (B).
Internal anion measurements.
Lysates of VAF recovered from White Point vents in 2001 and 2002 showed no detectable internal nitrate (Table 2) when measured by ion chromatography. Simultaneous measurements of cell lysates of large vacuolate Beggiatoa bacteria from the Carmel Canyon cold seep indicated an intracellular nitrate concentration of 16.6 ± 4 mM (n = 2) (Tables 2 and 3). For the White Point VAF, the internal levels of phosphate averaged 0.8 ± 0.3 mM (n = 2) over the entire cell volume, including the vacuole (Table 2). For the Carmel Canyon Beggiatoa sp. and an E. coli culture, internal phosphate concentrations, again averaged over the entire cell volume, were 0.18 ± 0.04 (n = 2) and 18.2 ± 0.09 (n = 3) mM, respectively.
TABLE 2.
Properties of White Point VAF and a control, vacuolate Carmel Canyon Beggiatoa sp.
| Sample | Date collected | Protein/biovolume (mg of protein/cm3) | Internal nitrate concn (mM)a,b | Internal phosphate concn (mM)b |
| VAF | 11 July 2001 | 6.8 | Not detected | 1.1 |
| VAF | 28 February 2002 | 8.4 | Not detected | 0.5 |
| Carmel Canyon Beggiatoa sp. | 24 September 2002 | 8.9 ± 1.1 | 16.6 ± 4 | 0.2 ± 0.04 |
a Not detected, 5 μM concentration for column and conductivity detector is implied. Necessary dilution of original pellet for (i) disruption of cells and (ii) column removal of interfering Cl− implies true detection threshold of approximately 800 μM internal NO3.
b Calculated by assuming anion distributed over cytoplasm and vacuole.
TABLE 3.
Evidence that uncultured, large, marine sulfur bacteria (Beggiatoa spp., Thioploca spp., Thiomargarita namibiensis, and attached filaments from White Point hydrothermal vents) are vacuolate and tabulation of internal nitrate concentrations
| Site and bacteriumb | Filament width (μm) | Vacuole present? | Presence/absence of central vacuole supported by: | Intracellular nitrate concn | Reference(s) or source | ||
| EMc | Protein/biovolume (mg of protein/cm3) | ||||||
| Narrow-filament controlaBeggiatoa sp. strains | |||||||
| MS-81-6 | 4 | No | Yes | 121 ± 17i | <0.3 μM | 20 | |
| MS-81-1c | 2 | No | Yes | ND | ND | 20 | |
| White Point, Calif., HTV; attached filaments | 4-112d | Yes | NDe | 7.6 ± 0.8 | Not detectedf (n = 2) | This study | |
| Carmel Canyon; Beggiatoa sp. | 20-76 | Yes | ND | 8.9 ± 1.1 | 16.6 ± 4 mM (n = 2) | This study | |
| Monterey Canyon; Beggiatoa sp. | 65-85 | Yes | ND | 24 | 160 ± 20 mM (n = 5) | 20 | |
| Guaymas Basin HTV; Beggiatoa sp. | 88-140 | Yes | Yes | 9.5 | 130 ± 10 mM (n = 3) | 27 | |
| Namibia; Thiomargarita namibiensis | 100-300 | Yes | ND | 4.5 | 100-800 mM | 33 | |
| OMZ, Peru and Chile | |||||||
| Thioploca araucae | 28-42 | Yes | Yes | ND | 150-500 mMg | 3, 34 | |
| Thioploca chileae | 12-22 | Yes | Yes | ND | |||
| Bay of Concepcion, Chile; Beggiatoa sp. | 35-40 | Yes | ND | ND | 15-116 mM | 38 | |
| Wadden Sea (Dangast/Jadebay); Beggiatoa sp. | 9-11 | Yes | ND | ND | 288 ± 80 mM | 21 | |
| Limfjorden, Denmark; Beggiatoa spp. | 5-40 | Yes | ND | ND | 156 ± 71h | 21 | |
| Tokyo Bay, Japan; Beggiatoa spp. | 9 | Yes | ND | ND | 105 ± 36 | 32 | |
a Narrow, nonvacuolate, pure-culture marine Beggiatoa strains are included for comparison.
b HTV, hydrothermal vents; OMZ, oxygen-minimum zone.
c EM, electron microscopy.
d White Point filament-specific fluorescent probe WPF445 hybridizes with filaments of ≥10 μm in diameter.
e ND, not determined.
f Not detected, below the detection limit of 5 μM for column and conductivity detector. Necessary dilution of original pellet for (i) disruption of cells and (ii) column removal of interfering Cl− implies true detection threshold of approximately 800 μM internal NO3.
g Data reported collectively for both Thioploca species.
h Intracellular nitrate concentration of filaments 9 to 12 μm in diameter.
i n = 12.
Biovolume, width, and protein determinations.
Vacuolate filaments ranged from 4 to 112 μm in diameter (Fig. 2). Unicells were not counted but were minimized by washing. Even though nonvacuolate filaments (those less than 4 μm in diameter) represented 54 to 66% of the total filaments counted in suspensions from White Point, they represented less than 0.1% of the total biovolume of the attached filaments for both years. Vacuolate filaments represented 99.98% of the total biovolume for 2001 and 99.9% of the total biovolume for 2002. The ratios of protein to biovolume for 2001 White Point VAF, 2002 White Point VAF, and 2002 Carmel Canyon vacuolate Beggiatoa sp. bacteria were 6.8, 8.4, and 8.9 ± 1.1 (n = 2) mg cm−3, respectively (Table 2). As a point of reference, the corresponding ratio for a pure culture of the nonvacuolate, control Beggiatoa sp. was 121 ± 17 mg cm−3 (Table 3). Protein/biovolume ratio data were used, relative to the value for the nonvacuolate Beggiatoa sp. control, to correct for the large contribution of vacuoles to biovolume. Even after this correction, White Point VAF contributed 99.7% (2001) and 98.6% (2002) of the total cell cytoplasm measured for all attached filaments. The Carmel Canyon Beggiatoa sp. bacteria represented 99.9% of the total biovolume of washed suspensions from Carmel Canyon, while the rest consisted of narrow (<2-μm-diameter), nonvacuolate filaments.
FITC.
Confocal microscopy images of White Point VAF stained with FITC (an amine-reactive dye that binds cellular protein) clearly demonstrated that protein is associated with cross walls and nascent cross walls, and this stained area almost certainly reflects the thin cytoplasmic layer surrounding the large vacuole (Fig. 1). Images of FITC-stained filaments less than 4 μm in diameter showed completely stained cytoplasm with no hint of the central clear area found in each cell of larger filaments (data not shown).
Cloning and sequencing of the 16S rRNA gene.
Conventional and cetyltrimethylammonium bromide genomic DNA extraction protocols (1a) failed to yield DNA that could be successfully amplified by PCR. A guanidine thiocyanate protocol (see Materials and Methods) was successful in extracting genomic DNA from White Point and Carmel Canyon cell pellets.
Mixed template DNA from the White Point pellet was first amplified with universal eubacterial primers 341f and 534r (Table 1), and a fragment of approximately 200 bp was successfully amplified. This fragment was cloned, and, following sequencing of randomly selected clones, sequences were screened by BLAST searches against those in the nucleotide database at the National Center for Biotechnology Information's website (http://www.ncbi.nlm.nih.gov/) and by the Ribosomal Database Project II online analysis program Sequence Match (http://rdp.cme.msu.edu/index.jsp). The sequence most closely aligning with sequences of known vacuolate sulfur-oxidizing bacteria (92% identity with Thiomargarita namibiensis) was chosen for further analysis, including construction of primers WPF445f and WPF464r (Table 1). These primers were used in conjunction with universal eubacterial primers to amplify virtually an entire 16S rRNA gene from White Point pellet mixed genomic DNA. Ten clones (amplified with 8fpl-WPF464r) (Table 1) and six clones (amplified with WPF445f-1492rpl) (Table 1) were fully or partially sequenced, respectively, with identical results. The other 200-bp clones from the initial amplification were most similar to marine members of the proteobacterial and bacteroides-flavobacterium groups. These were consistent with free-living and epibiotic contaminants and not analyzed further.
The 16S rRNA gene of the Carmel Canyon Beggiatoa sp. was successfully amplified from mixed template DNA with general eubacterial primers and primers (VSO656f and VSO673r) derived from a probe (VSO673) designed in this study to specifically target certain known vacuolate sulfur oxidizers (Monterey Canyon and Bay of Concepcion Beggiatoa spp., vacuolate Thioploca spp., and Thiomargarita namibiensis). Amplified fragments were cloned and randomly chosen for sequencing. All sequences were identical.
FISH.
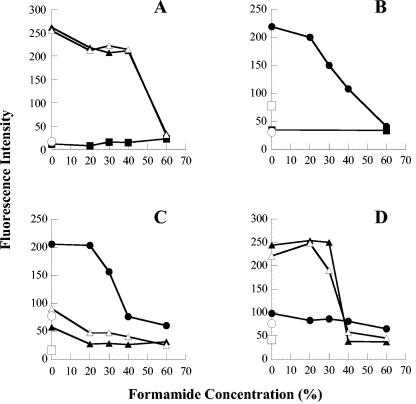
Based on White Point filament sequence data, probe WPF464, which differed in this region of the 16S rRNA gene by 4 bp from that of any other known vacuolate sulfur oxidizer, was constructed. The online analysis program Probe Match, from the Ribosomal Database Project II, was used to check for self-complementarities and for similarity to other sequences, allowing for two mismatches, with negative results. Hybridizations were carried out with the GAM42a, BET42a, Eub338, VSO673, G123T, G123T-C, and WPF464 probes (Table 1) over a wide range of stringent conditions (0 to 60% formamide). All White Point filaments with diameters of 10 μm or more strongly bound the general eubacterial probe (Eub338) (data not shown), the gamma proteobacterial probe (GAM42a), and the probe based on the sequence cloned from these same White Point filaments (WPF464). Fluorescence intensity remained strong under conditions of up to 30 or 40% formamide (Fig. 3A and D) and declined thereafter. Probes BET42a, VSO673, G123T, and G123T-C, which did not match the rRNA sequence derived from White Point filaments, did not hybridize significantly beyond the background autofluorescence (Fig. 3A and C). Narrower White Point VAF (4 to 10 μm in diameter) did not bind any probe very strongly, including Eub338 (Fig. 3A) (unpublished data).
FIG 3.
Relationship of fluorescence intensity (pixel strength maximum, 255) of bound probes to hybridization stringency. (A) White Point attached filaments hybridized with gamma and beta proteobacterial probes. Results with the GAM42a probe with filaments with diameters of <10 μm (▪), 10 to 29 μm (▴), and 30 to 70 μm (▵) are shown. The average signal for filaments of all widths with the BET42a probe (○) is also shown. Similarly, low fluorescence signals were recorded at 488 and 586 nm for no-probe controls (background fluorescence) with filaments of all widths. (B) Carmel Canyon Beggiatoa sp. hybridized with gamma and beta proteobacterial probes, including the GAM42a probe (•), the BET42a probe (▪), the no-probe control excited at 586 nm (○), and the no-probe control excited at 488 nm (□). (C) White Point attached filaments and Carmel Canyon Beggiatoa sp. hybridized with the VSO673 probe specific for large, nitrate-accumulating, vacuolate, sulfur-oxidizing bacteria. Results for White Point attached filaments with diameters of 10 to 29 μm (▴) and 30 to 70 μm (▵) are shown, as well as Carmel Canyon Beggiatoa sp. (•), the Carmel Canyon Beggiatoa sp. no-probe control (○), and the White Point attached filament no-probe control (10 to 70 μm in diameter) (□). (D) Results for White Point attached filaments and Carmel Canyon Beggiatoa sp. hybridized with the WPF464 probe specific for White Point attached filaments are shown. Symbols are the same as for panel C.
Carmel Canyon Beggiatoa sp. filaments strongly bound the Eub338 probe (data not shown), as well as the GAM42a probe and the vacuolate sulfur-oxidizing probe (VSO673) in the presence of 0 and 20% formamide, and the fluorescence intensity remained elevated in the presence of up to 40% formamide (Fig. 3B and C). By contrast, these nitrate-accumulating Beggiatoa filaments showed no significant hybridization with probes WPF464, BET42a, G123T, or G123T-C (Fig. 3B and D).
Thiothrix fructosivorans bacteria strongly bound probes Eub338, GAM42a, and G123T (data not shown) in the presence of formamide concentrations up to 30 or 40%. No significant hybridization beyond background autofluorescence was observed when the bacteria were hybridized with probes WPF464, BET42a, VSO673, or G123T-C.
Phylogenetic analysis.
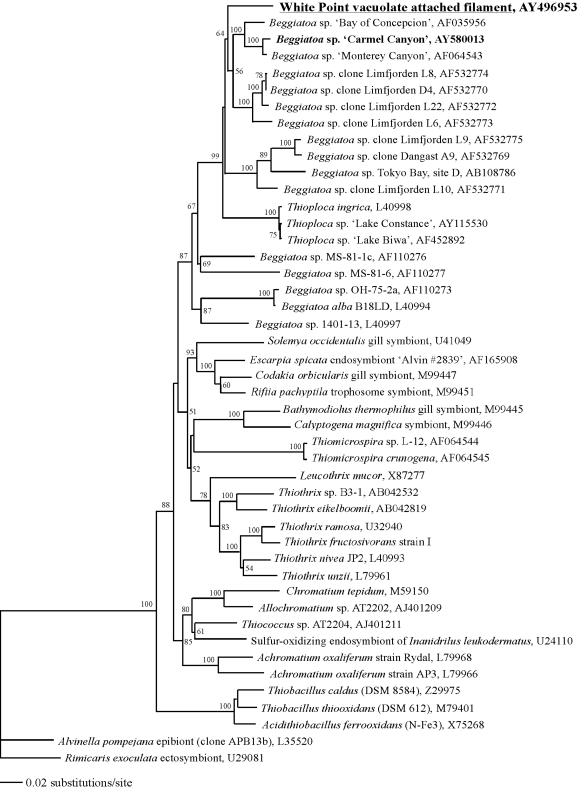
Analyses unambiguously placed the White Point VAF (diameters, ≥10 μm) within the clade of vacuolate bacteria in the gamma subdivision of the proteobacteria. The small-mask minimum evolutionary tree inferred with the Kimura two-parameter model is shown in Fig. 4. In this tree, as well as in trees generated by parsimony analysis (data not shown), the White Point VAF sequence forms a monophyletic group with Thioploca spp. and the vacuolate Beggiatoa spp. The bootstrap values for this branch are 99% (Fig. 4) and 92% for minimum-evolution and parsimony analyses, respectively. Narrow-filament strains of freshwater Thioploca spp. make up the most deeply branching group within the minimum-evolution analysis (Fig. 4). Using parsimony analysis, the White Point sequence forms the deepest branch within the clade of vacuolate bacteria, but the organisms comprising the clade were the same under either method of tree construction.
FIG 4.
Minimum evolutionary tree of the White Point VAF and diverse sulfur-oxidizing gamma proteobacteria, based on 16S rRNA gene sequences. Sequences of epsilon proteobacteria are included as an outgroup. The tree was constructed with an alignment of positions 126 to 1376 (E. coli numbering). Numbers on the nodes represent percentage bootstrap values greater than 50% (1,000 replicates). Accession numbers are shown.
Certain conspicuous vacuolate sulfur oxidizers analyzed by others, e.g., Thioploca chileae, Thioploca araucae, Thiomargarita namibiensis, and Beggiatoa spp. from Tokyo Bay (sites A and C), are represented by incomplete published sequences. This fact makes a robust phylogenetic analysis that includes all vacuolate sulfur bacteria impossible. Nonetheless, large-mask analyses (covering positions 358 to 828) using either minimum evolution or parsimony (data not shown) consistently placed these additional sequences, plus the sequence for White Point VAF, within the same clade described above. Only minor differences in internal branching patterns depended on the particular analysis employed (data not shown).
DISCUSSION
Others have previously described mats of attached filamentous sulfur bacteria for the White Point vents, where their density is high enough to serve as an important food source for grazers (8, 35). Based on the present study, as well as the findings of Jacq et al. (8), there appear to be at least three subpopulations of attached filaments at White Point: (i) narrow nonvacuolate filaments less than 4 μm in diameter; (ii) VAF 4 to 10 μm in diameter; and (iii) VAF greater than 10 μm in diameter (3). All filaments greater than 4 μm in diameter contained sulfur globules and appeared vacuolate when observed by light and fluorescence microscopies. Contrary to results of previous observations (8), rosettes and gonidia were observed in the narrower VAF (4 to 10 μm in diameter) but not in the wider ones (10 to 112 μm). Jacq et al. (8) examined the morphologies of narrow (6- to 9-μm-diameter) and wide (50- to 75-μm-diameter) White Point filaments by light and transmission electron microscopies and reported that significant differences in cell wall architecture occurred between the two observed width classes.
Based on FISH analysis with probes specific for gamma proteobacteria, all VAF greater than 10 μm in diameter appear to be gamma proteobacteria. VAF 4 to 10 μm in diameter did not bind any fluorescent probe very well, including the general eubacterial probe Eub338. It is possible that poor hybridization by this group of filaments reflects (i) low metabolic activity at the time of sampling and, hence, a low rRNA content; (ii) higher-ordered structures in the ribosomes that prevented probe hybridization (2); or (iii) unique cell wall layers that minimize permeation by probes. Although all VAF with ≥10-μm diameters bound the White Point filament-specific probe WPF464 with almost equal intensities, more-detailed cloning and sequencing might reveal additional strain differences among the extremes of filament diameters (10 to 112 μm). These probe-based findings support the view of Jacq et al. (8) that VAF with diameters of less than 10 μm are morphologically distinct from wider VAF. It is puzzling that these earlier researchers found no filaments in the 10- to 50-μm-diameter range, while we observed most of the biomass to be contributed by these VAF in collections from two separate years (Fig. 2).
Phylogenetic analysis using 16S rRNA gene sequences confirmed that VAF with diameters of ≥10 μm are part of the clade of vacuolate bacteria, which includes all other known vacuolate sulfur bacteria, e.g., Thioploca, Thiomargarita, and Beggiatoa spp. The VAF of the present study represent the first of any attached member of this clade to be described. All other representatives are unattached and capable of significant movements via gliding motility (Beggiatoa and Thioploca spp.) or presumed hydrodynamic transport-triggered resuspension of sediment induced by methane outgassing (Thiomargarita spp.) (3, 9, 16, 33). The clade of vacuolate bacteria (Fig. 4) contains only vacuolate marine strains, with the exception of the deeply branching cluster that includes three narrow (2- to 5.6-μm-diameter) freshwater Thioploca strains (13). Among these, Thioploca ingrica strains clearly contain vacuoles that comprise roughly 40% of their biovolume (1a, 17), while micrographs for the other strains show no evidence of vacuoles (13). The clade of vacuolate bacteria is part of the larger Beggiatoa cluster (87% bootstrap value) (Fig. 4) that also includes all sequences from narrow (1.5- to 5.0-μm-diameter), nonvacuolate pure cultures of Beggiatoa spp. from marine (strains MS-81-1c and MS-81-6) and freshwater (strains OH-75-2a, B18LD, and 1401-13) environments (25, 30). The large-vacuole Thiomargarita namibiensis (100 to 750 μm in diameter), Beggiatoa spp. from Tokyo Bay (sites A and C; 10 μm in diameter), and vacuolate Thioploca spp. (12 to 43 μm in diameter) are not included in Fig. 4 because only partial sequences were available (12, 16, 33). However, as reported earlier (see Results), large-mask-analysis trees placed these vacuolate, nonattached sulfur bacteria closest to the cluster composed of Bay of Concepcion, Monterey Canyon, and Carmel Canyon Beggiatoa spp., which is consistent with previous findings (1a, 12, 13, 21, 38). In summary, large- and small-mask analyses all support inclusion of White Point VAF bacteria in the clade of vacuolate bacteria of the gamma proteobacteria, which includes all vacuolate bacterial representatives of the genus Beggiatoa and all known Thioploca and Thiomargarita spp. To date, no member of this clade is available in a pure culture.
Although the White Point VAF bacteria show a strong morphological resemblance to cultured Thiothrix strains and were previously referred to as Thiothrix-like (8, 35), the significant evolutionary distance between the true Thiothrix cluster (bootstrap value, 83%) (Fig. 4) and the clade of vacuolate bacteria, which includes White Point filaments, argues against the possibility that a common ancestor exists as the cause of this common morphology. The symmetrical negative results of FISH studies (hybridizing VAF with a Thiothrix probe and vice versa) also support this view. In short, all available evidence is consistent with the view that vacuoles arose once in the gamma proteobacteria and that subsequent diversification of this ancestor, including convergent evolution of a Thiothrix-like morphotype, gave rise to the remarkable diversity of morphologies, sizes, and lifestyles currently found in the clade of vacuolate bacteria (Fig. 4).
All other members of the clade of vacuolate bacteria examined to date accumulate nitrate (presumed to be in the vacuoles) at concentrations of roughly 400 to 20,000 times that of the surrounding seawater. All known data are summarized (Table 3) to emphasize the extensive literature supporting this point. In the present study, repeated measurements of cell lysates via ion chromatography indicated that White Point VAF do not contain nitrate at levels above those in ambient seawater, while the positive contemporaneous results for Carmel Canyon Beggiatoa bacteria emphasize that technical issues cannot be the cause of the negative findings (Tables 2 and 3). Additionally, the essential cellular anion phosphate was present at levels similar to that for E. coli cytoplasm. If one makes the assumption that all detected intracellular phosphate was confined to the cytoplasm, the corresponding values are 18.2 ± 0.09 and 33.6 ± 10.3 mM for E. coli and the White Point VAF, respectively. A somewhat lower value of 5.7 ± 0.5 mM was calculated for the cytoplasm of the Carmel Canyon Beggiatoa sp., and all three concentrations are consistent with the dynamic range of bacteria (2a). White Point VAF cytoplasm contributed in excess of 98% of the total cell cytoplasm measured from cell pellets (see Results), and this result is strong evidence against the possibility that mass lysis accounted for our inability to detect intracellular nitrate in VAF. Thus, the VAF from White Point are the first example of a vacuolate sulfur bacterium that does not accumulate nitrate.
A vacuole is defined as “a fluid-filled space within the cytoplasm, bounded by a membrane” (1). For other genera in the clade of vacuolate bacteria, electron micrographs have confirmed the existence of a bounding membrane (9, 18, 33), and very high internal nitrate concentrations have been assumed to reflect vacuolar storage (3, 20, 21, 28, 33). The lack of similar storage in the White Point VAF highlights the importance of verifying the existence of vacuoles. Electron micrographs show virtually identical images of the thin cytoplasmic region that surrounds the large central vacuole in a White Point VAF (8) or a nitrate-accumulating Beggiatoa sp. from deep-sea vents (26). In both, the cytoplasm is dissected into many distinctive “balls” or electron-dense bodies that make it difficult to trace any continuous vacuolar or cell membrane. A protein/biovolume ratio that is only 6% of that determined for a nonvacuolate control but typical of values for sulfur bacteria confirmed to possess vacuoles (Table 3) provides strong supporting evidence. Additionally, since roughly half of the cellular protein is soluble, the protein-staining pattern observed (Fig. 1A) could arise only if an internal membrane limited the cytoplasm, thereby defining a vacuole. Likewise, as the above calculations indicate, cytoplasmic levels of phosphate in the VAF are an order of magnitude below typical levels in bacteria unless a vacuolar membrane is present and phosphate is confined to the cytoplasm.
At least two possible roles are proposed for vacuoles in the White Point filaments. All other vacuolate sulfur bacteria examined to date store nitrate as an electron acceptor, which allows them to remain metabolically active in an anoxic environment independent of an external oxidant for hours, days, or even months (3, 20, 33). One possibility, suggested here, is that attached White Point filaments represent an adaptation to a very brief temporal separation of the electron donor and acceptor. The habitat of the White Point VAF may expose filaments briefly, via wave surge, to alternating sulfidic (and possibly anoxic) vent water and fully oxygenated water that perhaps lacks soluble sulfide. Internal sulfur globules can easily supply reductant in the latter habitat, and it is possible that vacuolar oxygen, perhaps initially approaching equilibrium with the oxygenated portion of the bacterium's environment, may be sufficient to sustain metabolic activity during brief incursions into anoxic, sulfidic seawater. The other possible role, which is not mutually exclusive with transient O2 storage, would be to contribute to the apparent buoyancy of these filaments relative to other vacuolate filaments, such as those of Beggiatoa spp. Vacuolar fluid might be less dense due to a lower ion concentration or an abundance of low-density organic compounds. The physiological role(s) of these vacuoles is an obvious focus for ongoing studies.
Acknowledgments
This research was supported by funds from the U.S. National Science Foundation (grant NSF-9983119) and the Department of Energy (grant DE-FG03-OOER15077).
We are grateful to Patrick Whaling, the crew of the Pt. Lobos, and the pilots of the remotely operated vehicle Ventana for their assistance in sample collection. We also thank Kari Hagen and two anonymous reviewers for helpful comments.
REFERENCES
- Abercrombie, M., C. J. Hickman, and M. L. Johnson. 1971. A dictionary of biology, 5th ed., p. 272. Penguin Books Ltd., Harmondsworth, Middlesex, England.
- Ahmad, A., J. P. Barry, and D. C. Nelson. 1999. Phylogenetic affinity of a wide, vacuolate, nitrate-accumulating Beggiatoa sp. from Monterey Canyon, California, with Thioploca spp. Appl. Environ. Microbiol. 65:270-277. [DOI] [PMC free article] [PubMed]
- Amann, R. I. 1995. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, section 3.3.6, p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Bond, D. R., and J. B. Russell. 1998. Relationship between intracellular phosphate, proton motive force, and rate of nongrowth energy dissipation (energy spilling) in Streptococcus bovis JB1. Appl. Environ. Microbiol. 64:976-981. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed]
- Fossing, H., V. A. Gallardo, B. B. Jørgensen, M. Hüttel, L. P. Nielsen, H. Schultz, D. E. Canfield, S. Forster, R. N. Glud, J. K. Gundersen, J. Küver, N. B. Ramsing, A. Teske, B. Thamdrup, and O. Ulloa. 1995. Concentration and transport of nitrate by the mat-forming sulphur bacterium Thioploca. Nature 374:713-715.
- Graco, M., L. Farias, V. Molina, D. Guttiérrez, and L. P. Nielsen. 2001. Massive developments of microbial mats following phytoplankton blooms in a naturally eutrophic bay: implications for nitrogen cycling. Limnol. Oceanogr. 46:821-832.
- Guezennec, J., O. Ortega-Morales, G. Raguenes, and G. Geesey. 1998. Bacterial colonization of artificial substrate in the vicinity of deep-sea hydrothermal vents. FEMS Microbiol. Ecol. 26:89-99.
- Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98.
- Howarth, R., R. F. Unz, E. M. Seviour, R. J. Seviour, L. L. Blackall, R. W. Pickup, J. G. Jones, J. Yaguchi, and I. M. Head. 1999. Phylogenetic relationships of filamentous sulfur bacteria (Thiothrix spp. and Eikelboom type 021N bacteria) isolated from wastewater-treatment plants and description of Thiothrix eikelboomii sp. nov., Thiothrix unzii sp. nov., Thiothrix fructosivorans sp. nov. and Thiothrix defluvii sp. nov. Int. J. Syst. Bacteriol. 49:1817-1827. [DOI] [PubMed]
- Jacq, E., D. Prieur, P. Nichols, D. C. White, T. Porter, and G. G. Geesey. 1989. Microscopic examination and fatty acid characterization of filamentous bacteria colonizing substrata around subtidal hydrothermal vents. Arch. Microbiol. 152:64-71.
- Jannasch, H. W., D. C. Nelson, and C. O. Wirsen. 1989. Massive natural occurrence of unusually large bacteria (Beggiatoa sp.) at a hydrothermal deep-sea vent site. Nature 342:834-836.
- Jørgensen, B. B., and V. A. Gallardo. 1999. Thioploca spp.: filamentous sulfur bacteria with nitrate vacuoles. FEMS Microbiol. Ecol. 28:301-313.
- Kanagawa, T., Y. Kamagata, S. Aruga, T. Kohno, M. Horn, and M. Wagner. 2000. Phylogenetic analysis of and oligonucleotide probe development for Eikelboom Type 021N filamentous bacteria isolated from bulking activated sludge. Appl. Environ. Microbiol. 66:5043-5052. [DOI] [PMC free article] [PubMed]
- Kojima, H., and M. Fukui. 2003. Phylogenetic analysis of Beggiatoa spp. from organic rich sediment of Tokyo Bay, Japan. Water Res. 37:3216-3223. [DOI] [PubMed]
- Kojima, H., A. Teske, and M. Fukui. 2003. Morphological and phylogenetic characterizations of freshwater Thioploca species from Lake Biwa, Japan, and Lake Constance, Germany. Appl. Environ. Microbiol. 69:390-398. [DOI] [PMC free article] [PubMed]
- Larkin, J. M. 1989. Genus II. Thiothrix Winogradsky 1888, 39AL, p. 2098-2101. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holy (ed.), Bergey's manual of systematic bacteriology, vol. 3. Williams & Wilkins, Baltimore, Md.
- Maddison, D. R., and W. P. Maddison. 2002. MacClade 4: analysis of phylogeny and character evolution, version 4.05. Sinauer Associates, Sunderland, Mass.
- Maier, S., and V. A. Gallardo. 1984. Thioploca araucae sp. nov. and Thioploca chileae sp. nov. Int. J. Syst. Bacteriol. 34:414-418.
- Maier, S., and R. G. E. Murray. 1964. The fine structure of Thioploca ingrica and a comparison with Beggiatoa. Can. J. Microbiol. 11:645-655. [DOI] [PubMed]
- Maier, S., H. Völker, M. Beese, and V. A. Gallardo. 1990. The fine structure of Thioploca araucae and Thioploca chileae. Can. J. Microbiol. 36:438-448.
- Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600.
- McHatton, S. C., J. P. Barry, H. W. Jannasch, and D. C. Nelson. 1996. High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl. Environ. Microbiol. 62:954-958. [DOI] [PMC free article] [PubMed]
- Muβmann, M., H. N. Schulz, B. Strotmann, T. Kjζr, L. P. Nielsen, R. A. Rosselló-Mora, R. I. Amann, and B. B. Jørgensen. 2003. Phylogeny and distribution of nitrate-storing Beggiatoa spp. in coastal marine sediments. Environ. Microbiol. 5:523-533. [DOI] [PubMed]
- Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16s RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed]
- Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed]
- Nelson, D. C., B. B. Jørgensen, and N. P. Revsbach. 1986. Growth pattern and yield of a chemoautotrophic Beggiatoa sp. in oxygen-sulfide microgradients. Appl. Environ. Microbiol. 52:225-233. [DOI] [PMC free article] [PubMed]
- Nelson, D. C., J. B. Waterbury, and H. W. Jannasch. 1982. Nitrogen fixation and nitrate utilization by marine and freshwater Beggiatoa. Arch. Microbiol. 133:172-177.
- Nelson, D. C., C. O. Wirsen, and H. W. Jannasch. 1989. Characterization of large, autotrophic Beggiatoa spp. abundant at hydrothermal vents of the Guaymas Basin. Appl. Environ. Microbiol. 55:2909-2917. [DOI] [PMC free article] [PubMed]
- Nikolaus, R., J. W. Ammerman, and I. R. MacDonald. 2003. Distinct pigmentation and trophic modes in Beggiatoa from hydrocarbon seeps in the Gulf of Mexico. Aquat. Microb. Ecol. 32:85-93.
- Otte, S., J. G. Kuenen, L. P. Nielsen, H. W. Paerl, J. Zopfi, H. N. Schulz, A. Teske, B. Strotmann, V. A. Gallardo, and B. B. Jørgensen. 1999. Nitrogen, carbon, and sulfur metabolism in natural Thioploca samples. Appl. Environ. Microbiol. 65:3148-3157. [DOI] [PMC free article] [PubMed]
- Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156.
- Pringsheim, E. G. 1964. Heterotrophism and species concepts in Beggiatoa. Am. J. Bot. 8:898-913.
- Reinhard, J., C. Basset, J. Holton, M. Binks, P. Youinou, and D. Vaira. 2000. Image analysis method to assess adhesion of Helicobacter pylori to gastric epithelium using confocal laser scanning microscopy. J. Microbiol. Methods 39:179-187. [DOI] [PubMed]
- Sayama, M. 2001. Presence of nitrate-accumulating sulfur bacteria and their influence on nitrogen cycling in a shallow coastal marine sediment. Appl. Environ. Microbiol. 67:3481-3487. [DOI] [PMC free article] [PubMed]
- Schulz, H. N., T. Brinkhoff, T. G. Ferdelman, M. Hernández Mariné, A. Teske, and B. B. Jørgensen. 1999. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284:493-495. [DOI] [PubMed]
- Schulz, H. N., B. B. Jørgensen, H. A. Fossing, and N. B. Ramsing. 1996. Community structure of filamentous, sheath-building sulfur bacteria, Thioploca spp., off the coast of Chile. Appl. Environ. Microbiol. 62:1855-1862. [DOI] [PMC free article] [PubMed]
- Stein, J. 1984. Subtidal gastropods consume sulfur-oxidizing bacteria: evidence from coastal hydrothermal vents. Science 223:696-698. [DOI] [PubMed]
- Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- Teske, A., N. B. Ramsing, J. Küver, and H. Fossing. 1995. Phylogeny of Thioploca and related filamentous sulfide-oxidizing bacteria. Syst. Appl. Microbiol. 18:517-526.
- Teske, A., M. L. Sogin, L. P. Nielsen, and H. W. Jannasch. 1999. Phylogenetic relationships of a large marine Beggiatoa. Syst. Appl. Microbiol. 22:39-44. [DOI] [PubMed]
- Wagner, M., R. Amann, H. Lemmer, and K.-H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed]
- Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed]
- Williams, U., and R. F. Unz. 1985. Filamentous sulfur bacteria of activated sludge: characterization of Thiothrix, Beggiatoa, and Eikelboom type 021N strains. Appl. Environ. Microbiol. 49:887-898. [DOI] [PMC free article] [PubMed]
- Zopfi, J., T. Kjæ´r, L. P. Nielsen, and B. B. Jørgensen. 2001. Ecology of Thioploca spp.: nitrate and sulfur storage in relation to chemical microgradients and influence of Thioploca spp. on the sedimentary nitrogen cycle. Appl. Environ. Microbiol. 67:5530-5537. [DOI] [PMC free article] [PubMed]