Abstract
Few studies have been conducted on antimicrobial resistance in lactobacilli, presumably because of their nonpathogenic nature as anaerobic commensals. We assessed resistance in 43 type strains and isolates representing 14 species by using agar disk diffusion and MIC analysis in MRS medium. Most noteworthy were two general phenotypes displayed by nearly every strain tested: (i) they were more susceptible (up to 256-fold in some cases) to the deconjugated bile acid cholic acid than to the conjugate taurocholic or taurodeoxycholic acid, and (ii) they became susceptible to aminoglycosides when assayed on agar medium containing 0.5% fractionated bovine bile (ox gall). Two-dimensional MIC analyses of one representative strain, Lactobacillus plantarum WCFS1, at increasing concentrations of ox gall (0 to 30.3 mg/ml) displayed corresponding decreases in resistance to all of the aminoglycosides tested and ethidium bromide. This effect was clinically relevant, with the gentamicin MIC decreasing from >1,000 to 4 μg/ml in just 3.8 mg of ox gall per ml. In uptake studies at pH 6.5, [G-3H]gentamicin accumulation increased over control levels when cells of this strain were exposed to bile acids or reserpine but not when they were exposed to carbonyl cyanide m-chlorophenylhydrazone. The effect was dramatic, particularly with cholic acid, increasing up to 18-fold, whereas only modest increases, 3- and 5-fold, could be achieved with taurocholic acid and ox gall, respectively. Since L. plantarum, particularly strain WCFS1, is known to encode bile salt hydrolase (deconjugation) activity, our data indicate that mainly cholic acid, but not taurocholic acid, effectively permeabilizes the membrane to aminoglycosides. However, at pHs approaching neutral conditions in the intestinal lumen, aminoglycoside resistance due to membrane impermeability may be complemented by a potential efflux mechanism.
Lactobacilli play an important role in promoting gastrointestinal (GI) and vaginal health. Species of this genus are the predominate microbial constituents in the upper, nonsecreting, gastric epithelium in mammalian model systems. They also form subdominant populations in the lower GI tract regions of the cecum and colon (41, 42). In women, Lactobacillus species, specifically those of the Lactobacillus acidophilus complex, constitute the predominant microbes in the vaginal tract (31, 46). These GI and vaginal commensalisms confer significant health benefits upon the host organism through a barrier effect by preventing access to these regions by transient pathogens (9, 23). Indeed, a delicate quantitative balance of vaginal yeasts is maintained and limited by Lactobacillus species, presumably through peroxide production (24, 33). To this end, live microbes are being marketed and actively consumed as probiotic supplements to alleviate or prevent certain GI tract disorders and vaginal yeast and urinary tract infections (23, 33).
Apart from medicinal uses, lactobacilli are commonly used in the food industry although their consumption in foods is also touted to produce probiotic benefits (1, 40). They are used routinely in the making of a variety of consumables, including sourdough and wines, and as starters for yogurts, fermented milks, and cheeses. Their acidic nature and bacteriocin production make them particularly suited for use as preservatives to prevent the spoilage of meats (16, 29, 45). In addition, industrial use of lactobacilli has recently emerged as a means to produce lactic acid by fermentative biocatalysis. Consequently, lactobacilli contribute to a 30 to 50 billion dollar world market in fermentation-based bioprocesses (37).
Recent growing concern over antibiotic resistance has placed much-deserved attention on the investigation of commensal microbiota as a reservoir of resistance mechanisms (4). This concept is not a novel one but has been overlooked for decades because of the concern over controlling resistance in common pathogens. In contrast, lactobacilli have been consumed in foods for centuries and are not generally considered pathogens except under particular conditions in which the health of the host organism is compromised (7). However, the conversion to pathogenicity and multiple drug resistance (MDR) has occurred with another gram-positive commensal, Enterococcus (13, 25, 43). Instances of probiotic Lactobacillus species being isolated from infectious lesions, bacterial endocarditis, bloodstream infections (26), and apical periodontitis (14) suggest that pathogenicity may not be a benign issue with this genus. In fact, the European Union workshop on the safety of probiotics concluded that at least Lactobacillus rhamnosus warranted surveillance because of its repeated association with infections (2).
Unlike pathogenic microbes, lactobacilli are ubiquitous and are thus heavily subjected to any antimicrobial selection ingested by the consumer and used, intentionally or otherwise, by the food and commercial industries. According to conventional wisdom, MDR is favored by improper use of antimicrobials during infection or long-term exposure to low levels of such compounds (4), the latter of which is practiced prophylactically for growth promotion in animal husbandry (48) or for inducing hyperresistant mutations routinely in the laboratory. Nevertheless, this dynamic selection in commensals has not been fully explored, but doing so may lead to a better understanding of resistance evolution and dissemination. In addition, several host-derived factors influence microbial diversity and stability in the GI tract. Such factors include pH, temperature, oxidation-reduction potential, anaerobiosis, stasis, and bile acids (41), the latter of which are present at high concentrations and are substrates for several characterized gram-negative MDR efflux pumps (3, 32, 36). These pumps have been largely unstudied in gram positives compared with the well-characterized multiple pump systems in gram-negative organisms like Escherichia coli and Pseudomonas aeruginosa (32).
We therefore initiated a study of antimicrobial resistance in Lactobacillus species, which is addressed in the first part of this report. Drugs with intracellular targets were chosen and assayed with endogenous GI tract bile acids. These molecules caused aminoglycoside susceptibility, presumably by increasing membrane permeability. We also show in the latter part of this report that a putative efflux process may contribute to aminoglycoside resistance under near-neutral conditions.
MATERIALS AND METHODS
Culture techniques and strains.
The Lactobacillus strains used in this study were obtained from several worldwide culture collections, including the American Type Culture Collection (ATCC); the Belgian Coordinated Collection of Microorganisms (BCCM); the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM); the Japanese Collection of Microorganisms (JCM); Todd Klaenhammer, North Carolina State University (NCK); the Wageningen Centre for Food Sciences (WCFS1); and the principal investigator's personal collection (PI). Some strains were also obtained over a 3- to 4-month period from our in-house Microbiological Surveillance and Diagnostic Group. These strains (National Center for Toxicological Research [NCTR]) were isolated from colony-bred mice and rats and identified to the species level by 16S sequencing with a previously established primer set (47). All strains were preserved for long-term storage at −80°C in 10 to 15% glycerol stocks and revived as needed. The strains were routinely cultured on deMann-Rogosa-Sharpe (MRS) broth or agar medium (Becton Dickinson & Company) at 37 °C under an anaerobic atmosphere of 87% nitrogen, 8% hydrogen, and 5% carbon dioxide in an environmental chamber (Coy Laboratory Products, Inc.). Although these cultures were generally microaerophilic, there was substantial variation in oxygen sensitivity and viability with regard to short-term storage at 4 °C on MRS agar plates. Therefore, freshly revived subcultures were regularly used in experimentation.
Radioactivity, drugs, and miscellaneous chemicals.
[G-3H]gentamicin was obtained in solid form at a specific activity of 50 mCi/mmol (American Radiolabeled Chemicals, Inc.) and dissolved in distilled water to a final concentration of 0.1 mCi/ml for use in uptake assays. Antimicrobial susceptibility testing by the disk diffusion method was performed with Sensi-Disks (Becton, Dickinson & Company). Carbonyl cyanide m-chlorophenylhydrazone (CCCP; Fluka) and reserpine (Sigma-Aldrich Co.) were dissolved in dimethyl sulfoxide (final concentration, 100 mM) and chloroform (10 mg/ml), respectively, as described previously (27). Ciprofloxacin hydrochloride was obtained from Serologicals Proteins Inc. All other chemicals and drugs, including purified bile acids and fractionated bile (ox gall), were obtained from Sigma-Aldrich Co. In the case of ox gall, dried bovine bile powder was obtained that contained a minimum of 50% free and conjugated bile acids.
Antimicrobial drug susceptibility testing.
Lactobacilli were tested for drug susceptibilities by two different methods. In the first, overnight cultures of Lactobacillus species were spread as a lawn onto MRS agar plates (150 by 15 mm). Filter paper disks containing specified concentrations of drugs were placed on the surface of the agar medium with a BBL dispenser (Becton Dickinson & Company) with aseptic precautions. After 24 h of anaerobic incubation at 37°C, zones of growth inhibition around the disks were measured in millimeters and used to score susceptibilities based on breakpoints established by the National Committee for Clinical Laboratory Standards on Mueller-Hinton medium. Some MRS agar plates were supplemented with either ox gall or purified bile acids to determine their effect on resistance. In these cases, a plate containing only MRS was spread in tandem with the same overnight culture for a controlled comparison.
In the second method, MICs of several compounds were measured by the microdilution technique (20) in 96-well microtiter plates (Falcon; Becton Dickinson & Company). Serial twofold dilutions of drugs were prepared in 150 μl of MRS broth and inoculated with 5 μl of an overnight Lactobacillus culture. Visible signs of bacterial growth were recorded after 24 h of anaerobic incubation at 37°C. In some instances, two-dimensional MICs were prepared for each drug in an identical manner except that the drug dilutions were tested repeatedly in media containing successive twofold increases in the bile acid concentration.
Gentamicin uptake assay.
An uptake assay for gentamicin was performed essentially as described previously for bile acid uptake in E. coli except that the procedure was adapted for use with lactobacilli (21). A 24-h culture of Lactobacillus plantarum WCFS1 was harvested by centrifugation at 3,000 × g and washed in an equal volume of fresh, room temperature MRS broth (pH 6.5). The washed cell pellet was resuspended at 10−1 times the original culture volume in MRS broth, divided into 200-μl aliquots, and chilled on ice. Specified concentrations of certain compounds (CCCP, reserpine, and bile acids) were added as desired while the mixture remained on ice. The assay was initiated with a preincubation period of 7.5 min in a 37 °C water bath. After this period of time, 0.5 μCi (5 μl) of [G-3H]gentamicin was added to each aliquot, which was then incubated in the water bath for 3.5 min. The reaction was quenched by adding excess (1 ml) ice-cold 100 mM lithium chloride-100 mM potassium phosphate buffer (pH 5.8). The suspension was vortexed quickly, followed by immediate centrifugation at 3,000 × g in a prechilled, 4 °C Eppendorf centrifuge to pellet the cells. After a second ice-cold buffer wash, the cell pellet was digested in 1 ml of formamide at 65 °C and added to 10 ml of Ultima Gold scintillation cocktail (Perkin-Elmer, Inc.). The cell-associated radiolabel was quantified in a Packard 1600TR TRI-CARB liquid scintillation analyzer.
RESULTS
Antimicrobial resistance studies.
Resistance was scored for 43 isolates of Lactobacillus species by agar disk diffusion on MRS agar medium (Table 1). Although resistance breakpoints for Lactobacillus species have not been reported by the National Committee for Clinical Laboratory Standards, resistance scoring was possible by using established breakpoints on Mueller-Hinton medium since the study was essentially a controlled comparison between species of the same genus. With few exceptions, the strains displayed similar and reproducible resistance phenotypes regardless of the species designation (Table 1). Thus, they were resistant to aminoglycosides (streptomycin, kanamycin, and gentamicin) and quinolones (ciprofloxacin, nalidixic acid, and ofloxacin) but susceptible to chloramphenicol, bacitracin, rifampin, erythromycin, and tetracycline. Resistance to lincomycin and vancomycin was mixed and was not associated with any particular species, which suggests the presence of horizontally acquired resistance elements.
TABLE 1.
Lactobacillus species strains and antibiotic susceptibilitiesa on MRS agar medium
| Lactobacillus species and strain | Origin | Drugb
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STREP | KAN | GENT | CHLOR | BAC | RIF | ERY | TET | CIP | NAL | OFLX | LIN | VAN | ||
| L. acidophilus ATCCc 332 | Unknown | R | R | R | S | S | S | S | S | R | R | R | ND | I |
| L. acidophilus ATCC 4356 | Human | I/Rd | R | R | S | S | I | S | S | R | R | R | R | S |
| L. acidophilus ATCC 4357 | Human | R | R | I/R | S | S | S | R | S | S | R | S | I | S |
| L. acidophilus ATCC 53103 | Human feces | R | R | R | S | S | S | S | S | I | R | I | R | R |
| L. acidophilus ATCC 53544 | Infant | R | R | R | S | S | S | S | S | R | R | R | I | S |
| L. acidophilus PI KS-13 | Human intestine | R | R | R | S | S | S | S | S | R | R | ND | I | S |
| L. bifidus ATCC 11146 | Infant feces | R | R | R | S | S | S | S | S | R | R | R | S | R |
| L. delbrueckii ATCC 11842 | Yogurt | R | R | R | S | S | S | S | S | R | R | R | S | S |
| L. delbrueckii DSM 20080 | Yogurt | S | S | S | S | S | S | S | S | R | R | R | S | S |
| L. fermentum ATCC 11976 | Infant | S/I | I | S | S | S | S | S | S | R | R | ND | S | R |
| L. fermentum ATCC 23271 | Human intestine | R | R | I | S | S | S | S | S/I | R | R | ND | S | R |
| L. gasseri ATCC 19992 | Human feces | R | R | R | S | S | S | S | S | R | R | R | R | I |
| L. gasseri NCK 99 | Unknown | I/R | R | R | S | S | S | S | S | R | R | R | R | I/R |
| L. johnsonii ATCC 33200 | Human blood | R | R | R | S | S | S | I | I | R | R | ND | I | I |
| L. johnsonii PI 100-100 | Rat stomach | R | R | R | S | S | S | S | S | R | R | ND | R | S/I |
| L. maltaromicus BCCM 6903 | Raw milk | R | R | I | S | S/I | I | R | S | S | R | S | S | S/I |
| L. murinus NCTRe | Mouse and rat | R | R | R | S | S | S/I | S | S | S/I | R | S/I | ND | R |
| L. paracasei ATCC 27092 | Human feces | R | R | R | S | S | S | S | S | I | R | ND | S | R |
| L. paracasei JCM 1109 | Human intestine | R | R | R | S | S | S | S | S | R | R | I | I/R | R |
| L. plantarum ATCC 14917 | Pickled cabbage | R | R | I/R | S | I | I | I | S/I | R | R | R | R | R |
| L. plantarum WCFS1 | Human saliva | R | R | I | S | S | I | S | S/I | R | R | R | ND | R |
| L. reuteri ATCC 23272 | Human feces | I | I | I | S | S | S | S | S/I | R | R | R | S | R |
| L. ruminis ATCC 25644 | Human intestine | R | R | R | S | S | S | S | S | S | R | S | S | R |
| L. sake PI Lb5 | Turkey ham | R | R | R | S | I | S | S | S | R | R | R | ND | R |
| L. sake PI Lb6 | Turkey ham | R | R | R | S | S | S | S | S | R | R | R | ND | R |
| L. sake PI Lb15 | Turkey ham | R | R | R | S | S | I | I | R | R | R | R | ND | R |
| L. vaginalis PI 675a | Mouse vagina | R | R | I | S | S | S | S | I | R | R | R | ND | R |
Antibiotic profiles were determined by disc diffusion and scored as resistant (R), intermediate (I), or susceptible (S). ND, susceptibility not determined.
Drug name abbreviations (concentration used [micrograms per milliliter]): STREP, streptomycin (10); KAN, kanamycin (30); GENT, gentamicin (10); CHLOR, chloramphenicol (30); BAC, bacitracin (10); RIF, rifampin (5); ERY, erythromycin (2); TET, tetracycline (30); CIP, ciprofloxacin (5); NAL, nalidixic acid (30); OFLX, ofloxacin (5); LIN, lincomycin (2); VAN, vancomycin (30).
See Materials and Methods for type culture collection abbreviations.
The analysis was repeated with most strains producing identical results; however, in some cases slightly different profiles were obtained. These differences, however, occurred because the zone diameters approached the limits of the scoring range.
Representative profile of 17 isolates. Isolates were obtained from mouse and rat intestinal and nasopharyngeal swabs.
MIC analyses were performed to determine the levels of the observed resistance phenotypes reported in Table 1. The individual MICs obtained were shown to be highly repeatable, fluctuating at most by 2 dilutions (Table 2). Thus, the kanamycin and ciprofloxacin resistance observed by disk diffusion was actually at high levels, ranging from >100 to >2,000 μg/ml for both compounds. Likewise, resistance to erythromycin, tetracycline, and chloramphenicol was low in level and could be generally summarized as <20 μg/ml. Exceptions to these trends existed but were apparent in both disk diffusion and MIC analyses. For example, Lactobacillus delbrueckii DSM 20080 and Lactobacillus fermentum ATCC 11976 were the only strains susceptible to kanamycin by disk diffusion analysis and they also displayed the lowest MICs. Similarly, L. acidophilus ATCC 4357 and Lactobacillus maltaromicus BCCM 6903 were the only strains resistant to erythromycin by disk diffusion, and they also displayed the highest MICs. The results for ciprofloxacin were inconsistent between the two analyses and may be an agar medium artifact, as they conflicted even with data derived from a group of rodent isolates collected from colony-bred mice and rats.
TABLE 2.
MICa analysis of Lactobacillus species strains grown in MRS broth
| Lactobacillus species strain | MICb of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| KAN | ERY | TET | CIP | CHLOR | NOV | CA | TCA | TDCA | |
| L. acidophilus ATCC 332 | 128 | 0.5 | 1 | >1,024 | 16 | 2 | 2,000 | ND | 16,000 |
| L. acidophilus ATCC 4356 | 512 | 0.125 ± (1)c | 2 ± (1) | 256 | 8 ± (1) | 0.5 | 2,000 | 32,000 | 16,000 |
| L. acidophilus ATCC 4357 | 256 | 32 ± (1) | 4 | 1,024 | 4 ± (2) | 0.5 ± (1) | 2,000 ± (1) | 512,000 | >256,000 |
| L. acidophilus ATCC 53103 | >512 | 4 | 2 | >1,024 | 16 | >32 | 2,000 | ND | 256,000 |
| L. acidophilus ATCC 53544 | 256 | 0.125 | 4 | 128 | 8 | >32 | 2,000 | 32,000 | ND |
| L. acidophilus PI KS-13 | 128 ± (2) | 0.125 | 4 | 256 ± (2) | 4 | 0.5 | 1,000 ± (1) | 16,000 | 1,000 |
| L. bifidus ATCC 11146 | 128 | 0.0625 | 2 | >1,024 | 4 | 32 | 2,000 | ND | 8,000 |
| L. crispatus JCM 5810 | 256 ± (1) | <0.03125 ± (2) | 1 ± (2) | 256 ± (2) | 4 ± (2) | 0.5 ± (2) | 1,000 ± (1) | 32,000 | 16,000 |
| L. delbrueckii ATCC 11842 | 256 ± (1) | <0.03125 | 1 | 128 ± (2) | 4 | 0.25 ± (1) | 500 ± (1) | 4,000 | 1,000 |
| L. delbrueckii DSM 20080 | 16 | <0.03125 | 1 | 128 | 2 | >32 | 500 | 1,000 | ND |
| L. fermentum ATCC 11976 | 16 | <0.03125 | 2 | 64 | 2 | 2 | 2,000 | 32,000 | ND |
| L. fermentum ATCC 23271 | 128 | 0.125 | 16 | 128 | 4 | 8 | 1,000 | 256,000 | ND |
| L. gasseri ATCC 19992 | >512 | 0.125 ± (1) | 4 | 512 ± (1) | 8 | 2 ± (1) | 1,000 ± (1) | 16,000 | 2,000 |
| L. gasseri NCK 99 | 512 | 0.125 | 4 | 512 | 4 | 4 | 1,000 | 8,000 | ND |
| L. johnsonii ATCC 33200 | 512 | 0.125 ± (2) | 16 | 512 ± (1) | 8 ± (1) | 2 ± (2) | 1,000 ± (1) | 16,000 | 16,000 |
| L. johnsonii PI 100-100 | 256 ± (2) | 0.125 ± (2) | 4 ± (1) | 512 ± (1) | 4 | 0.5 ± (2) | 1,000 ± (1) | 16,000 | 4,000 |
| L. maltaromicus BCCM 6903 | 256 | 64 | 32 | 512 | 4 | 1 | 4,000 | ND | 256,000 |
| L. paracasei ATCC 27092 | 512 | 2 ± (2) | 8 ± (1) | >1,024 | 16 ± (1) | >32 | 1,000 | 4,000 | 1,000 |
| L. paracasei JCM 1109 | 256 | 0.5 | 1 | >1,024 | 4 | 0.25 | 1,000 | ND | 64,000 |
| L. plantarum ATCC 14917 | >512 | 2 | 64 | >1,024 | 8 | 1 | 4,000 | ND | >256,000 |
| L. plantarum WCFS1 | >1,024 | 2 | 64 | >2,048 | 8 | 1 | 2,000 | 512,000 | ND |
| L. reuteri ATCC 23272 | 128 | 1 | 64 | 256 | 16 | 4 | 2,000 | 256,000 | ND |
| L. ruminis ATCC 25644 | >512 | 0.25 | 1 | >1,024 | 4 | 0.125 | 1,000 | ND | 256,000 |
| L. sake PI Lb5 | 128 | 2 | 2 | 128 | 8 | 32 | 16,000 | 512,000 | ND |
| L. sake PI Lb6 | 128 | 1 | 2 | 128 | 4 | 32 | 16,000 | 512,000 | ND |
| L. sake PI Lb15 | >512 | 1 | 128 | 512 | 16 | >32 | 4,000 | 512,000 | ND |
| L. vaginalis PI 675a | 128 | 0.5 | 16 | 1,024 | 4 | 32 | 1,000 | 32,000 | ND |
MICs are reported in micrograms per milliliter. ND, susceptibility not determined.
For drug name abbreviations; see Table 1, footnote b. NOV, novobiocin; CA, cholic acid; TCA, taurocholic acid; TDCA, taurodeoxycholic acid.
MIC measurements were repeated for many strains and revealed some slight differences. These are reported as ± the number of dilution differences that were observed.
These rodent isolates were assayed for resistance by both methods to examine the diversity in resistance phenotypes, and specifically MICs, within animals housed under controlled conditions. All of the isolates were identified by 16S sequencing as Lactobacillus murinus and had the same resistance phenotype (Table 1). Collectively, they also displayed little MIC variability (data not shown), with the exception of that of chloramphenicol, which varied 32-fold (2 to 64 μg/ml). For example, the MICs of novobiocin varied fourfold (<0.0625 to 0.25 μg/ml), whereas these MICs for other Lactobacillus species (Table 2) varied by more than 256-fold. For the other drugs tested, the range varied by, at most, 2 dilutions from absolute values (in micrograms per milliliter) of 512 (kanamycin), 0.25 (erythromycin), 2 (tetracycline), 512 (ciprofloxacin), 2,000 (cholic acid), and 32,000 (taurocholic acid). These data were surprising when analyzed together with macrorestriction profiling of their genomic DNA, which contained a large degree of heterogeneity (R. Nayak and C. A. Elkins, unpublished data).
The MICs of some bile acids were also determined in this study (Table 2; see above for L. murinus). The resistance profiles of these natural detergents were consistent in every strain tested, demonstrating that lactobacilli are more resistant to conjugated bile acids than to un(de)conjugated bile acids. Hence, the MICs of conjugates such as taurocholic acid and taurodeoxycholic acid were higher than the MICs of cholic acid, an unconjugated bile acid (Table 2). In some cases, such as those of L. acidophilus ATCC 4357, L. fermentum ATCC 23271, and L. plantarum WCFS1, this difference was dramatic, varying 256-fold between the two classes of molecules. In addition, resistance to taurodeoxycholic acid tended to be slightly lower than resistance to taurocholic acid in the few strains for which the comparison was made (Table 2).
Effects of bile acids on intrinsic Lactobacillus resistances.
The effect of these natural detergents on resistance to several drugs was tested by disk diffusion (Table 3). Nearly every strain exhibited increased sensitivity to the aminoglycosides when grown in the presence of 0.5% ox gall, which supports a previous study conducted in a similar manner (11). Likewise in our study, resistance to several other drugs was also shown to be affected in a strain- and drug-dependent manner. In some cases, small decreases in zone diameters were observed, while in others small increases were apparent (Table 3). However, the largest of these differences was observed only with aminoglycosides (streptomycin, kanamycin, and gentamicin), which was reproduced with purified conjugated (taurocholic and taurodeoxycholic) and unconjugated (cholic and deoxycholic) bile acids at subinhibitory concentrations (data not shown). Incidentally, the effect was exacerbated with deoxycholic acid, which is the most hydrophobic of the bile acids tested.
TABLE 3.
Changes in antibiotic susceptibilitiesa of Lactobacillus species grown in the presence of bile acidsb
| Lactobacillus species and strain | Relative change in growth zonec
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STREP | KAN | GENT | CHLOR | BAC | RIF | ERY | TET | CIP | NAL | OFLX | VAN | |
| L. acidophilus ATCC 332 | 1.4 | 1.4 | 1.8 | — | 1.5 | — | (1.5) | — | — | — | — | — |
| L. acidophilus ATCC 4356 | 2 | 16 | 17 | — | — | (1.5) | — | (1.3) | — | — | — | — |
| L. acidophilus ATCC 4357 | — | 18 | 1.5 | (1.4) | — | — | — | — | — | — | — | — |
| L. acidophilus ATCC 53103 | 14 | 14 | 2.3 | — | — | — | — | — | — | — | — | — |
| L. acidophilus ATCC 53544 | 16 | 16 | 16 | 1.4 | — | — | — | — | — | — | — | — |
| L. acidophilus KS-13 | — | — | 1.4 | — | 1.9 | — | 1.3 | — | — | — | — | 1.6 |
| L. bifidus ATCC 11146 | 16 | 1.6 | 2.2 | — | — | (1.4) | 1.3 | — | — | — | — | 10 |
| L. delbrueckii ATCC 11842 | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG |
| L. delbrueckii DSM 20080 | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG | NG |
| L. fermentum ATCC 23271 | 11 | 10 | 2 | (1.3) | — | — | (20) | — | — | — | — | — |
| L. gasseri ATCC 19992 | 2.1 | 9 | 13 | — | — | — | — | (26) | — | — | — | 1.4 |
| L. gasseri NCK 99 | 1.6 | 17 | 16 | — | 1.5 | (1.3) | (1.3) | — | — | — | — | 1.4 |
| L. johnsonii ATCC 33200 | 14 | 13 | 1.8 | — | — | — | (20) | — | 2.9 | — | — | — |
| L. maltaromicus BCCM 6903 | 12 | 17 | 18 | (1.4) | — | — | — | (1.5) | — | — | — | — |
| L. paracasei ATCC 27092 | (8) | 11 | 2.3 | — | — | — | — | 1.2 | — | — | — | — |
| L. paracasei JCM 1109 | 27 | 20 | 2.3 | — | 1.4 | — | — | — | 1.4 | — | — | — |
| L. plantarum ATCC 14917 | 10 | 12 | 14 | 1.4 | 1.4 | (1.4) | — | — | — | — | — | — |
| L. plantarum WCFS1 | 12 | 14 | 1.4 | 1.2 | — | (1.3) | — | (1.2) | 11 | — | — | — |
| L. reuteri ATCC 23272 | — | — | 1.3 | — | — | — | — | — | — | — | — | — |
| L. ruminis ATCC 25644 | 1.7 | 15 | 1.8 | — | — | — | — | — | — | — | — | — |
| Surveillance strains | ||||||||||||
| L. murinus NCTR 2479 | 12 | 1.5 | 2 | — | — | — | — | — | 1.2 | — | — | — |
| L. murinus NCTR 2478 | 1.2 | 1.2 | 1.5 | (1.3) | (1.4) | (1.3) | — | (1.2) | — | — | (1.2) | — |
| L. murinus NCTR 2489 | 17 | 1.4 | 1.8 | 1.2 | — | — | — | — | — | — | — | — |
| L. murinus NCTR 2487 | 14 | 1.2 | 1.3 | (1.3) | (1.3) | (1.3) | — | (1.2) | — | — | (1.2) | — |
| L. murinus NCTR 976 | 12 | 12 | 1.4 | — | — | (2) | — | (1.2) | — | — | — | — |
| L. murinus NCTR 2744 | 16 | 18 | 1.5 | — | — | — | — | — | — | — | — | — |
| L. murinus NCTR 2745 | 1.7 | 16 | 1.7 | — | — | — | — | — | — | — | — | — |
| L. murinus NCTR 684 | 16 | 13 | 20 | — | — | — | — | 1.3 | — | — | (1.4) | — |
| L. murinus NCTR 688 | 17 | 17 | 1.6 | — | — | — | (1.2) | (1.2) | — | — | — | — |
| L. murinus NCTR 2507 | 12 | 11 | 1.5 | — | — | 1.3 | — | — | — | — | — | — |
Susceptibilities were evaluated by measuring (in millimeters) zones of growth inhibition in standard disc diffusion assays.
MRS plates were supplemented with 0.5% fractionated ox gall powder containing a complex mixture of bile acids.
Relative changes in growth zones were computed as ratios by dividing the size of the zone of inhibition on MRS plates containing bile acids by that of MRS control plates. For values of >10, there was no zone of inhibition on the MRS control plate and these values were normalized to 1 for compution of the ratio. A dash represents no change in zone diameter between the two plates. Values in parenthesis represent inverse ratios for which an increase in resistance (smaller zone diameter) was observed on bile acid-containing plates. NG, no growth observed on bile acid-containing plates. For drug name abbreviations and concentrations, see Table 1, footnote b.
One representative strain, L. plantarum WCFS1, was chosen to study changes in MICs as a function of bile concentration (Fig. 1). This strain is the only sequenced strain in our study (28) and displayed one of the highest levels of conjugated bile acid resistance. As anticipated, the MICs of the aminoglycosides gentamicin and kanamycin and the aminocyclitol spectinomycin decreased at increasing concentrations of ox gall as did the MIC of ethidium bromide (Fig. 1A). However, the differences observed with kanamycin and spectinomycin may actually be larger than that shown. The intrinsic MICs of these drugs without ox gall were not determined because they were above the highest concentrations tested in this study. Regardless, the MIC of gentamicin showed the largest clinically significant change, from 1,024 to 4 μg/ml in less than 3.8 mg of ox gall per ml. Thus, the observed bile acid-mediated changes in MICs represent a physiological situation since bile acid concentrations in the lower GI tract can reach as high as 20 mM (approximately 10 mg/ml) (3). In contrast, the MICs of tetracycline and erythromycin failed to demonstrate any changes upon exposure to ox gall and the MICs of chloramphenicol and ciprofloxacin decreased by 1 and 2 dilutions, respectively, but only at the highest concentration of ox gall (Fig. 1B). This concentration, >30 mg/ml, is approximately threefold more than would be encountered in the GI tract. In addition, the MIC of ciprofloxacin, like those of kanamycin and spectinomycin, was not determined but was at least 2,048 μg/ml, which is not a clinically significant concentration and therefore was not pursued further.
FIG. 1.
Two-dimensional MIC analysis of L. plantarum WCFS1. The MICs of the various drugs were determined at each concentration of ox gall. (A) Drug MICs that shift with increasing concentrations of ox gall. (B) Drug MICs largely unaffected by increasing concentrations of ox gall. Identical results were obtained under aerobic conditions; the MIC of ox gall was 256 mg/ml.
Effects of bile acids and energy inhibitors on [G-3H]gentamicin uptake.
L. plantarum WCFS1 became sensitive to aminoglycosides in the presence of 0.5% ox gall. [G-3H]gentamicin was used to determine if bile acids or energy and efflux pump inhibitors were affecting the uptake of this hydrophilic, cationic drug (Fig. 2). In the presence of 0.5% ox gall, gentamicin uptake in WCFS1 cells increased up to sevenfold over control levels. With CCCP (a proton motive force uncoupler) at 0.1 mM (1×) and 1 mM (10×) concentrations, gentamicin uptake was indistinguishable from control levels. With 0.1 mM reserpine (an inhibitor of several efflux pumps), uptake increased by approximately sevenfold but revealed a large degree of variability (Fig. 2). Therefore, gentamicin uptake was measured as a function of the reserpine concentration, which increased by a maximum of 18-fold over control levels (Fig. 2, inset). The first four data points produced modest effects on gentamicin accumulation before a drastic but stable change in uptake occurred at higher concentrations (>1% [vol/vol] exogenous, chloroform-dissolved reserpine) that, we suggest, may result from the solvent effects of chloroform on membrane permeability. However, at the concentrations of 10 and 20 μg/ml (0.02 and 0.03 mM) typically reported for reserpine inhibition studies, gentamicin accumulation increased 1.5- to 2.5-fold.
FIG. 2.
L. plantarum WCFS1 uptake of [G-3H]gentamicin in the presence of ox gall or energy inhibitors. Control values were obtained with WCFS1 cells devoid any exogenous agents. Error bars represent standard deviations of three replicates from the same bacterial suspension. Inset, uptake of [G-3H]gentamicin at different concentrations of chloroform-dissolved reserpine.
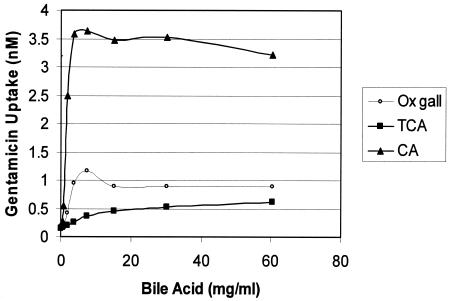
The increase in gentamicin uptake caused by a crude mixture of bile acids (ox gall) did not directly implicate bile acids as inhibitors of an efflux process. The presumed action of bile acids in membrane solubilization could simply promote leakage of gentamicin. Therefore, gentamicin uptake was measured as a function of ox gall, taurocholic acid, and cholic acid concentrations (Fig. 3). In each case, gentamicin uptake produced curves that differed significantly in magnitude. The conjugate taurocholic acid increased uptake by the smallest degree (threefold) but was more similar to ox gall (fivefold) than to cholic acid. This observation is easily explained since ox gall contains mostly conjugated bile acids. Interestingly, the maximum levels of gentamicin uptake achieved with cholic acid (18-fold) were similar to the maximum levels achieved with chloroform-dissolved reserpine, which indicates that cholic acid may permeabilize the membrane.
FIG. 3.
L. plantarum WCFS1 uptake of [G-3H]gentamicin at different concentrations of ox gall or purified bile acids. TCA, taurocholic acid; CA, cholic acid.
Time course analysis of effect of bile acid hydrolysis on gentamicin uptake.
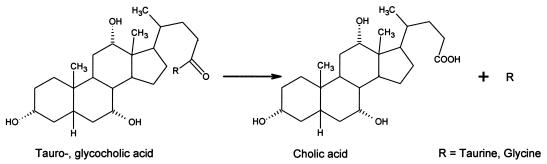
Microbial bile salt hydrolase (BSH) enzymes produce primary deconjugated bile acids from their conjugated counterparts (Fig. 4) (5) and are encoded by some Lactobacillus species (15, 19, 21, 28, 39), including L. plantarum WCFS1 (28, 39). The preincubation time in our uptake assay was varied to determine whether this activity, when combined with exogenous conjugated bile acid, affected gentamicin uptake in WCFS1 cells (Fig. 5). Cells exposed to 20 mg of taurocholic acid per ml showed increased gentamicin uptake as a function of time, whereas gentamicin uptake in WCFS1 cells devoid of exogenous conjugated bile acids was unaffected by the preincubation time. The observed increase in drug uptake peaked at 16 min at levels much lower than expected if complete conversion to cholic acid had occurred (cf. Fig. 3). This result was expected since the MIC of cholic acid, 2 mg/ml (Table 2), would prohibit full conversion of taurocholic acid by the endogenous BSHs.
FIG. 4.
Generalized BSH reaction. Conjugated bile acids are deconjugated to their primary bile acid counterparts in some GI tract prokaryotes.
FIG. 5.
Effect of BSH activity on [G-3H]gentamicin uptake by L. plantarum WCFS1. Cells were preincubated with and without 20 mg of taurocholic acid (TCA) per ml for increasing lengths of time before addition of radiolabeled gentamicin.
DISCUSSION
Recent studies of antimicrobial resistance in the genus Lactobacillus are limited to a few published reports (10, 12, 17). In those studies, resistance was assayed in MRS medium, which is complex and mildly selective in pH and supports luxuriant growth of a variety of lactobacilli. In our study, the resistance phenotypes of Lactobacillus species strains on MRS agar medium were generally similar for several drugs, were supported by MIC analyses in MRS broth culture except for ciprofloxacin, and were comparable to the findings previously reported (10, 12, 17). For a group of 17 rodent isolates, identified as L. murinus, the MICs were remarkably similar to each other when compared collectively with the MICs for other lactobacilli in this study (Table 2). However, macrorestriction profiling of L. murinus genomic DNA revealed a large degree of heterogeneity, suggesting that similarities in MICs may reflect physiological similarities rather than genetic ones, at least in this species.
In virtually every case in this study, lactobacilli were resistant to high levels of aminoglycoside antibiotics. These antibiotics have broad-spectrum activities; however, they are generally ineffective against gram-positive anaerobes (18). This is thought to occur because of membrane impermeability due to their characteristic multiple cationic charges. Thus, aminoglycosides have strong activity against aerobes because these bacteria generally have a high membrane potential, ΔΨ (intracellular negative), which facilitates their entry by an electrophoretic process or an unidentified transport mechanism. In anaerobe antibiotic resistance, impermeability can be explained by depression of Δψ by anaerobiosis (18), which is clearly exacerbated in acidophilic anaerobes like lactobacilli. Under these conditions, ΔpH dominates the proton motive force while Δψ contributes to a much lesser degree, if at all. For example, acidophiles at an external pH of <3 may actually produce a reverse membrane potential (intracellular positive) (49). We measured the pH of supernatants from an overnight culture of L. plantarum WCFS1, which dropped from 6.5 in fresh MRS to 3.85 in approximately 18 h (personal observations). With very little ΔΨ to drive aminoglycoside entry, we can rationalize membrane impermeability as an explanation for aminoglycoside MICs in milligram-per-milliliter quantities, as has been shown in this report.
Lactobacilli demonstrated a large variability in resistance to different species of bile acids (Table 2), an observation that is, thus far, unique to this genus. These differences correspond to the relative physical-chemical properties of taurocholic and cholic acids, hence pKas that influence solubility. Thus, lactobacilli are more resistant to largely soluble and negatively charged taurocholic acid (pKa, 1.4) than to a strongly hydrophobic and neutral cholic acid (pKa, 6.4), while moderately soluble taurodeoxycholic acid (pKa, 1.9) displayed intermediate activity. Taken in aggregate, our data suggest that bile acid resistance may be due to selective partitioning into leaflets of the membrane because of the strong influence of ΔpH and the strong anionic nature of tauroconjugates. Therefore, conjugates may be electrostatically biased into facing the acidic extracellular milieu with their sulfate groups. Neutral bile acids, like cholic acid, are free to carry out their natural action as strong detergents and compromise the entire membrane bilayer. This view supports our recent description of a taurocholic-cholic acid antiporter from Lactobacillus johnsonii 100-100 that is encoded in tandem with a BSH enzyme (22). Furthermore, as Kurdi et al. pointed out in their study (30), deconjugates can accumulate intracellularly to levels predicted by the Henderson-Hasselbalch equation in a spontaneous fashion because of their nature as weak, hydrophobic acids.
Gentamicin MICs in two-dimensional analyses (Fig. 1A) were affected far greater by ox gall than were those of the other aminoglycosides tested in this study. Accordingly, gentamicin should have a better ability to cross the membrane than kanamycin or spectinomycin. This observation is supported by the structures of these three aminoglycosides and their interactions with lipid layers. Thus, in one study, binding of different aminoglycosides to phosphatidylinositol was related to the number of amino groups carried by the drug and is largely dependent upon electrostatic interactions. In fact, the calculated interaction energy of gentamicin, −8.5 kcal/mol, was more favorable than for the other aminoglycosides tested in their study (8). In another study dealing with molecular hydrophobicity potentials and accessibility to water, gentamicin was shown to be associated with hydrophobic envelopes when assembled with phosphatidylinositol, whereas kanamycin was surrounded by hydrophilic envelopes (34). In our study, these properties in concert with bile acids resulted in a decrease in the MIC of gentamicin for strain WCFS1 from 1,024 to 4 μg/ml. Since human therapeutic levels of gentamicin range from approximately 5 to 12 μg/ml with target peak levels of 20 μg/ml in plasma (6), physiological concentrations of endogenous bile acids may produce a clinically significant impact on commensal Lactobacillus populations.
Several antimicrobial agents other than aminoglycosides were subjected to two-dimensional MIC analyses (Fig. 1). Similar to aminoglycosides, ethidium bromide was affected by increasing concentrations of ox gall (Fig. 1A); however, the planar structure of this intercalating agent may be responsive to membrane perturbations, or alternatively or in addition, this molecule may be subject to an efflux process inhibited by bile acids. In contrast, agents with hydrophobic moieties such as chloramphenicol, tetracycline, erythromycin, and ciprofloxacin were not affected by ox gall at physiological concentrations (Fig. 1B). These antimicrobials, however, have much less difficulty penetrating the membrane and would not be affected by bile, at least at levels that could be detected in MIC analyses. This may explain why the increase in the tetracycline resistance of WCFS1 observed in the disk diffusion assay (Table 3) was not observed in MIC analyses.
The lumen of the intestine is neutral to slightly alkaline (41). The gentamicin uptake assays in this study were performed at pH 6.5, a condition that we speculate approximates this environment because of moderate local pH depressions within buried Lactobacillus communities. Under these nearly neutral conditions, we observed with aminoglycoside uptake (i) a membrane-permeabilizing effect with high concentrations (vol/vol) of chloroform (Fig. 2, inset), (ii) an increase in uptake with cholic acid to the maximum levels observed with chloroform (Fig. 3), and (iii) a modest threefold increase in uptake with taurocholic acid (Fig. 3). We can conclude that bile (ox gall)-mediated sensitivity to aminoglycosides likely results from increased membrane permeation by deconjugated bile acids like cholic acid. Nonetheless, conjugated bile acids are typically the predominant species produced in liver hepatocytes and released into the intestinal lumen (44). Significant levels of deconjugated species are found in the lower small and large bowels because of the action of microbial BSH enzymes (41). Because of the preincubation inherent in the uptake assays, the modest effects observed with taurocholic acid and the largely conjugated constituents of ox gall may result from deconjugated bile acids produced by such enzymes. Therefore, we further demonstrated that BSH activity encoded by WCFS1 can modulate this permeability. These data, taken collectively with the resistance phenotypes in the first part of this study, support the conclusion from a different study showing that BSH activity and conjugated bile acid resistance are unrelated properties in lactobacilli (35). However, we can conclude, on the basis of our data, that the dramatic difference between the levels of resistance to conjugated bile acids and their primary bile acid counterparts can account for this previous conclusion.
That Lactobacillus membranes are impermeable to aminoglycosides at low pHs eliminates an alternative hypothesis suggesting that bile-mediated sensitivity interferes with a putative MDR efflux process. However, we observed an effect on gentamicin uptake with low concentrations of reserpine (Fig. 2, inset). At pHs approaching neutral, Δψ may make a contribution to the proton motive force and permit entry of small quantities of aminoglycosides, as was evident in the controls reported in the gentamicin uptake assays (Fig. 2). It is known that a related bacterium, Lactococcus lactis, encodes an efflux pump with broad specificity that is sensitive to reserpine. This pump, LmrA, has been shown to cause aminoglycoside efflux, producing modest levels of resistance (two- to threefold) when heterologously expressed in E. coli (38). WCFS1 is known to encode an LmrA homolog (28), which may offer an explanation for these observations. Uptake assays with directed-knockout mutants is a subject for future study to assess the contribution of this and other efflux processes to intrinsic resistance in lactobacilli, especially at different pHs.
Acknowledgments
We thank Hiroshi Nikaido for providing several helpful comments during the preparation of the manuscript and Carl Cerniglia, Mark Hart, and R. Doug Wagner for critically reviewing the manuscript. We also thank Michiel Kleerebezem of the Wageningen Centre for Food Sciences, Wageningen, The Netherlands, for the generous gift of L. plantarum WCFS1 and Dwayne C. Savage for access to his Lactobacillus type culture collection. Finally, we acknowledge Sherry Hopper and Tabitha Stewart for help with some of the disk diffusion susceptibility assays and Lillie Sims, Roger Steele, and Christine Summage-West for providing the in-house (NCTR) isolates.
REFERENCES
- 1.Abbott, A. 2004. Gut reaction. Nature 427:284-286. [DOI] [PubMed] [Google Scholar]
- 2.Adams, M. R., and P. Marteau. 1995. On the safety of lactic acid bacteria from food. Int. J. Food Microbiol. 27:263-264. [DOI] [PubMed] [Google Scholar]
- 3.Altman, P. L., and D. S. Dittman. 1968. Metabolism, p. 262. Federation of American Societies for Experimental Biology, Bethesda, Md.
- 4.Andremont, A. 2003. Commensal flora may play key role in spreading antibiotic resistance. ASM News 69:601-607. [Google Scholar]
- 5.Baron, S. F., and P. B. Hylemon. 1997. Biotransformation of bile acids, cholesterol, and steroid hormones, p. 470-510. In R. I. Mackie and B. A. White (ed.), Gastrointestinal microbiology, vol. I. Gastrointestinal ecosystems and fermentations. International Thomson Publishing, New York, N.Y. [Google Scholar]
- 6.Bartal, C., A. Danon, F. Schlaeffer, K. Reisenberg, M. Alkan, R. Smoliakov, A. Sidi, and Y. Almog. 2003. Pharmacokinetic dosing of aminoglycosides: a controlled trial. Am. J. Med. 114:194-198. [DOI] [PubMed] [Google Scholar]
- 7.Borriello, S. P., W. P. Hammes, W. Holzapfel, P. Marteau, J. Schrezenmeir, M. Vaara, and V. Valtonen. 2003. Safety of probiotics that contain lactobacilli or bifidobacteria. Clin. Infect. Dis. 36:775-780. [DOI] [PubMed] [Google Scholar]
- 8.Brasseur, R., G. Laurent, J. M. Ruysschaert, and P. Tulkens. 1984. Interactions of aminoglycoside antibiotics with negatively charged lipid layers. Biochemical and conformational studies. Biochem. Pharmacol. 33:629-637. [DOI] [PubMed] [Google Scholar]
- 9.Cerniglia, C. E., and S. Kotarski. 1999. Evaluation of veterinary drug residues in food for their potential to affect human intestinal microflora. Regul. Toxicol. Pharmacol. 29:238-261. [DOI] [PubMed] [Google Scholar]
- 10.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 1998. Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 61:1636-1643. [DOI] [PubMed] [Google Scholar]
- 11.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 2000. Effect of conjugated bile salts on antibiotic susceptibility of bile salt-tolerant Lactobacillus and Bifidobacterium isolates. J. Food Prot. 63:1369-1376. [DOI] [PubMed] [Google Scholar]
- 12.Charteris, W. P., P. M. Kelly, L. Morelli, and J. K. Collins. 2001. Gradient diffusion antibiotic susceptibility testing of potential probiotic lactobacilli. J. Food Prot. 64:2007-2014. [DOI] [PubMed] [Google Scholar]
- 13.Chavers, L. S., S. A. Moser, W. H. Benjamin Jr., S. E. Banks, J. R. Steinhauer, A. M. Smith, C. N. Johnson, E. Funkhouser, L. P. Chavers, A. M. Stamm, and K. B. Waites. 2003. Vancomycin-resistant enterococci: 15 years and counting. J. Hosp. Infect. 53:159-171. [DOI] [PubMed] [Google Scholar]
- 14.Chavez De Paz, L. E., G. Dahlen, A. Molander, A. Moller, and G. Bergenholtz. 2003. Bacteria recovered from teeth with apical periodontitis after antimicrobial endodontic treatment. Int. Endod. J. 36:500-508. [DOI] [PubMed] [Google Scholar]
- 15.Christiaens, H., R. J. Leer, P. H. Pouwels, and W. Verstraete. 1992. Cloning and expression of a conjugated bile salt hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl. Environ. Microbiol. 58:3792-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 17.Danielsen, M., and A. Wind. 2003. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 82:1-11. [DOI] [PubMed] [Google Scholar]
- 18.Davis, B. D. 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkins, C. A., S. A. Moser, and D. C. Savage. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147:3403-3412. [DOI] [PubMed] [Google Scholar]
- 20.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elkins, C. A., and D. C. Savage. 1998. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J. Bacteriol. 180:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elkins, C. A., and D. C. Savage. 2003. CbsT2 from Lactobacillus johnsonii 100-100 is a transport protein of the major facilitator superfamily that facilitates bile acid antiport. J. Mol. Microbiol. Biotechnol. 6:76-87. [DOI] [PubMed] [Google Scholar]
- 23.Elmer, G. W. 2001. Probiotics: “living drugs. ” Am. J. Health Syst. Pharm. 58:1101-1109. [DOI] [PubMed] [Google Scholar]
- 24.Fredricsson B., K. Englund, C. E. Nord, L. Weintraub. 1992. Could bacterial vaginosis be due to the competitive suppression of lactobacilli by aerobic microorganisms? Gynecol. Obstet. Investig. 33:119-123. [DOI] [PubMed] [Google Scholar]
- 25.Gilmore, M. S., and J. J. Ferretti. 2003. The thin line between gut commensal and pathogen. Science 299:1999-2002. [DOI] [PubMed] [Google Scholar]
- 26.Ishibashi, N., and S. Yamazaki. 2001. Probiotics and safety. Am. J. Clin. Nutr. 73S:465S-470S. [DOI] [PubMed] [Google Scholar]
- 27.Jonas, B. M., B. E. Murray, and G. M. Weinstock. 2001. Characterization of emeA, a norA homolog and multidrug resistance efflux pump, in Enterococcus faecalis. Antimicrob. Agents Chemother. 45:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. Klein Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Nierop Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konings, W. N., J. Kok, O. P. Kuipers, and B. Poolman. 2000. Lactic acid bacteria: the bugs of the new millennium. Curr. Opin. Microbiol. 3:276-282. [DOI] [PubMed] [Google Scholar]
- 30.Kurdi, P., H. W. Van Veen, H. Tanaka, I. Mierau, W. N. Konings, G. W. Tannock, F. Tomita, and A. Yokota. 2000. Cholic acid is accumulated spontaneously, driven by membrane ΔpH, in many lactobacilli. J. Bacteriol. 182:6525-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lachlak, N., E. Ageron, O. Zampatti, G. Michel, and P. A. D. Grimont. 1996. Composition of the Lactobacillus acidophilus complex isolated from vaginal flora. Microbiologica 19:123-132. [PubMed] [Google Scholar]
- 32.Li, X.-Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 33.Mastromarino, P., P. Brigidi, S. Macchia, L. Maggi, F. Pirovano, V. Trinchieri, U. Conte, and D. Matteuzzi. 2002. Characterization and selection of vaginal Lactobacillus strains for the preparation of vaginal tablets. J. Appl. Microbiol. 93:884-893. [DOI] [PubMed] [Google Scholar]
- 34.Mingeot-Leclercq, M. P., P. M. Tulkens, and R. Brasseur. 1992. Accessibility of aminoglycosides, isolated and in interaction with phosphatidylinositol, to water. A conformational analysis using the concept of molecular hydrophobicity potential. Biochem. Pharmacol. 44:1967-1975. [DOI] [PubMed] [Google Scholar]
- 35.Moser, S. A., and D. C. Savage. 2001. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 67:3476-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patnaik, R., S. Louie, V. Gavrilovic, K. Perry, W. P. C. Stemmer, C. M. Ryan, and S. del Cardayré. 2002. Genome shuffling of Lactobacillus for improved acid tolerance. Nat. Biotechnol. 20:707-712. [DOI] [PubMed] [Google Scholar]
- 38.Poelarends, G. J., P. Mazurkiewicz, M. Putman, R. H. Cool, H. W. van Veen, and W. N. Konings. 2000. An ABC-type multidrug transporter of Lactococcus lactis possesses an exceptionally broad substrate specificity. Drug Resist. Updates 3:330-334. [DOI] [PubMed] [Google Scholar]
- 39.Pridmore, D. R., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A.-C. Pittet, M.-C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanders, M. E., and T. R. Klaenhammer. 2001. The scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84:319-331. [DOI] [PubMed] [Google Scholar]
- 41.Savage, D. C. 1985. Gastrointestinal microflora of rodents, p. 85-117. In E. J. Ruitenberg and P. W. J. Peters (ed.), World animal science, vol. C2. Laboratory animals. Laboratory animal models for domestic animal production. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 42.Savage, D. C. 2001. Microbial biota of the human intestine: a tribute to some pioneering scientists. Curr. Issues Intest. Microbiol. 2:1-15. [PubMed] [Google Scholar]
- 43.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 44.Stevens, C. E. 1988. Comparative physiology of the digestive system. Cornell University Press, Ithaca, N.Y.
- 45.Stiles, M. E. 1996. Biopreservation by lactic acid bacteria. Antonie Van Leeuwenhoek 70:331-345. [DOI] [PubMed] [Google Scholar]
- 46.Vásquez, A., T. Jakobsson, S. Ahrné, U. Forsum, and G. Molin. 2002. Vaginal Lactobacillus flora of healthy Swedish women. J. Clin. Microbiol. 40:2746-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wegener, H. C. 2003. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 6:439-445. [DOI] [PubMed] [Google Scholar]
- 49.White, D. 1995. Membrane bioenergetics: the proton potential, p. 49-82. In D. White (ed.), The physiology and biochemistry of prokaryotes. Oxford University Press, New York, N.Y.