Abstract
Chitin is an abundant biopolymer whose degradation is mediated primarily by bacterial chitinases. We developed a degenerate PCR primer set to amplify a ∼900-bp fragment of family 18, group I chitinase genes and used it to retrieve these gene fragments from environmental samples. Clone libraries of presumptive chitinase genes were created for nine water and six sediment samples from 10 aquatic environments including freshwater and saline lakes, estuarine water and sediments, and the central Arctic Ocean. Putative chitinase sequences were also retrieved from the Sargasso Sea metagenome sequence database. We were unable to obtain PCR product with these primers from an alkaline, hypersaline lake (Mono Lake, California). In total, 108 partial chitinase gene sequences were analyzed, with a minimum of 5 and a maximum of 13 chitinase sequences obtained from each library. All chitinase sequences were novel compared to previously identified sequences. Intralibrary sequence diversity was low, while we found significant differences between libraries from different water column samples and between water column and sediment samples. However, identical sequences were retrieved from samples collected at widely distributed locations that did not necessarily represent similar environments, suggesting homogeneity of chitinoclastic communities between some environments.
Chitin is the second-most abundant biopolymer on the planet (15). In aquatic systems alone, chitin production has been estimated at 1011 metric tons per year (23). Chitin is composed of repeating units of the monomer N-acetylglucosamine (GlcNAc) and contains carbon and nitrogen in a ratio of 8:1. Chitin degradation is a key step in the cycling of nutrients in the environment (15, 32), and microorganisms are the primary agents of chitin degradation. Due to its polymeric nature, chitin must undergo at least partial hydrolysis prior to assimilation by microbial cells (6); this is accomplished by the enzyme chitinase (EC 3.2.1.14) (8, 18). Chitinases hydrolyze the bonds between GlcNAc residues, typically yielding oligomeric or dimeric products capable of being transported across the cellular membrane where they can be metabolized further (3, 23). Bacterial chitinases are often associated with the outer membrane or are secreted as extracellular enzymes (23, 24). The extracellular location of chitinases suggests that they must be adapted to function under the physicochemical conditions present in the surrounding environment. Thus, unique environmental conditions (e.g., high salinity, pH, or extreme temperatures) may select for proteins with unique sequences and thus biochemical properties.
Chitinases are classified as either family 18 or 19 glycosyl hydrolases based on amino acid sequence similarity (19). These two families are truly distinct; they share no similarity at the amino acid level and have different three-dimensional structures (10) and mechanisms of action (22). The vast majority of bacterial chitinases fall within family 18 and can be further organized into five different groups (I to V) based on conservation of amino acid residues within the catalytic domain (38). Group I chitinases are widely distributed among members of diverse proteobacterial lineages (7). Groups II to IV contain chitinases from more-narrowly restricted lineages. Group V is a collection of chitinases that do not fall into one of the other four groups (38).
Bacterial chitinase genes have been retrieved from diverse terrestrial environments, including alkaline soils (40), sandy soils (46), and pastures (25, 27). However, equivalent studies of chitinases in aquatic systems are relatively rare (8, 24, 33). Furthermore, no studies have compared chitinases across a broad range of distinct environments. Comparison of chitinase genes retrieved from similar, but geographically isolated, environments could yield insight into the biogeography of functional genes. In addition, comparisons of gene sequences retrieved from environments with distinct chemical and physical characteristics (water column versus sediments, estuaries, freshwater and saline lakes, temperate coastal waters, the Sargasso Sea, and the Arctic Ocean) may yield insights into how environmental conditions select for enzymes with novel properties.
In this study, we used a degenerate primer set to retrieve putative chitinase genes from 10 aquatic systems with distinct environmental characteristics. The results suggest that similar environments yield similar chitinase gene sequences. Furthermore, unique signature sequences were retrieved from one set of samples that may translate into fundamental differences in enzyme properties.
MATERIALS AND METHODS
Community DNA.
Locations sampled in this study and a summary of the environmental conditions at these locations are given in Table 1. Samples of surficial (0 to 1 cm) sediments were collected with a plastic spatula or by hand, placed in a glass jar, and stored on ice for transport to the laboratory. Water samples were collected with a Niskin sampler (Mono Lake, Soap Lake, and Walker Lake samples), a clean plastic bottle or bucket (estuarine and coastal water samples), or from a submarine as described by Bano and Hollibaugh (1) (Arctic Ocean samples).
TABLE 1.
Location, summary characteristics, and references for further descriptions of the environments where samples used in this study were collected
| Location | Latitude/longitude | Depth (m)a | Salinity (ppt) | pH | Sample date (mo/yr) | Reference | Clones sequencedb |
|---|---|---|---|---|---|---|---|
| Arctic Ocean Station 1.33.1 | 70°53.083′ N, 141°49.1′ W | 5 | 32 | 8 | 9/1997 | 1 | 10 (9) |
| Arctic Ocean Station 1.33.4 | 70°53.033′ N, 141°50.033′ W | 31 | 33 | 8 | 9/1997 | 1 | 10 (7) |
| Mono Lake (California) | 38°00.388′ N, 119°01.64′ W | 5 | 80 | 9.8 | 8/2001 | 30 | 10 (0) |
| Soap Lake (Washington) | 47°24.3′ N, 119°29.85′ W | 21 | 18 | 9.5 | 9/2003 | 31 | 10 (7) |
| Soap Lake (Washington) | 47°24.3′ N, 119°29.85′ W | 23 | 142 | 9.9 | 9/2003 | 31 | 10 (6) |
| San Francisco Bay (California) | 37°54.667′ N, 122°19.783′ W | 0 | ∼33 | 8 | 6/2003 | 17 | 10 (8) |
| San Joaquin River (California) | 37°40.34′ N, 121°15.55′ W | 0 | <1 | ∼7 | 6/2003 | 36 | 10 (5) |
| Sapelo Island (Georgia) | 31°25.05′ N, 81°17.75′ W | 0 | ∼20 | 8 | 4/2003 | 5 | 10 (9) |
| Bodega Bay (California) | 38°18.3′ N, 123°3.95′ W | 0 | 33 | 8 | 11/2003 | 47 | 10 (10) |
| Mono Lake (California) | 38°00.388′ N, 119°01.64′ W | SED | 80 | 9.8 | 8/2001 | 30 | 10 (0) |
| Tomales Bay (California) | 38°13.133′ N, 123°56.833′ W | SED | ∼33 | 8 | 6/2003 | 34 | 10 (9) |
| Topaz Lake (Nevada) | 38°41.583′ N, 119°31.167′ W | SED | <1 | 7 | 5/2003 | 41 | 10 (7) |
| San Francisco Bay (California) | 37°54.667′ N, 122°19.783′ W | SED | ∼33 | 8 | 6/2003 | 17 | 15 (13) |
| Sapelo Island (Georgia) | 31°25.05′ N, 81°17.75′ W | SED | ∼20 | 8 | 4/2003 | 5 | 15 (13) |
| Walker Lake (Nevada) | 38°43′ N, 118°43′ W | SED | 12 | 9.8 | 5/2003 | 4 | 10 (5) |
| Soap Lake mixolimnion | 47°24.3′ N, 119°29.85′ W | SED | 18 | 9.5 | 9/2003 | 31 | 0 |
| Soap Lake monimolimnion | 47°24.3′ N, 119°29.85′ W | SED | 142 | 9.9 | 9/2003 | 31 | 0 |
Entries either give the depth from which a water sample was collected or indicate sediment samples (SED).
Number of clones showing sequence similarity to previously identified chitinases is given in parentheses.
Samples were either frozen or immediately processed in the lab upon return from the field. Microbial biomass was collected from water samples by pressure filtration through Millipore Sterivex cartridge filters (ca. 50 kPa; 0.22-μm pore size). Excess water was expelled from the capsule. The cartridges were filled (1.8 ml) with a buffer containing 50 mM Tris (pH 8.3), 40 mM EDTA, and 0.75 M sucrose, capped, frozen on dry ice, shipped to the laboratory, and stored at −70°C until processed.
Community DNA was extracted from sediment samples and purified using an Ultraclean Soil DNA Kit (MoBio Laboratories Inc., Solana Beach, Calif.) following the manufacturer's instructions. Extraction and purification of DNA from cartridge filters were essentially as described by Ferrari and Hollibaugh (13). Briefly, 40 μl of lysozyme (50 mg ml−1) was added to each cartridge, and the cartridges were incubated for 60 min at 37°C. Fifty microliters of proteinase K (20 mg ml−1) and 100 μl of a 20% (wt/vol) solution of sodium dodecyl sulfate were added to each cartridge, and the cartridges were incubated at 55°C for 2 h. DNA was purified from 800 μl of the lysate by sequential extraction with 800 μl of phenol-chloroform-isoamyl alcohol (25:24:1), chloroform-isoamyl alcohol (24:1), and finally n-butanol. The aqueous phase was removed, placed in a Centricon-100 concentrator (Amicon, Bedford, Mass.), mixed with 500 μl of TE buffer (10 mM Tris and 1 mM EDTA, pH 8.0), and centrifuged at 1,000 × g for 10 min. Next, 500 μl of TE buffer was added to the Centricon-100 concentrator, and the mixture was centrifuged for another 10 min. Successful extraction of high-molecular-weight DNA was verified for all samples by electrophoresis on 1% agarose gels.
Primer design.
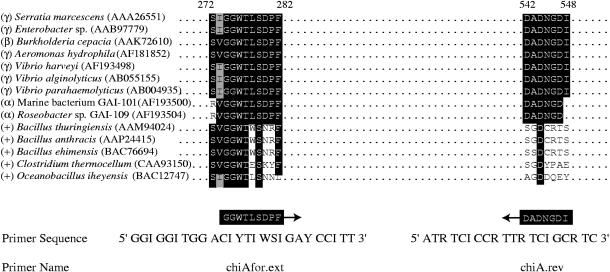
The degenerate primer chiAfor.ext was based on conserved residues identified in chitinases from diverse proteobacteria (Fig. 1). Protein sequences were aligned using the PILEUP tool of the Wisconsin package, version 10.2 (Accelrys, San Diego, Calif.). chiAfor.ext was used in conjunction with chiA.rev, a primer developed by Cottrell et al. (9). This primer set successfully amplified the chitinase gene from Vibrio harveyi.
FIG. 1.
Design of degenerate primers for family 18, group I chitinase genes. Alignments of chitinase amino acid sequences from organisms representing diverse phylogenetic lineages were used to design the degenerate primers. Symbols represent bacterial taxonomic groups: γ, γ-proteobacteria; β, β-proteobacteria; α, α-proteobacteria; and +, gram-positive bacteria. GenBank accession numbers are provided in parentheses. Position designations relative to the Serratia marcescens chitinase sequence (P07254) are shown above the alignment. Conserved residues are shown in black, similar residues in grey. I, inosine base; Y, C or T; W, A or T; S, G or C; and R, A or G. The degeneracy for both primers in this study is 16-fold. The references for chiAfor.ext and chiA.rev are this study and reference 9, respectively.
PCR and cloning.
PCR primers chiAfor.ext and chiA.rev were used to amplify putative chitinase gene fragments from community DNA. PCR was run with the following conditions on an MJ Research PTC-200 Peltier thermal cycler: denaturation at 94°C for 1 min, annealing at 58°C for 1 min, and an extension step at 72°C for 1 min. This sequence was repeated 35 times followed by a 10-min final extension step at 72°C.
Products of the appropriate size (∼900 bp) were recovered from a 1.5% agarose gel using the QiaQuik gel extraction kit (QIAGEN, Valencia, Calif.) and cloned into the pCR 2.1 vector (Invitrogen Corp., Carlsbad, Calif.) following the manufacturer's protocols. Clone libraries were generated for all samples that yielded a PCR product of the expected size. Colonies were selected randomly, and then plasmids were isolated from Escherichia coli host cells with a Qiaprep Spin Miniprep kit (QIAGEN). Insert size was verified by digestion with EcoRI, and then inserts of the correct size were sequenced using an ABI PRISM 310 genetic analyzer and a BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) using primers that recognized the cloning vector (M13 forward and reverse). Reads of approximately 550 bp of nucleotide sequence were obtained in each direction. Sequences were edited and assembled using the AssemblyLign program (Oxford Molecular, 1998). The forward and reverse reactions resulted in a complete sequence for the amplified region of the chitinase gene with ∼200 bp of overlap. Regions corresponding to the primer binding sites were removed from the sequences prior to analysis.
Phylogenetic analyses.
Sequences were analyzed using the Wisconsin Package v. 10.2 (Accelrys) and homology searches (BLASTX) were carried out at the network server of the National Center for Biotechnology Information. Phylogenetic trees were constructed with the PHYLIP package using evolutionary distances (Jukes-Cantor or Kimura) and the neighbor-joining method (12). A maximum-likelihood tree was also constructed using the phylogenetic analysis program PAUP (39) to verify the results from the Jukes-Cantor algorithm.
Database sequences.
Putative chitinase sequences were retrieved from the Sargasso Sea metagenome database (SSMD) (http://www.ncbi.nih.gov/BLAST/Genome/EnvirSamplesBlast.html) (43) by interrogation (BLASTX) using one sequence from each of the five clusters of our tree (refer to Fig. 2; WLS-07 [accession no. AY674163], TLS-08 [AY674150], BBW-04 [AY674077], AOW55-10 [AY674066], and SLW21-07 [AY674140]). Homology searches were then carried out against the entire GenBank database using each of the SSMD potential chitinase sequences. Criteria for inclusion in our phylogenetic analysis were as follows: (i) the sequenced portion of the gene had to contain the entire region of the gene analyzed in this study and (ii) the putative genes had to be capable of being aligned to our existing library of chitinases using the PILEUP tool of the Wisconsin package. Accession numbers of all potential chitinases from the SSMD have been recorded in a spreadsheet that can be accessed at the Mono Lake Microbial Observatory web site (http://www.monolake.uga.edu/research.htm; “Ancillary Data” section, “Sargasso_Sea_Chitinases.xls” file).
FIG. 2.
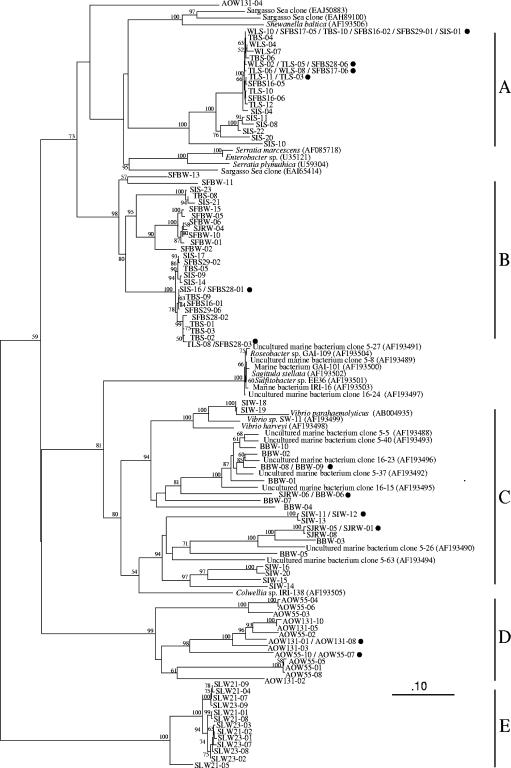
Neighbor-joining tree (partial sequence, ∼800 bp) showing phylogenetic relationships between family 18, group I chitinase nucleotide sequences. Clone designations are as follows: AOW55, Arctic Ocean, 55-m depth; AOW131, Arctic Ocean, 131-m depth; BBW, Bodega Bay water column; SIS, Sapelo Island sediments; SIW, Sapelo Island water column; SFBS, San Francisco Bay sediments; SFBW, San Francisco Bay water column; SJRW, San Joaquin River water column; SLW21, Soap Lake, 21-m depth; SLW23, Soap Lake, 23-m depth; TBS, Tomales Bay sediments; TLS, Topaz Lake sediments; and WLS, Walker Lake sediments. Water column samples for which no depth is given were collected at the surface (nominal depth, 0.1 m). Each sequence from a given library is also provided with a numerical designation. Branches containing identical sequences are indicated with a filled circle. The scale bar indicates Jukes-Cantor distance. Bootstrap values of >50% (for 100 iterations) are shown at branch nodes. The tree is unrooted with the chitinase gene from Bacillus circulans (AF154827) as the outgroup. GenBank accession numbers for reference sequences are provided in parentheses.
Nucleotide sequence accession numbers.
The sequences determined during this study have been submitted to GenBank under the following accession numbers: AY674058 to AY674165.
RESULTS AND DISCUSSION
We were unable to amplify chitinase genes from alkaline and estuarine environmental samples using primer sets previously described in the literature (9). Upon inspection and after examining published chitinase sequences, the problem seemed to be related to the sequence of the forward primer. To remedy the problem, we developed a new, degenerate forward primer based on the published forward primer sequence (9) but modified to be consistent with sequences from diverse proteobacteria (sequence divergence prevented design of a primer set that included chitinases from gram-positive bacteria; Fig. 1). We used the redesigned forward primer in combination with the published reverse primer (9) to target family 18, group I chitinase genes. Amplification with this primer set yielded a PCR product of the expected size for all samples except those from Mono Lake water and sediment and Soap Lake sediments. Despite repeated attempts to optimize PCR conditions and alter DNA extraction protocols, only nontarget amplification products were obtained from Mono Lake samples (both water and sediments) and Soap Lake sediments never yielded products of the correct size.
A total of 160 inserts was sequenced from 15 clone libraries with inserts from at least 10 randomly selected clones sequenced from each library. Homology searches suggested that the inserts in 52 of the clones were not chitinase genes (Table 1). Nontarget sequences were retrieved from all environments examined in this study (including 20 from Mono Lake). These typically lacked significant similarity to any database sequence and were not analyzed further. We checked a subset of our remaining sequences (13 total; all of the deeply branching, unique sequences in Fig. 2, for example SIS-10, SFBW-13, and SFBW-11) for possible chimera formation by using BLAST on 200 bp from each end of the sequence against the database to ensure that they returned the same top hits. None of the sequences we examined failed this test; however, some of the 52 discarded sequences may have been chimeras. All 108 putative chitinase genes retrieved were unique when compared to sequences presently in the GenBank database. At the nucleotide level, the sequences were between 57 to 94% identical to previously identified chitinase genes. At the amino acid level, the sequences were 44 to 98% identical and 52 to 98% similar to current (July 2004) GenBank entries.
Phylogenetic analysis (Jukes-Cantor) placed the chitinase nucleotide sequences into five major clusters, designated clusters A to E (Fig. 2). A maximum-likelihood tree of the nucleotide sequences (data not shown) was essentially identical to this tree. A phylogenetic tree (Kimura) was also constructed using deduced amino acid sequences (data not shown). The topologies of the nucleotide and amino acid trees were similar, with the composition of the clusters being the same for all trees. Cluster A contained sequences from the sediments collected at Sapelo Island, Georgia; San Francisco Bay, California; Tomales Bay, California; Topaz Lake, Nevada; and Walker Lake, Nevada. Cluster B contained sequences retrieved from sediments collected at Sapelo Island, Georgia; San Francisco Bay, California; Tomales Bay, California; and Topaz Lake, Nevada. In addition, sequences retrieved from the San Francisco Bay and San Joaquin River water samples formed a distinct subcluster within cluster B. Cluster C contained sequences retrieved from the Sapelo Island, San Joaquin River, and Bodega Bay water column samples. Cluster D consisted of sequences retrieved from Arctic Ocean water samples. These sequences segregated into subclusters that typically corresponded to sample depth. Cluster E contained sequences retrieved exclusively from the two Soap Lake, Washington, water column samples.
We identified 43 potential family 18, group I chitinase sequences (maximum E value of 8e-4) in the SSMD. The region possessing the signature motif, [DG]-G-[LIV]-[DG]-[IV]-[DH]-W-[EG], of the family 18, group I chitinase sequences (38), was present in 13 (30%) of these sequences. These putative chitinases appear to be diverse in origin, as the most similar sequences in GenBank were obtained from γ-proteobacteria (23%), gram-positive bacteria (51%), Bacteroides (2%) bacteria, arthropods (9%), mammals (5%), fungi (5%), and Caenorhabditis elegans (5%). The majority of the SSMD putative chitinases either did not have any overlap with the region of the gene analyzed in this study (58%) or contained only a portion of the region (30%). The remaining five (12%) SSMD sequences contained the entire region of the chitinase gene delimited by the primers we used; however, only three of these sequences were similar enough to be included in the tree (Fig. 2). The three SSMD sequences included in the tree fell outside of the clusters (A to E) defined by sequences retrieved from our samples. Two SSMD sequences (EAJ50883 and EAH89100) clustered with a family 18, group I chitinase reference sequence from Shewanella baltica and were most closely related to our cluster A (Fig. 2). The third SSMD sequence (EAI65414) grouped with one Enterobacter and two Serratia chitinase sequences. Given the overall dominance of Shewanella-like sequences in the Sargasso Sea metagenome library (42), it is not surprising that we retrieved Shewanella-like chitinase sequences from it. We were surprised that we did not find sequences similar to those from our Arctic Ocean samples, since the 16S rRNA gene libraries from these samples contained sequences similar to those retrieved from Sargasso Sea samples (1).
Some of the chitinase sequences retrieved from different samples were identical (Fig. 2). For example, a sequence from the Sapelo Island library (SIS-01) was identical to three San Francisco Bay sequences (SFBS16-02, SFBS17-05, and SFBS29-01). Both of these samples are intertidal sediments from salt marshes dominated by Spartina alterniflora (Sapelo Island) or Salicornia virginica (San Francisco Bay). The estuaries have similar temperatures and salinity ranges, which would lead to the expectation that they harbor similar microflora, but they are geographically isolated. We are unaware of other reports of identical functional gene sequences having been retrieved from isolated environments; however, this may simply be due to the smaller database for functional genes, as closely related (16) or identical (1, 2) 16S rRNA genes have been retrieved from distant locations.
Interestingly, some sequences retrieved from sediments collected in freshwater Topaz Lake were identical to sequences retrieved from estuarine sediments of San Francisco Bay and from sediments of alkaline, saline Walker Lake (i.e., TLS-05, SFBS28-06, and WLS-02). Furthermore, another San Francisco Bay sequence (SFBS17-06) was identical to a clone from Topaz Lake (TLS-06) and from Walker Lake (WLS-08). This was a surprising finding, as these environments range in salinity from <1 (Topaz Lake) up to ∼30 ppt (San Francisco Bay) and in pH from ∼7 (Topaz Lake) to 9.8 (Walker Lake) (Table 1).
One factor that these sequences have in common is that they were all retrieved from sediment samples. This suggests that physicochemical properties common to sediments (surfaces, hypoxia and anoxia, elevated organic carbon concentrations, and likely elevated chitin concentrations since shed arthropod exoskeletons sink) override other environmental factors (temperature, salinity, and pH) in determining the distribution of functional gene sequences. Clearly there is a limit to this generalization because Mono Lake chitinases (water column, sediment, and isolates) were not amplified by the primer set used in this study, even though enzyme assays demonstrated chitinase activity (G. R. LeCleir, unpublished data). DNA extracted from Soap Lake sediment also failed to yield PCR product with our primer set. In contrast to chitinase sequences retrieved from sediment communities, sequences retrieved from water column samples collected at different locations segregated into separate clusters (Fig. 2). Furthermore, within cluster D, sequences retrieved from mixed-layer (55 m) and halocline (131 m) samples collected at the same station tended to fall into separate subclusters. The bacterial assemblages associated with these water masses have been characterized previously and were found to be distinct from one another (1, 2) and from those of temperate coastal water assemblages (1). Because the composition of Soap Lake water differs significantly from either seawater or freshwater, the bacterial assemblages from the lake might also be expected to be phylogenetically distinct. Biodiversity studies of other saline, alkaline lakes have verified that the composition of bacterial assemblages differs from those in other aquatic environments and also that the same suites of organisms are found in lakes from widely separated locations (11, 21, 36).
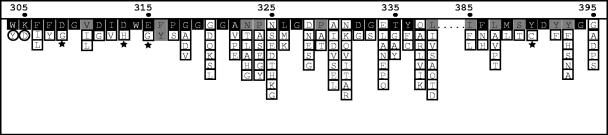
Alignment of family 18 glycosyl hydrolases shows that a number of residues essential for catalytic activity are conserved (29). The majority of chitinase sequences identified in this study (94%) contain a conserved motif encompassing the catalytic site, [DG]-G-[LIV]-[DG]-[IV]-[DH]-W-[EG], corresponding to positions 308 to 315 of the Serratia marcescens ChiA protein (29) (Fig. 3). Two additional residues, a tyrosine and an aspartate at positions 390 and 391, respectively, are also conserved in most of our sequences. However, seven of the sequences we obtained contained substitutions at one of these conserved positions. All of these substitutions result from single-base-pair changes: six A→G transitions and one G→C transversion. Both SLW23-03 and AOW131-04 contain a glycine instead of an aspartate at position 308. WLS-07 contains histidine rather than aspartate at position 313. Interestingly, this same substitution is found in narbonin, a protein found in plants with high similarity to chitinase but with no known enzymatic function (42). WLS-08, TLS-06, and SFBS17-6 have a glycine instead of glutamate at position 315. This glutamate residue has been shown to be the essential catalytic proton donor in structurally characterized bacterial chitinases (45). Finally, clone SFBS16-01 contains a cysteine rather than the completely conserved tyrosine at position 390. Collectively, these seven sequences may represent pseudogenes. Alternatively, they may correspond to genes that encode enzymes with unique properties, including different activities and mechanisms of action, or they may encode proteins with no known enzymatic function that share sequence similarity with chitinase (i.e., narbonin). They may also simply be the result of PCR (44), cloning (28, 35), or sequencing errors, although the sequence reads were unambiguous at these positions. In the absence of biochemical data for the expressed protein, it is difficult to evaluate the significance of these substitutions.
FIG. 3.
Conserved residues including and surrounding the catalytic domain of proteobacterial chitinases. Residues are coded according to degree of conservation as follows: black, >75%; gray, 50 to 75%; and no color, <50%. Positions that are altered in chitinase sequences retrieved in this study are indicated by symbols. Stars indicate residues found in a limited number of sequences and that are described in the text. Residues found exclusively in sequences retrieved from Soap Lake samples are circled. Numbers and dots indicate residue positions relative to the Serratia marcescens gene chiA (P07254).
All of the sequences retrieved from the two Soap Lake libraries contain aspartate rather than lysine at residue 305 (D305K), as well as a more conserved substitution at position 304 (tyrosine for tryptophan; Fig. 3). The D305K substitution has only been found in a novel chitinase recently identified in the marine bacterium Microbulbifer degradans 2-40 (20). The M. degradans chitinase has two catalytic domains, each with distinct activities towards polymeric chitin. Despite significant homology between domains at the amino acid level, the D305K substitution is present only in one of the domains, designated GH18C (BK001042). Overall, the Soap Lake sequences share approximately 45% identity and 55% similarity to the GH18C domain. It is speculative to infer the physiological or biochemical implications of these substitutions with only sequences in hand. Nonetheless, this finding raises the possibility that the Soap Lake chitinases may have properties similar to those identified in the M. degradans protein.
Chitinases from polar microorganisms appear to have adaptations required to function well in cold environments, as recently demonstrated for two chitinase alleles, ChiA (CAB62382) and ChiB (CAB62499), from an Arthrobacter strain isolated from Antarctic sediment (26). The increased heat lability of these chitinases is believed to be a consequence of structural changes that give the enzymes greater flexibility at lower temperatures, permitting conformational changes necessary for catalysis (14). Similar sequence modifications might be expected in genes from other cold-adapted microbes, regardless of their phylogenetic affiliation, leading to unique sequences for Arctic Ocean genes, as we have found (Fig. 2).
The form and source of chitin found in the environment may also select for specific genes in different environments. There are three major types of chitin, designated α, β, and γ (32). Each has unique physical attributes and chemical properties. Chitin can also vary by the degree of acetylation and the presence of cross-linked structural components (37). The composition of the chitin matrix and its associated molecules is typically organism dependent (15). Other molecules associated with the chitin matrix often select for specific enzymes and control rates of chitin hydrolysis (32, 37). Therefore, predominance of different structural variants of chitin in the environments we examined may dictate elaboration of what appear to be environmental-specific proteins that are in reality required for efficient hydrolysis of the predominant form of chitin.
Acknowledgments
We thank Nasreen Bano for laboratory assistance and Charles Budinoff for help with phylogenetic analyses. We are grateful to Robert Jellison, Mandy Joye, Vladimir Samarkin, Steve Carini, Jen Fisher, Sandra Roll, Catherine Albaugh, and the Soap Lake Microbial Observatory for assistance with sample collection. We thank three anonymous reviewers of the manuscript for their helpful comments.
This work was supported by the Mono Lake Microbial Observatory grant NSF MCB 9977886.
REFERENCES
- 1.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bano, N., S. Ruffin, B. Ransom, and J. T. Hollibaugh. 2004. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl. Environ. Microbiol. 70:781-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassler, B. L., C. Yu, Y. C. Lee, and S. Roseman. 1991. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24276-24286. [PubMed] [Google Scholar]
- 4.Beutel, M. W., A. J. Horne, J. C. Roth, and N. J. Barratt. 2001. Limnological effects of anthropogenic desiccation of a large, saline lake, Walker Lake, Nevada. Hydrobiologia 466:91-105. [Google Scholar]
- 5.Chalmers, A. G. 1997. The ecology of the Sapelo Island National Estuarine Research Reserve. Office of Coastal Resource Management, National Oceanic and Atmospheric Administration, Washington, D.C.
- 6.Chrost, R. J. 1992. Significance of bacterial ectoenzymes in aquatic environments. Hydrobiologia 243:61-70. [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., J. A. Moore, and D. L. Kirchman. 1999. Chitinases from uncultured marine microorganisms. Appl. Environ. Microbiol. 65:2553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottrell, M. T., D. N. Wood, L. Y. Yu, and D. L. Kirchman. 2000. Selected chitinase genes in cultured and uncultured marine bacteria in the alpha- and gamma-subclasses of the proteobacteria. Appl. Environ. Microbiol. 66:1195-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, G. J., S. P. Tolley, B. Henrissat, C. Hjort, and M. Schulein. 1995. Structures of oligosaccharide-bound forms of the endoglucanase V from Humicola insolens at 1.9 angstrom resolution. Biochemistry 34:16210-16220. [DOI] [PubMed] [Google Scholar]
- 11.Duckworth, A. W., W. D. Grant, B. E. Jones, and R. vanSteenbergen. 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol. Ecol. 19:181-191. [Google Scholar]
- 12.Felsenstein, J. 1993. Phylip (Phylogeny Inference Package), 3.5 ed. University of Washington, Seattle.
- 13.Ferrari, V. C., and J. T. Hollibaugh. 1999. Distribution of microbial assemblages in the Central Arctic Ocean Basin studied by PCR/DGGE: analysis of a large data set. Hydrobiologia 401:55-68. [Google Scholar]
- 14.Gerday, C., M. Aittaleb, J. L. Arpigny, E. Baise, J. P. Chessa, G. Garsoux, I. Petrescu, and G. Feller. 1997. Psychrophilic enzymes: a thermodynamic challenge. Biochim. Biophys. Acta 1342:119-131. [DOI] [PubMed] [Google Scholar]
- 15.Gooday, G. W. 1990. The ecology of chitin degradation. Adv. Microb. Ecol. 11:387-430. [Google Scholar]
- 16.Gordon, D. A., and S. J. Giovannoni. 1996. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 62:1171-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarini, J. M., J. E. Cloern, J. Edmunds, and P. Gros. 2002. Microphytobenthic potential productivity estimated in three tidal embayments of the San Francisco Bay: a comparative study. Estuaries 25:409-417. [Google Scholar]
- 18.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard, M. B., N. A. Ekborg, L. E. Taylor II, R. M. Weiner, and S. W. Hutcheson. 2004. Chitinase B of “Microbulbifer degradans” 2-40 contains two catalytic domains with different chitinolytic activities. J. Bacteriol. 186:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iseli, B., S. Armand, T. Boller, J. M. Neuhaus, and B. Henrissat. 1996. Plant chitinases use two different hydrolytic mechanisms. FEBS Lett. 382:186-188. [DOI] [PubMed] [Google Scholar]
- 23.Keyhani, N. O., and S. Roseman. 1999. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473:108-122. [DOI] [PubMed] [Google Scholar]
- 24.Kirchman, D. L., and J. White. 1999. Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat. Microb. Ecol. 18:187-196. [Google Scholar]
- 25.Krsek, M., and E. M. H. Wellington. 2001. Assessment of chitin decomposer diversity within an upland grassland. Antonie Leeuwenhoek 79:261-267. [DOI] [PubMed] [Google Scholar]
- 26.Lonhienne, T., K. Mavromatis, C. E. Vorgias, L. Buchon, C. Gerday, and V. Bouriotis. 2001. Cloning, sequences, and characterization of two chitinase genes from the Antarctic Arthrobacter sp. strain TAD20: isolation and partial characterization of the enzymes. J. Bacteriol. 183:1773-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metcalfe, A. C., M. Krsek, G. W. Gooday, J. I. Prosser, and E. M. H. Wellington. 2002. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl. Environ. Microbiol. 68:5042-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paabo, S., and A. C. Wilson. 1988. Polymerase chain reaction reveals cloning artefacts. Nature 334:387-388. [DOI] [PubMed] [Google Scholar]
- 29.Papanikolau, Y., G. Prag, G. Tavlas, C. E. Vorgias, A. B. Oppenheim, and K. Petratos. 2001. High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis. Biochemistry 40:11338-11343. [DOI] [PubMed] [Google Scholar]
- 30.Patten, D. T., F. P. Conte, W. E. Cooper, J. Dracup, S. Dreiss, K. Harper, G. L. Hunt, Jr., P. Kilham, H. E. Klieforth, J. M. Melack, and S. A. Temple. 1987. The Mono Basin ecosystem: effects of changing lake level. National Academy Press, Washington, D.C.
- 31.Peyton, B. M., and D. R. Yonge. 2002. Biodegradation of non-point source pollutants in Soap Lake, Washington. Project completion report. State of Washington Water Research Report WRR-11. State of Washington Water Research Center, Pullman.
- 32.Poulicek, M., F. Gaill, and G. Goffinet. 1998. Chitin biodegradation in marine environments. ACS Symp. Ser. 707:163-210. [Google Scholar]
- 33.Ramaiah, N., R. T. Hill, J. Chun, J. Ravel, M. H. Matte, W. L. Straube, and R. R. Colwell. 2000. Use of a chiA probe for detection of chitinase genes in bacteria from the Chesapeake Bay. FEMS Microbiol. Ecol. 34:63-71. [DOI] [PubMed] [Google Scholar]
- 34.Smith, S. V., W. J. Wiebe, J. T. Hollibaugh, S. J. Dollar, S. W. Hager, B. E. Cole, G. W. Tribble, and P. A. Wheeler. 1987. Stoichiometry of C, N, P, and Si fluxes in a temperate-climate embayment. J. Mar. Res. 45:427-460. [Google Scholar]
- 35.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stepanauskas, R., M. A. Moran, B. A. Bergamaschi, and J. T. Hollibaugh. 2003. Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquat. Microb. Ecol. 31:85-98. [Google Scholar]
- 37.Svitil, A. L., S. M. N. Chadhain, J. A. Moore, and D. L. Kirchman. 1997. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol. 63:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svitil, A. L., and D. L. Kirchman. 1998. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: implications for the ecology and evolution of 1,4-beta-glycanases. Microbiology 144:1299-1308. [DOI] [PubMed] [Google Scholar]
- 39.Swofford, D. L. 2002. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4.0b10. Sinauer Associates, Sunderland, Mass.
- 40.Tsujibo, H., T. Kubota, M. Yamamoto, K. Miyamoto, and Y. Inamori. 2003. Characterization of chitinase genes from an alkaliphilic actinomycete, Nocardiopsis prasina OPC-131. Appl. Environ. Microbiol. 69:894-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.U.S. Geological Survey. 2001. National Water Information System (NWISWeb) data. [Online.] http://waterdata.usgs.gov/nwis/. Accessed 12 February 2004.
- 42.vanScheltinga, A. C. T., M. Hennig, and B. W. Dijkstra. 1996. The 1.8 angstrom resolution structure of hevamine, a plant chitinase/lysozyme, and analysis of the conserved sequence and structure motifs of glycosyl hydrolase family 18. J. Mol. Biol. 262:243-257. [DOI] [PubMed] [Google Scholar]
- 43.Venter, J. C., K. Remington, J. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 44.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe, T., K. Kobori, K. Miyashita, T. Fujii, H. Sakai, M. Uchida, and H. Tanaka. 1993. Identification of glutamic acid-204 and aspartic acid-200 in chitinase-A1 of Bacillus circulans Wl-12 as essential residues for chitinase activity. J. Biol. Chem. 268:18567-18572. [PubMed] [Google Scholar]
- 46.Williamson, N., P. Brian, and E. M. H. Wellington. 2000. Molecular detection of bacterial and streptomycete chitinases in the environment. Antonie Leeuwenhoek 78:315-321. [DOI] [PubMed] [Google Scholar]
- 47.Wing, S. R., L. W. Botsford, L. E. Morgan, J. M. Diehl, and C. J. Lundquist. 2003. Inter-annual variability in larval supply to populations of three invertebrate taxa in the northern California Current. Estuar. Coast. Shelf Sci. 57:859-872. [Google Scholar]